- Academic Editor
Infection diagnosis and antibiotic sensitivity testing are important aspects of clinical microbiology that are in dire need of improvement owing to the inadequate current standards in the early detection of bacterial response to antibiotics. The increasing antimicrobial resistance is a serious global threat to human health. Current resistance-detecting methods, using the phenotypic antibiotic sensitivity test, which measures bacterial growth as affected by antibiotics, have long analysis times. Therefore, new and rapid methods are needed to detect antibiotic resistance. Here, we review the methods used to detect antibiotic resistance in bacteria, including that caused by biofilm development, and we look at the development of rapid methods for evaluating antimicrobial resistance (AMR).
Currently, the world is faced with a rapid spread of antimicrobial resistance (AMR). Yet, more and more drugs become ineffective against nosocomial and community-acquired infections. The emergence of resistance in microorganisms is a natural response to the widespread use of antimicrobials in clinical practice. Diseases caused by multidrug-resistant (MDR) bacteria are severe and most often require hospitalization. The lack of effective antibiotic therapy is both a worldwide threat to all countries without exception and a threat to each person who encounters pathogens in one way or another [1, 2].
Combating bacterial antibiotic resistance now includes the following directions: (1) Designing new drugs able to effectively suppress pathogens. (2) Finding ways to slow down the spread of resistance, in particular by reducing antibiotic consumption. (3) Developing methods to “turn off” the resistance. Unfortunately, it is next to impossible to transition to the rational use of antimicrobials. Routinely informing health professionals and patients about antibiotic resistance and its global consequences could be the first step in curbing its spread [3]. Because efforts to combat bacterial resistance by blocking its mechanisms are ineffective, the development of new antibacterial agents is a priority in the current war against drug resistance [4].
Antimicrobials are used to prevent and treat infections in humans, animals, and plants. They generally include antibiotic, antiviral, antifungal, and antiparasitic agents. The overuse of antibiotics and their improper use give rise to MDR strains. The increasing antibiotic resistance is regarded worldwide as a threat to national security. The World Health Organization (WHO) considers it of the highest priority, as evidenced by WHO Global Strategy for Containment of Antimicrobial Resistance. A United Nations report calls antibiotic resistance a “serious threat” to global health [5, 6, 7, 8, 9]. The actual magnitude of worldwide antibiotic resistance remains unknown [10]; therefore, the development of methods to evaluate the sensitivity of bacteria to antibacterial drugs is of great importance.
Here, we review the methods used currently to detect antibiotic resistance in bacteria, including that caused by biofilm development, and we look at the development of rapid methods for evaluating AMR.
Antibiotics are a broad group of drugs of natural, synthetic, or semisynthetic
origin that are selectively toxic to bacteria or other single-celled
microorganisms. The main antibiotic groups are
A major source of antibiotic entry into the environment is treated wastewater. Because antibiotics do not undergo metabolic transformation, they pass through wastewater treatment systems [13, 14, 15]. Soils are polluted by antibiotics coming from the use of manure, sludge, and wastewater [16], but some antibiotics are used directly on farmland to control pathogens [17]. Sometimes antibiotic concentrations may be as high as or even greater than therapeutic levels, which in turn increases bacterial resistance. The recent widespread and often uncontrolled use of antimicrobials has resulted in a rapid spread of multidrug-resistant microorganisms [18]. Fig. 1 (Ref. [13]) shows the main pathways by which antibiotics enter the environment.
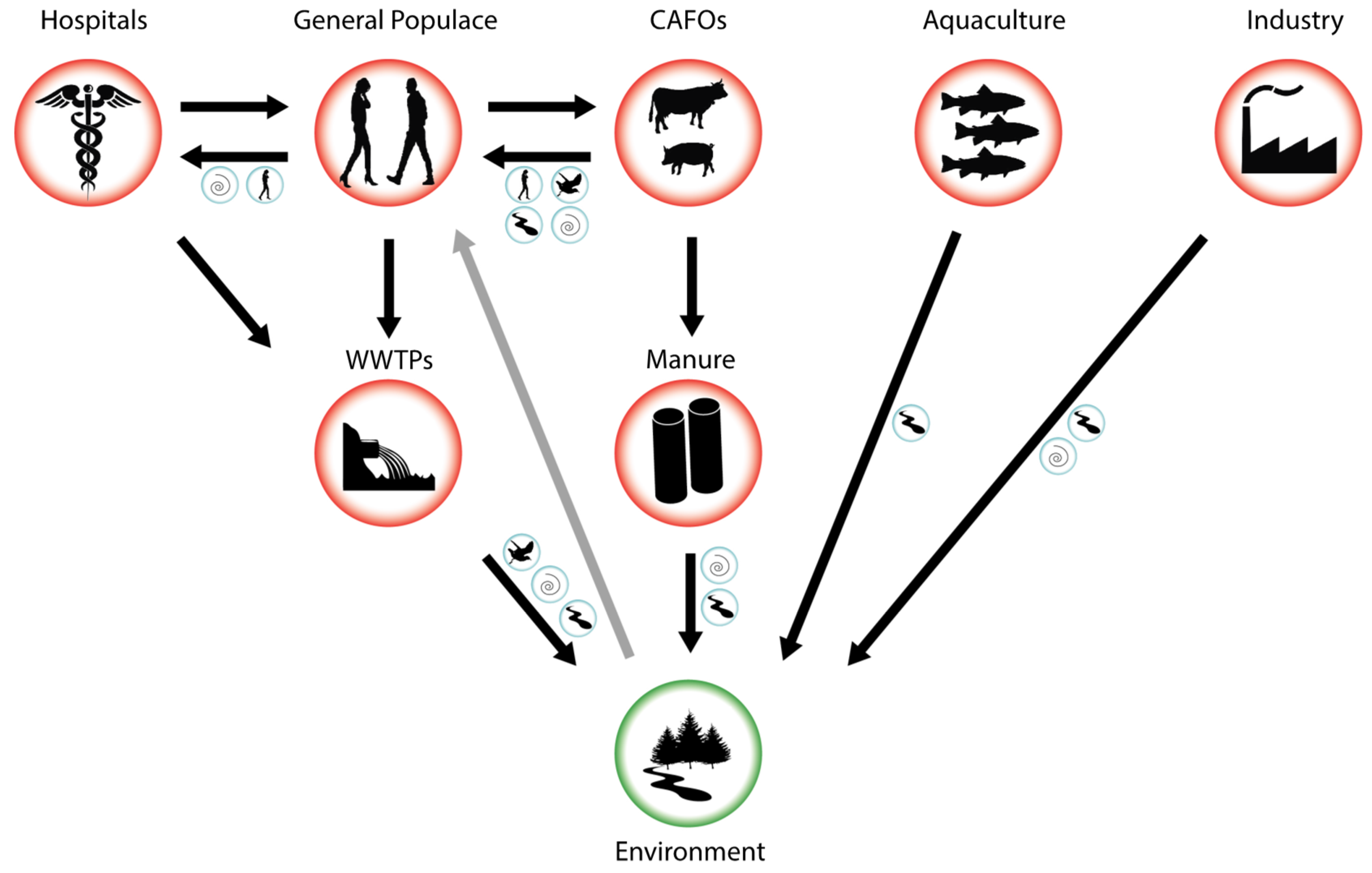 Fig. 1.
