- Academic Editor
-
-
-
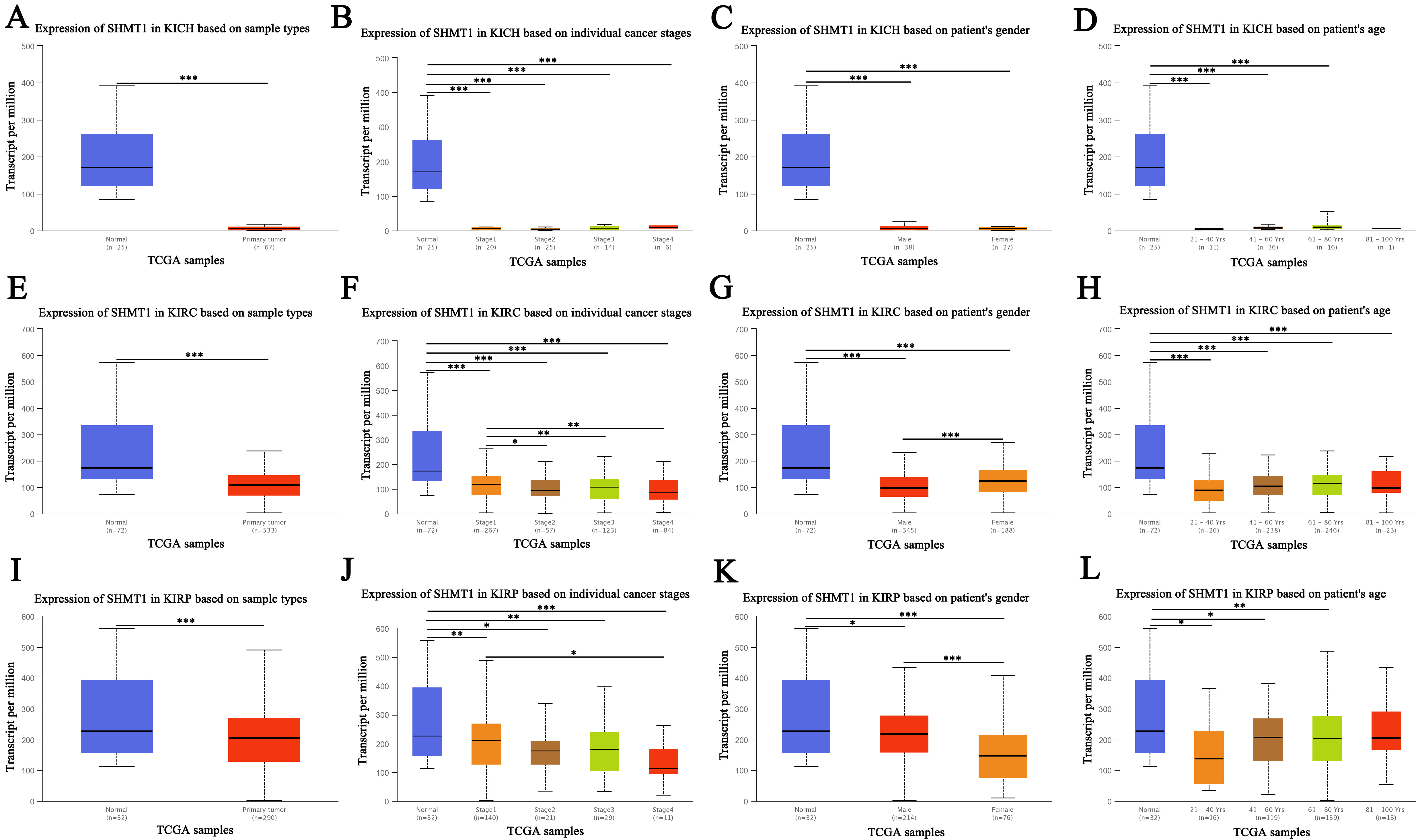
Background: Serine hydroxymethyltransferase (SHMT) is a serine-glycine-one-carbon metabolic enzyme in which SHMT1 and SHMT2 encode the cytoplasmic and mitochondrial isoenzymes, respectively. SHMT1 and SHMT2 are key players in cancer metabolic reprogramming, and thus are attractive targets for cancer therapy. However, the role of SHMT in patients with renal cell carcinoma (RCC) has not been fully elucidated. We aimed to systematically analyze the expression, gene regulatory network, prognostic value, and target prediction of SHMT1 and SHMT2 in patients with kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), and kidney renal papillary cell carcinoma (KIRP); elucidate the association between SHMT expression and RCC; and identify potential new targets for clinical RCC treatment. Methods: Several online databases were used for the analysis, including cBioPortal, TRRUST, GeneMANIA, GEPIA, Metascape, UALCAN, LinkedOmics, and TIMER. Results: SHMT1 and SHMT2 transcript levels were significantly down- and upregulated, respectively, in patients with KICH, KIRC, and KIRP, based on sample type, individual cancer stage, sex, and patient age. Compared to men, women with KIRC and KIRP showed significantly up- and downregulated SHMT1 transcript levels, respectively. However, SHMT2 transcript levels were significantly upregulated in the patients mentioned above. KIRC and KIRP patients with high SHMT1 expression had longer survival periods than those with low SHMT1 expression. In patients with KIRC, the findings were similar to those mentioned above. However, in KICH patients, the findings were the opposite regarding SHMT2 expression. SHMT1 versus SHMT2 were altered by 9% versus 3% (n = 66 KICH patients), 4% versus 4% (n = 446 KIRC patients), and 6% versus 7% (n = 280 KIRP patients). SHMT1 versus SHMT2 promoter methylation levels were significantly up- and downregulated in patients with KIRP versus KIRC and KIRP, respectively. SHMT1, SHMT2, and their neighboring genes (NG) formed a complex network of interactions. The molecular functions of SHMT1 and its NG in patients with KICH, KIRC, and KIRP, included clathrin adaptor, metalloendopeptidase, and GTPase regulator activities; lipid binding, active transmembrane transporter activity, and lipid transporter activity; and type I interferon receptor binding, integrin binding, and protein heterodimerization, respectively. Their respective Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were involved in lysosome activity, human immunodeficiency virus 1 infection, and endocytosis; coronavirus disease 2019 and neurodegeneration pathways (multiple diseases); and RIG-I-like receptor signaling pathway, cell cycle, and actin cytoskeleton regulation. The molecular functions of SHMT2 and its NG in patients with KICH, KIRC, and KIRP included cell adhesion molecule binding and phospholipid binding; protein domain-specific binding, enzyme inhibitor activity, and endopeptidase activity; and hormone activity, integrin binding, and protein kinase regulator activity, respectively. For patients with KIRC versus KIRP, the KEGG pathways were involved in cAMP and calcium signaling pathways versus microRNAs (MiRNAs) in cancer cells and neuroactive ligand-receptor interactions, respectively. We identified the key transcription factors of SHMT1 and its NG. Conclusions: SHMT1 and SHMT2 expression levels were different in patients with RCC. SHMT1 and SHMT2 may be potential therapeutic and prognostic biomarkers in these patients. Transcription factor (MYC, STAT1, PPARG, AR, SREBF2, and SP3) and miRNA (miR-17-5P, miR-422, miR-492, miR-137, miR-30A-3P, and miR-493) regulations may be important strategies for RCC treatment.