-
- Academic Editor
-
-
-
Background: Structural variations (SVs) are common
genetic alterations in the human genome. However, the profile and clinical
relevance of SVs in patients with hereditary breast and ovarian cancer (HBOC)
syndrome (germline BRCA1/2 mutations) remains to be fully elucidated.
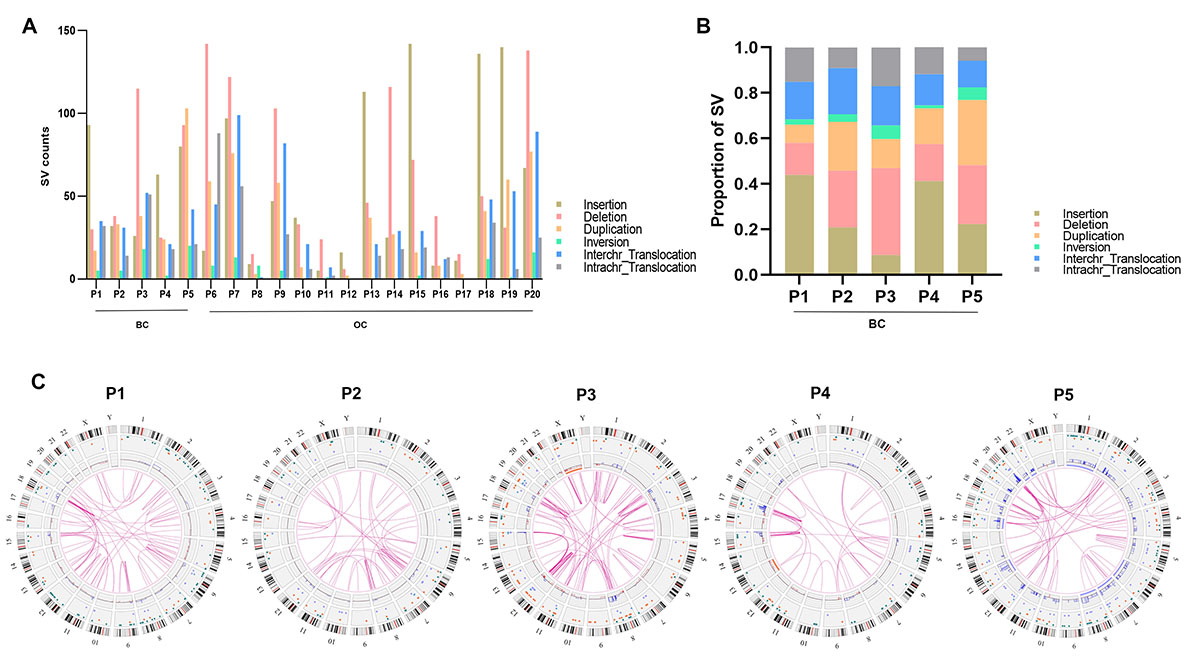
Methods: Twenty HBOC-related cancer samples (5 breast and 15 ovarian
cancers) were studied by optical genome mapping (OGM) and next-generation
sequencing (NGS) assays. Results: The SV landscape in the 5 HBOC-related
breast cancer samples was comprehensively investigated to determine the impact of
intratumor SV heterogeneity on clinicopathological features and on the pattern of
genetic alteration. SVs and copy number variations (CNVs) were common genetic
events in HBOC-related breast cancer, with a median of 212 SVs and 107 CNVs per
sample. The most frequently detected type of SV was insertion, followed by
deletion. The 5 HBOC-related breast cancer samples were divided into SV