Fig. 1.Pathways for antibiotic entry into the environment. CAFOs, concentrated animal feeding operations; WWTP, wastewater treatment plants [13].
The worldwide consumption of antibiotics is hundreds of thousands of tons per year. In 2013, for example, the total antibiotic consumption in the United Kingdom was 27.4 defined daily doses per 1000 inhabitants per day [14]. In the United States, antibiotic resistance affects 2 million people a year and causes at least 23,000 deaths [19]. A report published by a panel assembled by the UK Department of Health states that between 2014 and 2016, about 700,000 people worldwide die each year from infectious diseases caused by antimicrobial-resistant pathogens [20]. According to the 2019 data, about 68,000 people per year in the United States and Europe die cumulatively from diseases caused by resistant bacteria [21, 22]. If this trend continues and antiresistance measures remain ineffective, that number could rise to 10 million people per year by 2050 [20].
According to the European Center for Disease Prevention and Control, about 25,000 people die annually in Europe directly from drug-resistant infections. However, anecdotal evidence suggests that the actual death rate is much higher, possibly 500,000 people per year [13].
Antibiotic resistance also greatly affects the global economy, with the United States alone spending about $35 billion annually to treat resistant infections [23]. Importantly, the rate of antibiotic discovery has declined in recent decades because of technical and economic problems, which has led to the so-called antibiotic crisis [24].
Increasing antibiotic concentrations in the environment may give rise to resistant bacteria and may potentially contribute to the emergence of new resistance determinants [25]. Aquatic microbial consortia are foci of the horizontal transfer of the AMR genes. In addition, antibiotic resistance genes that persist in the environment may be recruited by human bacterial pathogens through horizontal gene transfer, which causes human health risks [26, 27]. Resistance of nonhospital microorganisms to antimicrobials is a growing concern in the scientific community owing to the public health crisis, and antibiotic resistance genes are considered new environmental contaminants [28].
Antimicrobial agents inhibit bacterial cell wall construction, disrupt cell
membrane structure and function, and block the synthesis of vital biopolymers,
including RNA and DNA [29, 30]. Antibiotics are called bactericidal if they cause
cell death, for example, by disrupting cell wall construction. Bacteriostatic
antibiotics are those that only inhibit bacterial growth, disrupting the
synthesis of proteins and nucleic acids. Some antibacterials are effective
against a narrow range of bacteria; for example, glycopeptides are active only
against gram-positive strains. Other antibiotics, such as
Evolution has led bacteria to develop a number of defense mechanisms to inactivate antibiotics, and this has led to the emergence of multidrug-resistant microorganisms [29]. Bacterial resistance to antibiotics can be natural or acquired. Natural resistance is characterized by the absence in bacteria of appropriate targets of antibiotic action, low permeability of the cell wall, or the possibility of enzymatic inactivation of the antibiotic. This type of resistance is species-specific [31]. Acquired resistance may arise as a result of the selection of antibiotic-exposed bacteria, either through mutations in chromosomal or plasmid DNA or through horizontal transfer of resistance genes via plasmids or transposons [29, 32].
The main MDR mechanisms include:
Antibiotic resistance can be transmitted through different mechanisms [38, 39]. Human bacterial pathogens, too, undergo evolutionary changes in human-changed environments, which affects their antibiotic resistance [32, 40, 41, 42, 43, 44].
Obviously, bacteria combat antibiotic effects by acquiring preexisting resistance determinants. De novo mutations play a part in the acquisition of drug resistance by microorganisms, but horizontal gene transfer through transduction, transformation, and conjugation is of primary importance in the spread of antibiotic resistance determinants [45]. This is achieved through the coordinated action of mobile genetic elements capable of moving within or between DNA molecules, which include transposons, gene cassettes/integrons, and plasmids. Together, these elements are central for the acquisition and spread of resistance genes [46]. This process encompasses not only nosocomial strains but also microorganisms in natural settings. For example, subinhibitory antibiotic concentrations promote the spread of antibiotic resistance determinants in a bacterial population through horizontal transfer of genetic material [47, 48]. Nonhospital antibiotic-resistant bacteria pose a potential threat, because they can act as reservoirs for the maintenance and spread of resistance genes [49].
One mechanism that allows bacteria to survive in the presence of antibiotics is specialized transport systems located in the bacterial cell membrane. These efflux systems are protein complexes involved in the secretion of toxic substances from the cell [50]. Bacterial efflux systems are divided into five families: the ABC transporter (ATP-binding cassette) family, the major facilitator superfamily (MFS), the multidrug and toxic compound extrusion (MATE) family, the small multidrug resistance (SMR) family, and the resistance nodulation division (RND) family [51]. The most clinically relevant family is the RND one. These efflux systems are widespread among bacteria, and their genes are almost always present on the chromosome [52]. The MacA–MacB–TolC efflux system of Escherichia coli BL21 (DE3) protects cells against macrolide antibiotics and is involved in the secretion of heat-stable enterotoxin II [53]. The MacAB efflux system protects cells of Salmonella enterica serovar Typhimurium American Type Culture Collection (ATCC) 14028 from macrolide antibiotics [54]. In Klebsiella pneumoniae, the MacAB efflux system protects cells from the synthetic tetracycline eravacycline [55].
The genes coding for efflux systems are not newly acquired as a result of horizontal transfer; rather, they are part of the core genome [56]. Considering that active human use of antibiotics began less than a century ago, one can see that the function of efflux systems is not limited to the removal of antibiotics from the cell.
Evidence for the role of efflux systems in biofilm formation came from a study on the biofilm-forming ability of deletion mutants of S. enterica serovar Typhimurium [57]. The authors showed that the deletion of the efflux system genes acrB, acrD, acrEF, emrAB, macAB, mdfA, mdsABC, mdtABC, mdtK, and tolC results in reduced biofilm formation, as compared with that in the wild-type strain.
Almost every bacterial genome has several efflux systems involved in the elimination of the same antibiotics from the cells. Moreover, many of them are also capable of removing nonantibiotic compounds, such as dyes and detergents. For example, there are at least 11 efflux systems with overlapping functions in the genome of Salmonella typhimurium. By using one of these systems, S. typhimurium MasAb, involved in the efflux of macrolide antibiotics [58], it has been shown that this system is necessary to protect bacteria from oxidative stress [59, 60].
In 2017, WHO published a list of “priority pathogens”, whose antibiotic resistance requires the immediate development of new antibacterials. This list includes Serratia spp. In many hospitalized patients, S. marcescens causes central nervous system diseases (meningitis), urinary tract infections, respiratory diseases, endocarditis, and sepsis. Because antibiotic resistance decreases bacteria’s drug potential, a list was compiled of the currently most dangerous pathogens, which was designated by the acronym ESKAPE (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.). Importantly, P. aeruginosa, an opportunistic human pathogen and a causative agent of severe nosocomial infections, is on the WHO list of “critical priority” microorganisms [61], whose spread has to be contained with new antimicrobials [62]. Moreover, recent studies have shown that nonpathogenic Pseudomonas strains, despite their low virulence, can cause bacteremia in humans with compromised immune systems [63]. Different bacterial species isolated from infected wounds have shown one or more mechanisms of resistance to each of the major classes of antimicrobials [64, 65]. The wound surface provides a favorable environment for microbial colonization, proliferation, and infection [66, 67]. Major bacterial pathogens associated with wound infection and possessing MDR properties include S. aureus, E. coli, K. pneumoniae, Streptococcus pyogenes, P. aeruginosa, A. baumannii, Proteus species, Streptococcus, and Enterococcus [32, 68, 69, 70, 71].
