-
- Academic Editors
-
-
-
†These authors contributed equally.
This is an open access article under the CC BY 4.0 license.
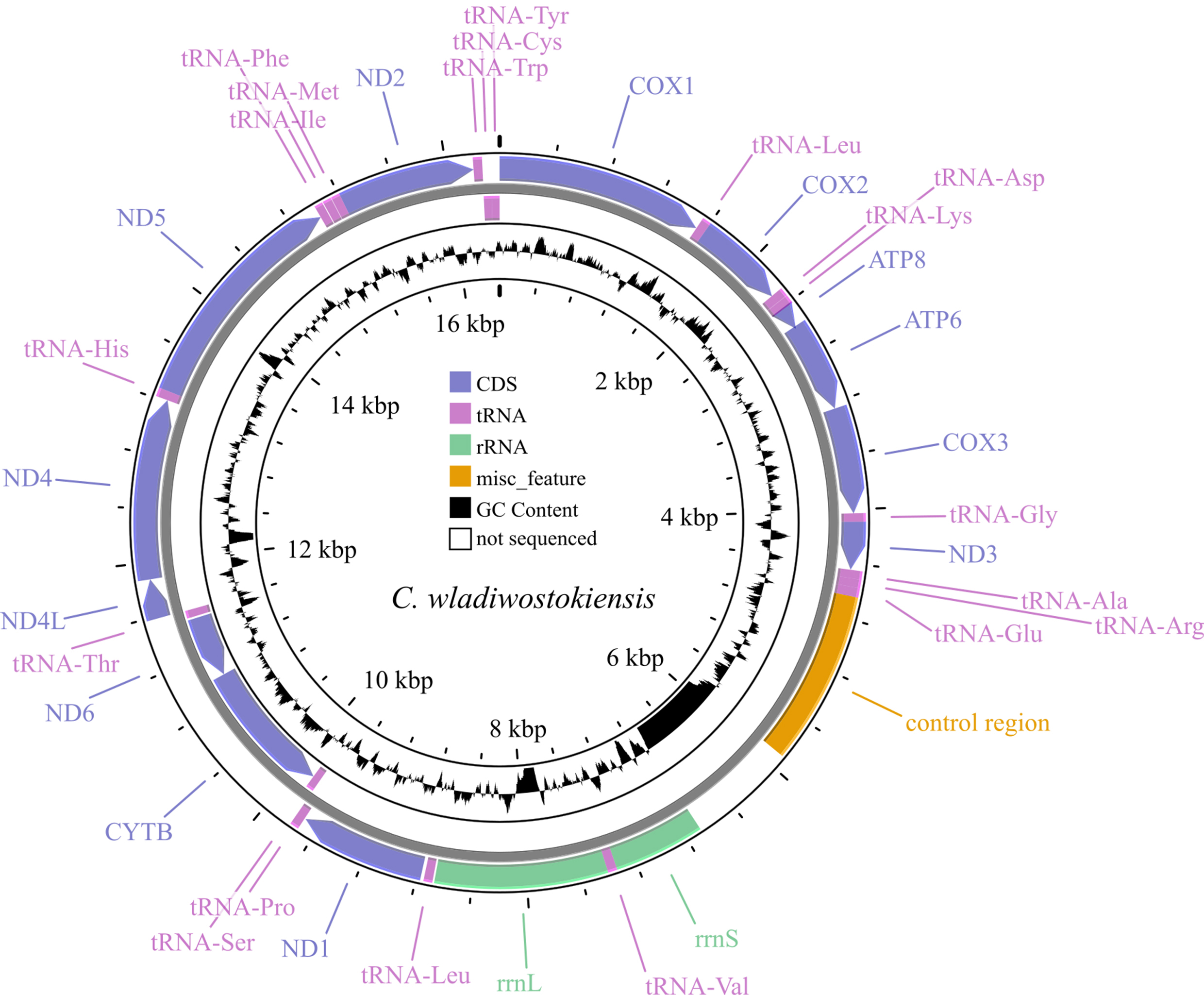
Background: The mitochondrial genome is a powerful tool for exploring and confirming species identity and understanding evolutionary trajectories. The genus Cambaroides, which consists of freshwater crayfish, is recognized for its evolutionary and morphological complexities. However, comprehensive genetic and mitogenomic data on species within this genus, such as C. wladiwostokiensis, remain scarce, thereby necessitating an in-depth mitogenomic exploration to decipher its evolutionary position and validate its species identity. Methods: The mitochondrial genome of C. wladiwostokiensis was obtained through shallow Illumina paired-end sequencing of total DNA, followed by hybrid assembly using both de novo and reference-based techniques. Comparative analysis was performed using available Cambaroides mitochondrial genomes obtained from National Center for Biotechnology Information (NCBI). Additionally, phylogenetic analyses of 23 representatives from three families within the Astacidea infraorder were employed using the PhyloSuite platform for sequence management and phylogenetic preparation, to elucidate phylogenetic relationships via Bayesian Inference (BI), based on concatenated mitochondrial fragments. Results: The resulting genome, which spans 16,391 base pairs was investigated, revealing 13 protein-coding genes, rRNAs (12S and 16S), 19 tRNAs, and a putative control region. Comparative analysis together with five other Cambaroides mitogenomes retrieved from GenBank unveiled regions that remained unread due to challenges associated with the genome skimming technique. Protein-coding genes varied in size and typically exhibited common start (ATG) and stop (TAA) codons. However, exceptions were noted in ND5 (start codon: GTG) and ND1 (stop codon: TAG). Landscape analysis was used to explore sequence variation across the five available mitochondrial genomes of Cambaroides. Conclusions: Collectively, these findings reveal variable sites and contribute to a deeper understanding of the genetic diversity in this genus alongside the further development of species–specific primers for noninvasive monitoring techniques. The partitioned phylogenetic analysis of Astacidea revealed a paraphyletic origin of Asian cambarids, which confirms the data in recent studies based on both multilocus analyses and integrative approaches.