†These authors contributed equally.
Background: Ankylosing spondylitis (AS) is a chronic inflammatory
autoimmune disease that affects axial joints such as the spine. Early diagnosis
is essential to improve treatment outcomes. The purpose of this study is to
uncover underlying genetic diagnostic features of AS. Methods: We
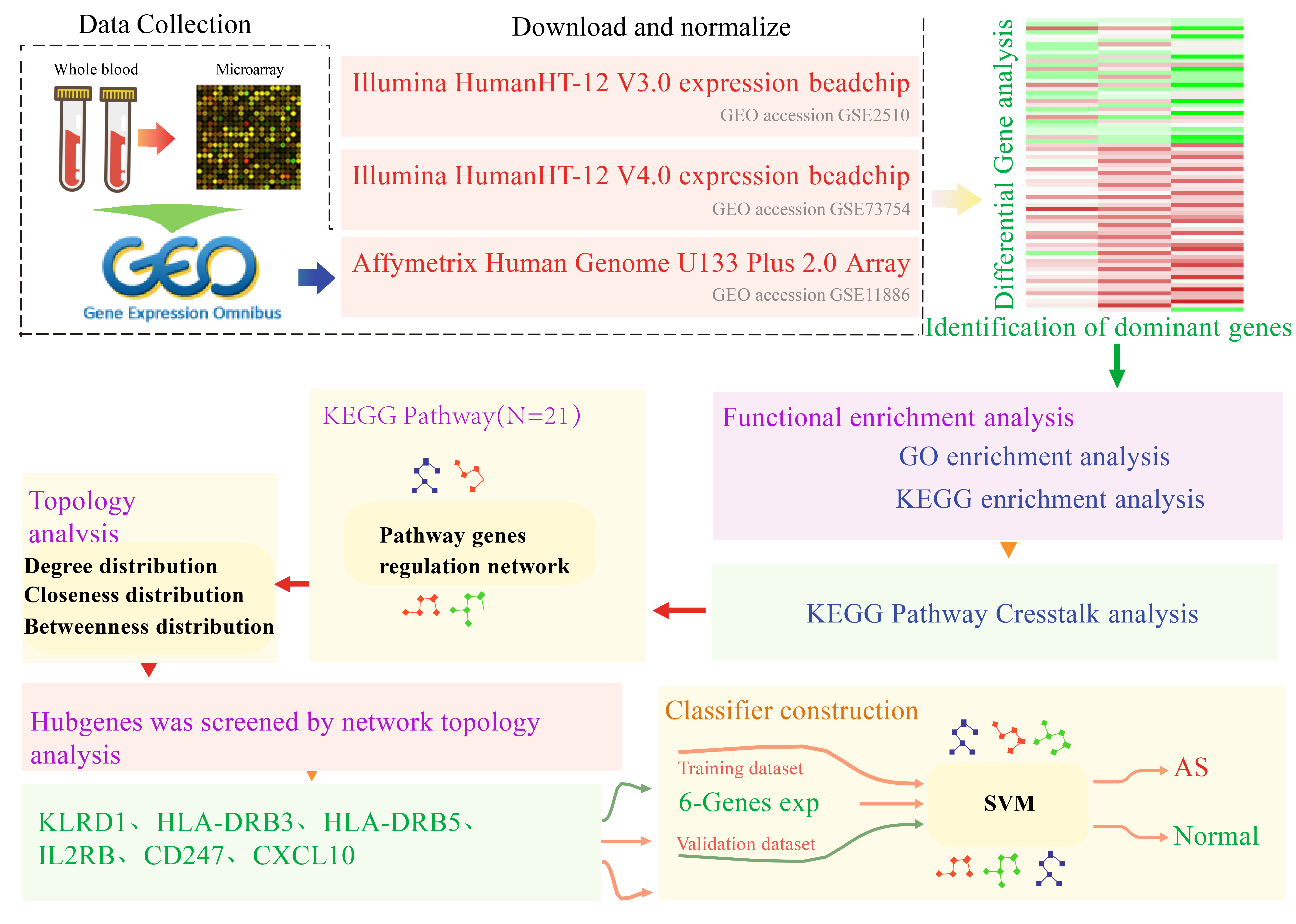
downloaded gene expression data from the Gene Expression Omnibus (GEO) database
for three studies of groups of healthy and AS samples. After preprocessing and
normalizing the data, we employed linear models to identify significant
differentially expressed genes (DEGs) and further integrated the differential
genes to acquire reliable differential transcriptional markers. Gene functional
enrichment analysis was conducted to obtain enriched pathways and regulatory gene
interactions were extracted from pathways to further elucidate pathway networks.
Seventy-three reliably differentially expressed genes (DEGs) were integrated by
differential analysis. Utilizing the regulatory relationships of the 21 Kyoto
Encyclopedia of Genes and Genomes (KEGG) Pathway genes that were enriched in the
analysis, a regulatory network of 622 genes was constructed and its topological
properties were further analyzed. Results: Functional enrichment
analysis found 73 DEGs that were strongly associated with immune pathways like
Th17, Th1 and Th2 cell differentiation. Using KEGG combined with DEGs, six hub
genes (KLRD1, HLA-DRB3, HLA-DRB5, IL2R