In this context, one topical area is the development of methods to monitor the sensitivity of bacteria to antibiotics. Antibiotic resistance monitoring systems are designed to develop methods for evaluating the sensitivity of bacteria within a short time to provide timely care to patients and predict the spread of resistance among microorganisms and the emergence of new resistance mechanisms.
The main goal of antibiotic therapy is to determine the antimicrobial sensitivity (AMS) of bacteria. Standard microbiological methods, such as determination of the minimum inhibitory concentration (MIC) of an antibiotic in beef-extract broth or on solid nutrient media, are most commonly used to evaluate antibiotic effects on bacteria. The determination of the antimicrobial activity of antibiotics is based on their ability to inhibit the growth of microorganisms and is expressed as a logarithmic dependence of the size of the growth inhibition zones of the test bacteria on the antibiotic concentration. This dependence should be linear.
Methods for determining the AMS of microbes can be divided as follows [72]:
Manual systems in clinical microbiology
Automated systems in clinical microbiology
Emerging technologies
The MIC is defined as the minimum antibiotic concentration that prevents visible microbial growth on agar or in a broth dilution test [73]. Antibiotic sensitivity results are evaluated with a special ready-made table that contains borderline values of the bacteriostasis zone diameters for resistant, moderately resistant, and sensitive strains and also MIC values for resistant and sensitive strains.
Sensitive strains are those whose growth is inhibited by drug concentrations detectable in the sera of patients receiving usual doses of antibiotics. Moderately resistant strains are those whose growth is suppressed by concentrations that are detected in the sera of patients receiving maximum drug doses. Resistant microorganisms are those whose growth is not inhibited by the concentrations produced in the body when maximum doses are used [74, 75]. The data obtained for each bacterium–antibiotic pair are predetermined according to the recommendations of the Clinical Laboratory Standards Institute (CLSI) in the United States and the European Committee for Antibiotic Sensitivity Testing (EUCAST) in Europe [72]. These values provide clinicians with the necessary information to select the appropriate antibacterial agent.
Conventional microbiological methods, though highly sensitive, are time-consuming [76]. Conventional AMS testing is essentially limited to observations of cell colony growth, which may last for several days (Fig. 2, Ref. [72]). It is important to note that while waiting for AMS test results, patients continue to receive broad-spectrum antibiotics, which increase the likelihood of antibiotic resistance. This can be avoided by prescribing antibiotics early in infection, thereby decreasing the use of broad-spectrum antibiotics [72]. Every hour of delay in prescribing the right antibiotic in patients with septic shock reduces their chances of survival by 7.6% [77].
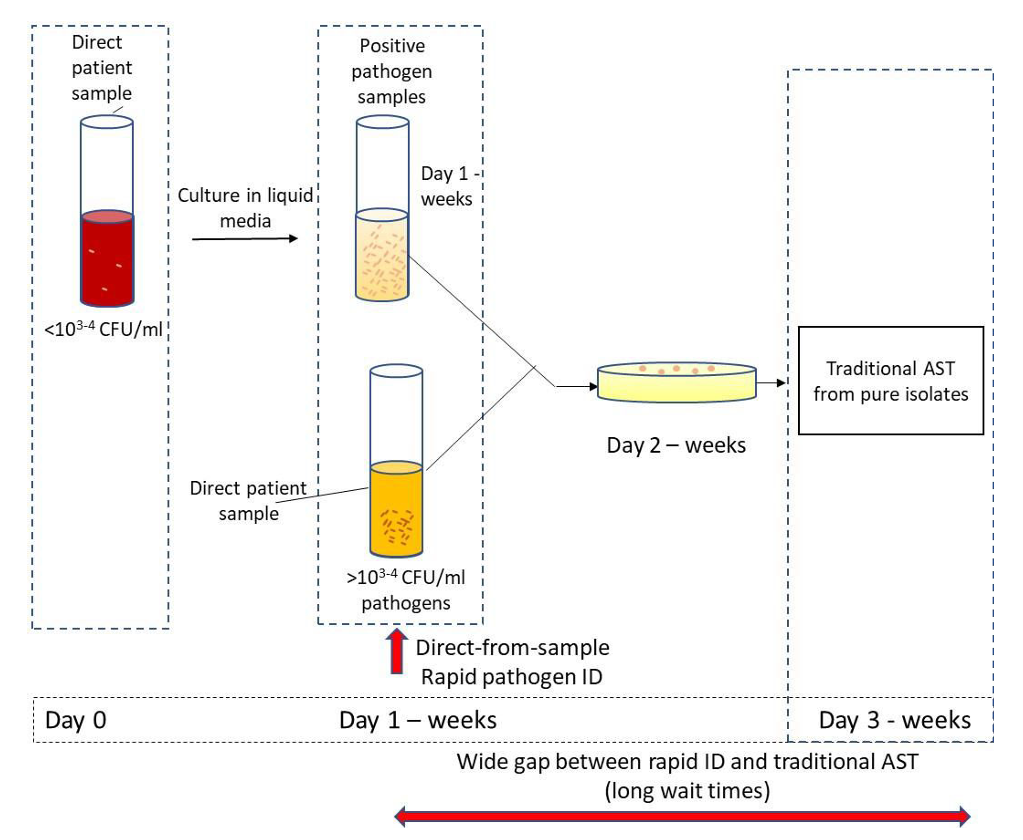 Fig. 2.
Fig. 2.Antimicrobial sensitivity (AMS) testing. Sample processing and AMS testing in the clinical laboratory [72].
Therefore, it is vital to design rapid methods to analyze the AMS of bacteria. In this direction, sensory systems are being developed that are based on electrophysical analysis.
Electrophysical analysis of microbial cells is very promising for the rapid testing of AMS, because it generates information about metabolically active cells within a short time. For example, Johnson et al. [78] used a quartz resonator, nonmotile E. coli, and antibiotics (polymyxin B and ampicillin) to record the mechanical vibrations of bacteria before and after their exposure to the antibiotics. In conjunction with cell imaging before and after exposure to the antibiotics and with postexperiment counting of colony-forming units (CFU), the results showed that AMS can be tested within 7–15 min.
Guliy et al. [79] used a lithium niobate–based piezoelectric resonator with a lateral electric field to evaluate the effects of amoxicillin and polymyxin on microbial cells. The analysis was done directly in the liquid phase. It was shown that when an antibiotic is added, the frequency dependences of the real and imaginary parts of the electrical impedance of the resonator loaded with a suspension of sensitive cells differ strongly from the dependences for the resonator loaded with a no-antibiotic control cell suspension. The analysis time was not greater than 10 min.
Kaittanis et al. [80] showed that iron oxide nanosensors, either coated with dextran supplemented with concanavalin A or coated with silica conjugated directly to concanavalin A, can be useful for the fast (1) measurement of polysaccharides, (2) evaluation of metabolic activity, and (3) determination of AMS in blood (Fig. 3, Ref. [80]). It is expected that these polysaccharide nanosensors will be used in AMS determination in the clinic or in the field and also in pharmaceutical research and development.
