- Academic Editor
-
-
-
Background: Fallopia aubertii (L. Henry) Holub is a perennial
semi-shrub with both ornamental and medicinal value. The mitochondrial genomes of
plants contain valuable genetic traits that can be utilized for the exploitation
of genetic resources. The parsing of F. aubertii mitochondrial genome
can provide insight into the role of mitochondria in plant growth and
development, metabolism regulation, evolution, and response to environmental
stress. Methods: In this study, we sequenced the mitochondrial genome of
F. aubertii using the Illumina NovaSeq 6000 platform and Nanopore
platform. We conducted a comprehensive analysis of the mitochondrial genome of
F. aubertii, which involved examining various aspects such as gene
composition, repetitive sequences, RNA editing sites, phylogeny, and organelle
genome homology. To achieve this, we employed several bioinformatics methods
including sequence alignment analysis, repetitive sequence analysis, phylogeny
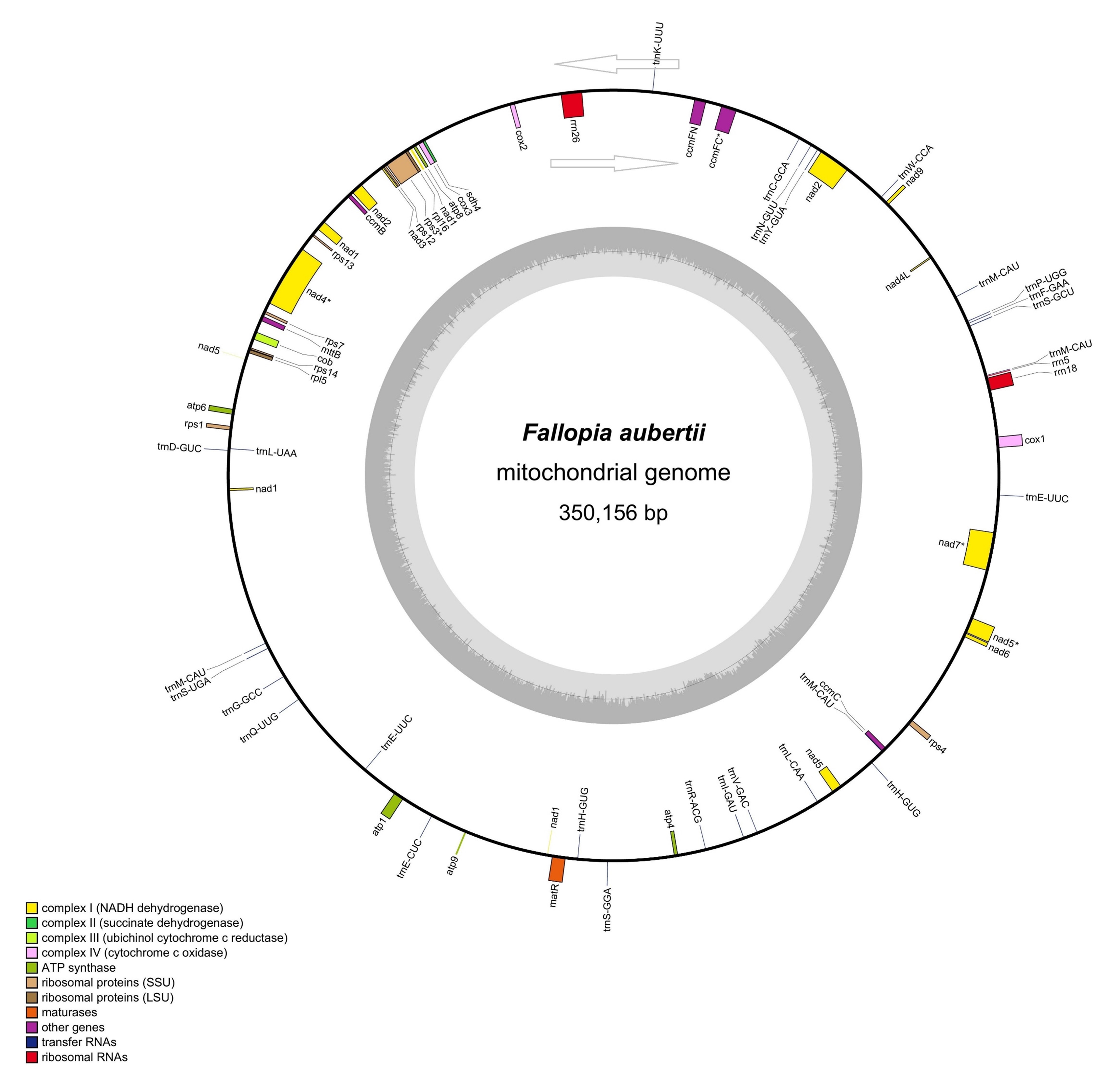
analysis, and more. Results: The mitochondrial genome of F.
aubertii has 64 genes, including 34 protein-coding genes (PCGs), three rRNAs,
and 27 tRNAs. There were 77 short tandem repeat sequences detected in the
mitochondrial genome, five tandem repeat sequences identified by Tandem Repeats
Finder (TRF), and 50 scattered repeat sequences observed, including 22 forward
repeat sequences and 28 palindrome repeat sequences. A total of 367 RNA coding
sites were predicted in PCGs, with the highest number (33) found within
ccmB. Ka/Ks values estimated for mitochondrial genes of
F. aubertii and three closely related species representing
Caryophyllales were less than 1 for most of the genes. The maximum likelihood
evolutionary tree showed that F. aubertii and Nepenthes