Nanomechanical biosensors belong to the subfamily of microelectromechanical systems (MEMSs), which can convert biological processes into measurable mechanical motion. For example, nanomechanical sensors are advantageous for the detection of bacteria and their AMS [81]. Pathogens (and their growth and metabolic activities) are detected on the sensor surface, which causes sensor deformation (static response) and/or a change in the frequency resonance (dynamic response; Fig. 4 (Ref. [81])). Three components were combined into a single nanomechanical device to detect bacteria in real time and measure their AMS (Fig. 5A–E) [81].
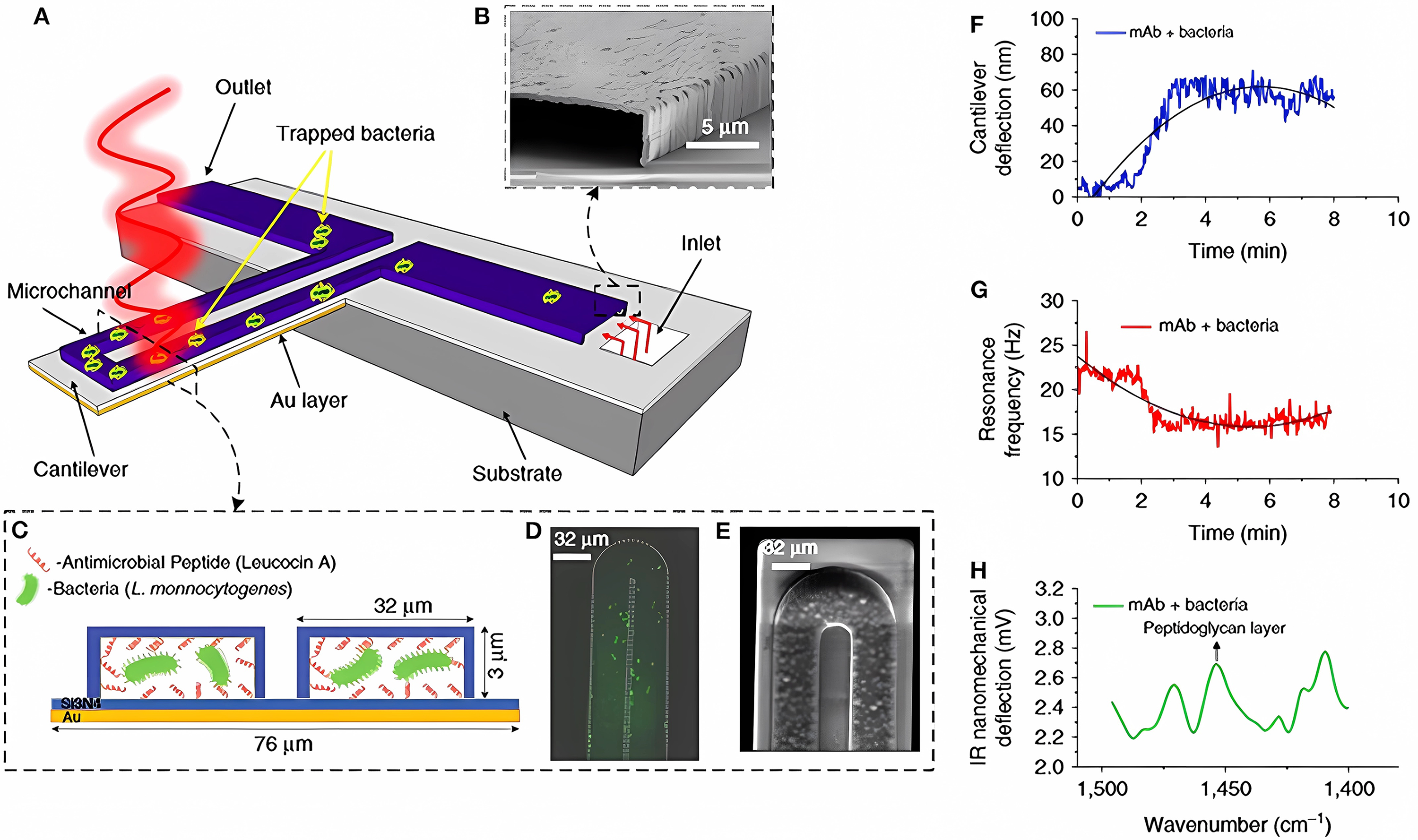 Fig. 5.
Fig. 5.Detection of bacteria with multimodal nanomechanical sensors. (A) Microfluidic gold-coated cantilever. (B) Cross-sectional SEM image of the inlet channel. (C) Cross-section of the functionalized microchannel with adsorbed bacteria. (D) Fluorescent image and (E) SEM image of the tip of the microfluidic cantilever containing bacteria. (F) Temporal changes in cantilever deflection, as caused by the trapped bacteria. (G) Temporal changes in the resonance frequency, as caused by the trapped bacteria. (H) Infrared absorption spectra of the trapped bacteria, obtained by measuring the cantilever deflection induced by illumination with infrared light [81].
A gold-coated silicon nitride microcantilever containing an integrated microfluidic channel trapped the bacteria and measured deflection changes (owing to the surface tension changes caused by cell binding; Fig. 5F) and changes in the resonance frequency (owing to the cell mass; Fig. 5G). In addition, infrared spectra of the trapped bacteria were measured by excitation with infrared radiation, which induced cantilever deflection proportional to the infrared absorption of the cell (Fig. 5H). This multivariate analysis allows the detection and identification of bacterial strains in situ and the accurate differentiation between damaged and nondamaged cells.
Longo et al. [82] found that bacterial metabolism can be measured by observing static-mode fluctuations of a cantilever coated with bacteria in an aqueous environment. The detection is based on temporal changes in the fluctuation (background noise) of the transducer, rather than on classical static deviation or on changes in the resonance frequency. Using viable S. aureus, Longo et al. [82] showed that in the presence of ampicillin and kanamycin, the fluctuation deviation is strongly reduced, because the antibiotics cause the bacteria to die. The MIC data were consistent with those obtained by standard methods of AMS determination.
Antibiotics affect bacterial physiology on many levels. Rather than simply compensating for direct cellular defects, bacteria respond to antibiotics by altering their morphology, macromolecular composition, metabolism, and so on [83]. Inevitably, all these processes influence each other, resulting in a complex response. The cell-surface damage is accompanied by the redistribution of ions and charges, which can be recorded with an electro-optical sensor. An optical method was applied to the determination of bacterial sensitivity to ampicillin and kanamycin, with the analysis time being not greater than 20 min [84]. The method is based on recording changes in the electro-optical characteristics of microbial cells exposed to antibiotics.
Jain et al. [85] put forward a unique method of AMS determination that combines the disk-diffusion method with microwave radiation for noncontact and noninvasive probing and monitoring. Their study described the use of a split-ring microwave resonator to monitor the effect of different erythromycin concentrations on E. coli grown on a solid agar medium (Fig. 6, Ref. [85]).
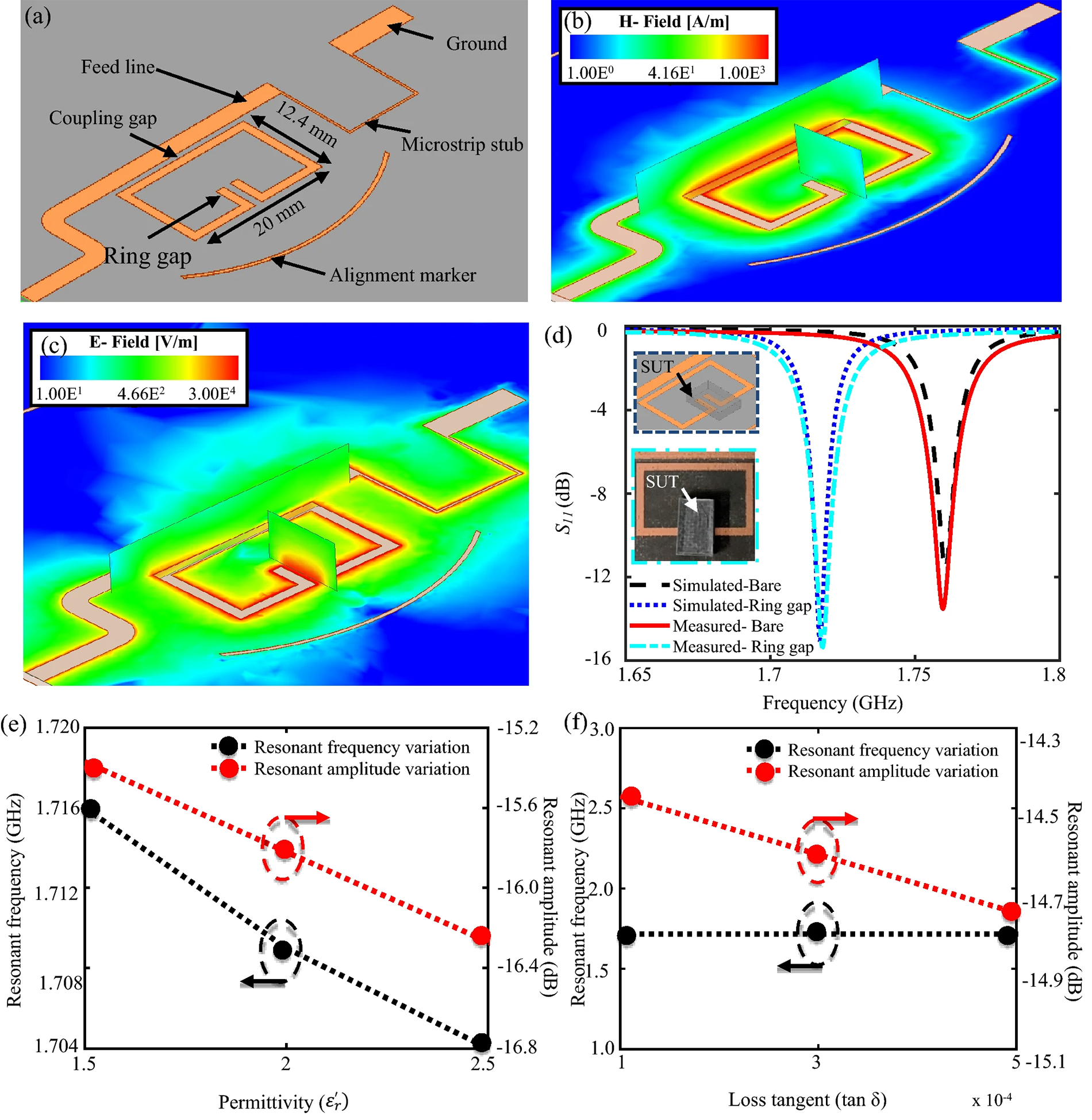 Fig. 6.
Fig. 6.Modeled one-port split-ring resonator. (a) Modelled one-port split-ring resonator. (b) Magnetic feld distribution on the resonator. (c) Electric feld distribution on the resonator, with the sensitive zone found in the ring gap. (d) Simulated versus measured reflection coefficient in the absence and presence of the test sample. Effect of variation in (e) permittivity and (f) loss tangent of the sample on the resonator’s resonant amplitude and frequency [85].
The change in the amplitude decreased to low values, indicating a delay in bacterial growth at high antibiotic concentrations. The sensor showed convincing AMS results in less than 6 h and is, therefore, promising for the expansion of work process automation in clinical settings.
The same team of authors [86] described a microwave and microfluidic biosensor for the rapid, noncontact, and noninvasive testing of E. coli concentration and growth in media of different pH to improve efficacy in clinical microbiology. The thin interface layer between the microfluidic channel and the microwave resonator greatly increased the detection sensitivity. The presence of different concentrations of bacteria in solutions of different pH was determined by screening for changes in the resonance amplitude and frequency characteristics of the microwave system. The sensor showed an almost immediate response to changes in the bacterial concentration and a maximum sensitivity of 3.4 MHz, as compared with the logarithmic value of the bacterial concentration. The changes in the sensor’s variables (resonance frequency and amplitude) were used to monitor the growth of antibiotic-exposed bacteria for 500 min (Fig. 7, Ref. [86]).
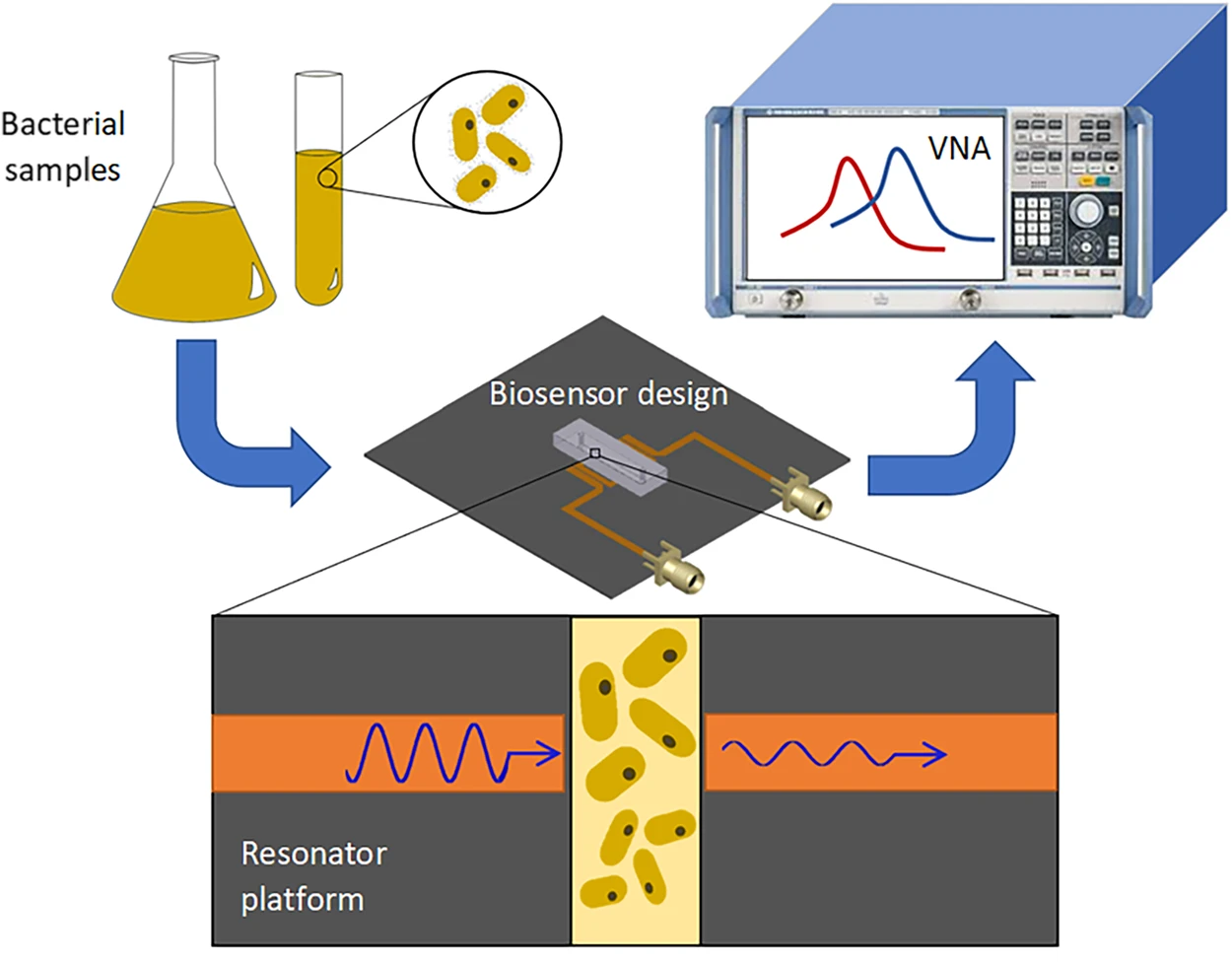 Fig. 7.
Fig. 7.Detection of bacteria concentrations and proliferation. Bacteria are diluted to a desired concentration and are introduced into the microwave–microfluidic platform. The electrical signal of the resonator is analyzed through a vector network analyzer (VNA) to obtain the resonant profile for the bacteria [86].
Bennett et al. [87] described an alternative method that detects the phenotypic resistance of bacteria to antibiotics within 45 min. It uses a laser and detector system to detect in media single bacterial cells as they pass through the laser focus (Fig. 8, Ref. [87]). This provides a simple readout of AMS by detecting the growth (resistant) or death (sensitive) of bacteria much faster than standard methods. The method was applied in both laboratory and clinical settings, with E. coli as an example.
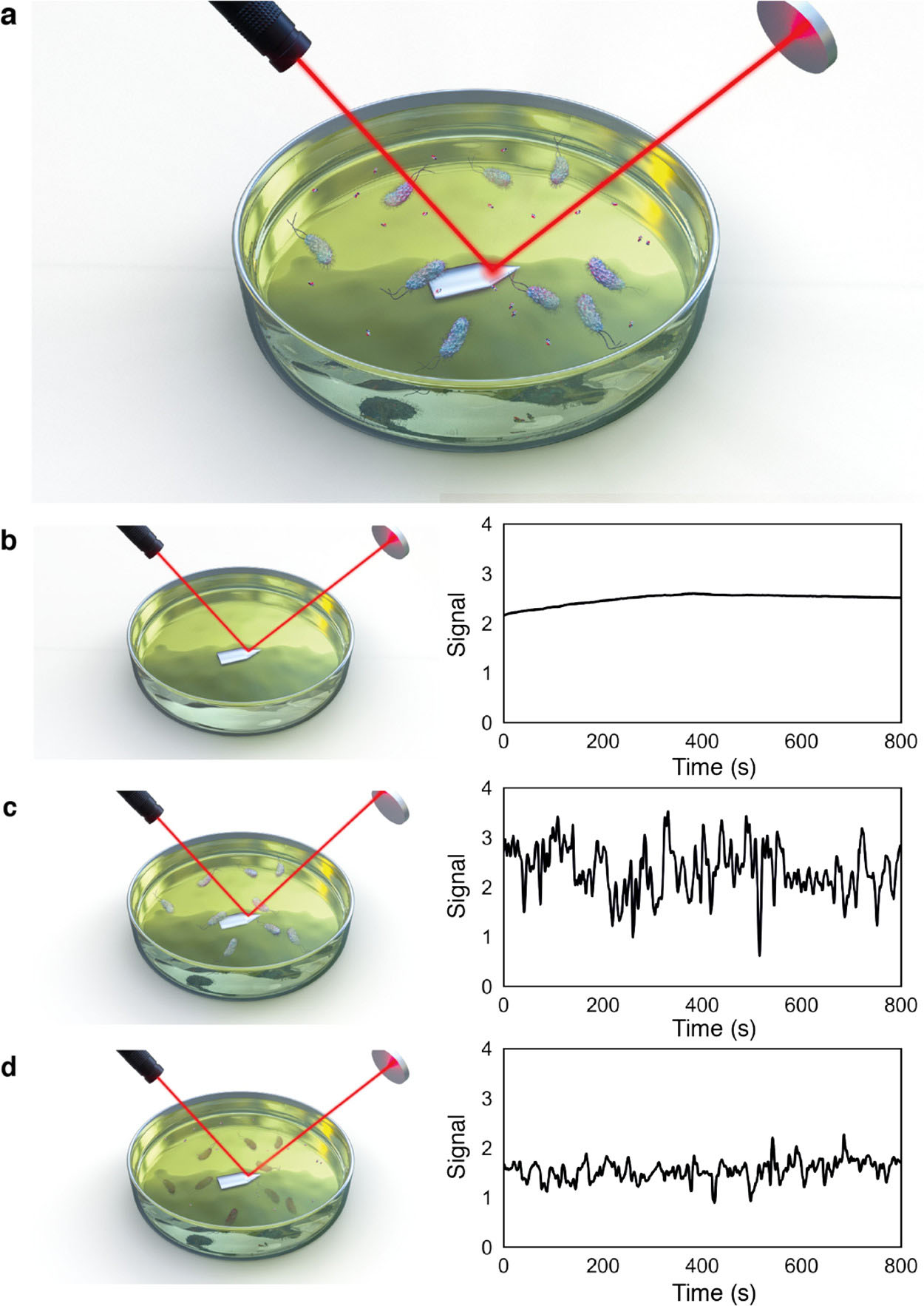 Fig. 8.
Fig. 8.Principle of the rapid optical AST method. (a) Bacteria are inoculated into antibiotic-containing growth media, with laser reflecting off the cantilever surface onto a photodiode detector. Bacteria in solution move through the laser beam (observed as peaks in the photodiode signal). The photodiode signal, measured from the media solution, decreases after the antibiotic is added for sensitive strains. (b–d) Photodiode signal (b) without bacterial inoculant, (c) with bacteria in solution, and (d) 45 min after addition of the antibiotic [87].
The fact of microbial cells being sensitive to a particular antibiotic does not guarantee its effectiveness in treatment. Lack of antibiotic efficacy is often associated with the formation of bacterial biofilms, which are difficult for antibiotics to penetrate [88, 89]. Biofilm-associated bacteria differ in many ways from their planktonic counterparts; a typical property of biofilms is their reduced sensitivity to antimicrobial agents [90]. Because biofilm infections and problems with their treatment seriously threaten human health, more and more recent research has addressed the formation of biofilms and the strategies to combat them. The International Center for Disease Control and Prevention and the American National Institutes of Health (NIH) estimated that biofilms are implicated in 65% of bacterial infections and in more than 80% of chronic infections [91]. At least 80% of biofilm-forming pathogens are associated with persistent infections [92, 93]. The most common exogenous microorganisms are staphylococci and streptococci [94]. It is now understood that about 40–80% of bacteria on Earth can form biofilms [95]. Biofilms may contain bacteria of one or more species. Most bacteria and fungi can form biofilms [96], such as P. aeruginosa [97, 98], Staphylococcus epidermidis [99], Candida albicans [100], A. baumannii [101], Helicobacter pylori [102], S. aureus [103], Listeria monocytogenes [104, 105], Vibrio cholerae [106], and S. enterica [107]. According to the European Antimicrobial Resistance Surveillance Network (EARS-Net) protocol, pathogens subject to infection control surveillance include E. coli, K. pneumoniae, P. aeruginosa, Acinetobacter spp., S. aureus, S. pneumoniae, E. faecalis, and E. faecium. These pathogens have been chosen because they often cause invasive infections, and if resistance develops, treatment options may be severely limited. Also, these pathogens can spread in nonhospital settings and in health care facilities. In 2016, Salmonella spp. were listed as subject to Central Asian and Eastern European Surveillance of Antimicrobial Resistance (CAESAR) surveillance [108]. Of note, approximately 80% of the wound infections reported in the United States may be associated with biofilms [91, 109]. Bacteria growing in a biofilm are thought to be between 10 and 1000 times more tolerant to antibiotics than their planktonic counterparts [110, 111]. The biofilm acts as a physical barrier that reduces the rate of penetration of antibiotics, antibodies, and granulocytic cell populations. In addition, low doses of antibiotics may promote biofilm formation and may be responsible for biofilm-specific AMR [112]. In the largest and most comprehensive study to date on the global burden of AMR, an international team of researchers estimated that in 2019, more than 1.2 million people died from drug-resistant infections. Of the 1.27 million deaths [uncertainty interval (UI), 95%; 0.911 million to 1.71 million) directly attributable to AMR, 929,000 (73%) were caused by six pathogens: E. coli, S. aureus, K. pneumoniae, S. pneumoniae, A. baumannii, and P. aeruginosa. These same pathogens were responsible for 3.57 million (72%) of the 4.95 million deaths (UI, 95%; 3.62 million to 6.57 million) that were associated with AMR [113]. The first comprehensive analysis of the global impact of AMR estimated that in 2019, resistance itself caused 1.27 million deaths—more deaths than HIV/AIDS or malaria—and that AMR infections were implicated in 4.95 million deaths. Overall, the investigators obtained 471 million individual records to estimate, through statistical modeling, the disease burden associated with and attributable to AMR for 12 major infectious syndromes for all regions of the world, including countries with no data [114].
Biofilm research is one of the most challenging areas of modern medicine, because the results of classical antibiotic susceptibility tests cannot be used to predict therapeutic success with biofilms.
The MICs of antibiotics are usually determined by using planktonic bacteria, and the values obtained do not correspond to the concentrations necessary to prevent, suppress, or destroy biofilms [115]. A major challenge in the treatment of biofilm-associated infections is to develop an appropriate standardized method to test the AMS of biofilms [116, 117]. Because the existing antibiotics have been developed against free-floating (planktonic) bacteria, the treatment of biofilm-related infections fails quite often.
Because AMS differs substantially between planktonic and biofilm bacteria, additional parameters have been introduced to evaluate antibiotic efficacy. These include (a) minimal biofilm inhibitory concentration (MBIC; [118]), (b) minimal biofilm eradication concentration (MBEC; [118]), (c) biofilm bactericidal concentration (BBC; [115]), and (d) biofilm-prevention concentration (BPC; [115]). Some researchers define the MBEC as the lowest antimicrobial concentration that kills 99.9% of the bacteria in biofilms, as compared with the control [119]. Other research groups consider the MBEC to be similar to the BBC and compare it to the minimum bactericidal concentration (MBC) for planktonic cultures [115, 120]. The inhibitory effect of antimicrobials on biofilm formation is usually evaluated by using the MBIC, i.e., the lowest concentration at which there is no time-dependent increase in the average number of viable cells within biofilms [115]. By contrast, the BPC is defined as the concentration at which the initial culture density decreases so much that biofilms do not form [115].
Many laboratory methods are used to evaluate the AMS of biofilm bacteria, such
as microtiter plate assays, the Calgary biofilm device, substrate suspension
reactors, and flow cell systems. These are commonly used to evaluate the AMS of
biofilms in vitro [115, 118, 120, 121]. The MBEC of biofilms is tested with
kits such as Assay
Sensor-based methods are being developed as well. For example, Hendolin et al. [123] described the real-time monitoring of the state of bacterial biofilms (including that after exposure of cells to antibiotics) on the surface of a piezoelectric resonator by measuring its resonance frequency. However, the weak point of this technique is that the detection system is influenced by temperature, whose fluctuations may also change the resonance frequency and, consequently, distort the analysis results.
A microfluidic redox-reactive nanoFET (field-effect-transistor) biosensor for extracellular bacterial metabolic analysis was described in [124]. A piezoelectric quartz tuning fork ring-down system was used to determine the ciprofloxacin activity against P. aeruginosa biofilm in situ [125].
Yeor-Davidi et al. [124] monitored the metabolic activity of biofilms by using a nanosensor and by analyzing glucose metabolites in solutions with high ionic strength, such as bacterial media, without sample pretreatment (Fig. 9, Ref. [124]). Biofilms were treated with antibiotics varying in their mechanism of action and were compared with untreated samples. Further study of biofilms during antibiotic treatment with silicon nanowire–FET devices clarified the process occurring in biofilms. In addition, the search for appropriate treatment to eliminate biofilms could be tested with the new nanosensor to monitor the formation of microbial communities after treatment with antibiotics differing in their mechanism of action. For such antibiotics (tetracycline and ampicillin), the biofilm response was different in each case. Treatment with ampicillin reduced glucose uptake early in treatment; a short treatment period did not harm biomass to the point at which glucose uptake ceased. Prolonged incubation with ampicillin led to partial lysis of biomass, and therefore only 60% of the glucose was consumed. In the case of tetracycline, short-term treatment started with a slow increase in glucose consumption until the metabolite was completely consumed; long-term incubation damaged the biofilm and caused a change in metabolic activity, but it did not eliminate the bacteria completely [124].
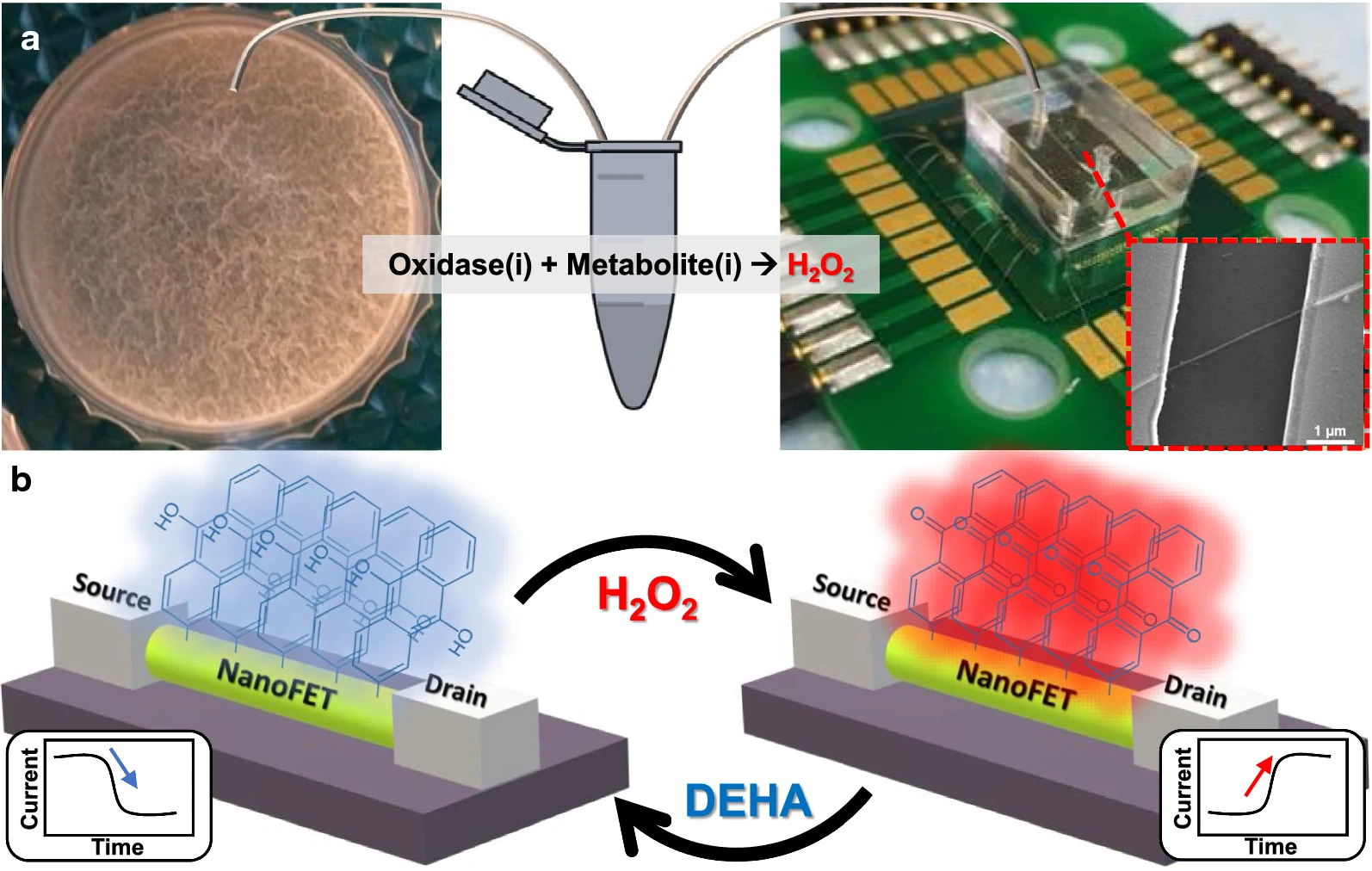 Fig. 9.
Fig. 9.Microfluidic redox-reactive nanoFET biosensor for extracellular bacterial metabolic analysis. (a) Silicon wafer chip has a 600-nm thermal oxide layer, which contains 200 potential redox-reactive silicon nanowire field-effect-transistor (FET) devices sharing a common gate. The nanoFETs are covered with a polydimethylsiloxane microfluidic channel connected via tubing to an Eppendorf tube with a small bacterial medium sample mixed with an oxidase. The forming Bacillus subtilis biofilms are shown in the left panel. Inset: scanning electron microscope image of a single redox-reactive nanoFET consisting of 20-nm p-type silicon nanowire connected to the source and drain electrodes. The nanoFETs chip is wire-bonded to the PCB holder, which is connected to the electrical recording system. (b) Operation mechanism of the redox-reactive nanoFET biosensor. The redox-reactive nanoFET biosensor is reversely reduced or oxidized in the presence of N,N-diethylhydroxylamine or water, respectively. When the redox reactive device is oxidized, the conductivity of the device increases, and 9,10-anthraquinone moieties are formed on the nanoFET surface (right panel). On the other hand, when the redox reactive device is reduced, the conductivity of the device decreases and 9,10-dihydroxyanthracene moieties are formed on the nanoFET surface (left panel) [124].
Blanco-Cabra et al. [126] presented a microfluidic platform with an integrated counter-pin probe (BiofilmChip) for the attachment of bacteria of clinical origin, even directly from clinical samples. The grown biofilms can be monitored by confocal microscopy or by electrical impedance spectroscopy. The device is suitable for studying polymicrobial communities and measuring the effect of antimicrobials on biofilms without disruption owing to manipulation, which better simulates actual clinical situations.
Despite the considerable variety of existing methods for evaluating the AMS of biofilm bacteria, further development in this direction is needed. Analysis of modern technologies shows that the main problems with these methods are the collection and preparation of samples, the duration of analysis, and the elimination of false positive results. Therefore, new methods to determine the AMS of bacteria are needed in microbiology and medicine.
Since 2010, through a trilateral alliance, the World Organization for Animal Health, WHO, and the Food and Agriculture Organization of the United Nations have been increasing awareness of AMR among countries. It is recognized that the underlying principle of addressing AMR lies within the One Health approach. The Trilateral Alliance is committed to taking steps to ensure that national action plans against AMR include the development and strengthening of effective surveillance, monitoring, and regulation of the storage, use, and sale of antimicrobials for humans and animals in accordance with the national context and the international commitments. This requires comprehensive multisectoral action linking human, animal, and environmental health. Increasing awareness and knowledge about AMR is necessary to seek cooperation from national representatives of the health, agriculture, and animal health sectors and to encourage behavioral change. Because the AMR driving forces are deeply rooted in human and animal health care and in the food and agriculture systems, reform is needed that requires sustainable funding for all activities outlined in the national action plans.
A Global Steering Group (GSG) on AMR was organized at the recommendation of the Inter-Agency Coordination Group on AMR to activate global political forces and strengthen leadership in the fight against AMR. The GSG includes representatives of member states, civil society and the private sector.
The mission of the GSG is to work globally with governments, institutions, civil society, and the private sector in a One Health approach to provide guidance and advocate for priority policy actions to prevent drug-resistant infections by ensuring responsible and sustainable access to and use of antimicrobials.
The goal of the agreed global action plan against AMR is to “to ensure, for as long as possible, continuity of successful treatment and prevention of infectious diseases with effective and safe medicines that are quality-assured.”
Its five strategic objectives are:
Objective 1: Improve awareness and understanding of AMR through effective communication, education and training.
Objective 2: Strengthen the knowledge and evidence base through surveillance and research.
Objective 3: Reduce the incidence of infection through effective sanitation, hygiene and infection prevention measures.
Objective 4: Optimize the use of antimicrobial medicines in human and animal health.
Objective 5: Develop the economic case for sustainable investment that takes account of the needs of all countries, and increase investment in new medicines, diagnostic tools, vaccines and other interventions [10]. In addition, a special place in the fight against AMR is occupied by the development of methods for the rapid analysis of the AMR of both planktonic and biofilm bacteria. Ideally, this problem can be solved by using comprehensive systems that allow the monitoring of both planktonic and biofilm bacteria within a short period. Such systems will avoid false assay results.
Antibiotics are used widely to prevent and treat bacterial infections. In veterinary medicine, they are also commonly used to promote animal growth. Nontransformed antibiotic residues are excreted by humans and animals into the environment, which leads to adverse environmental effects [127, 128].
Contamination of terrestrial and aquatic ecosystems by pharmaceutical waste is ubiquitous, with antibiotic concentrations in the environment sometimes exceeding their therapeutic levels [129, 130, 131].
Rivers are important links between urban and rural ecosystems [55]. Antimicrobials enter aquatic environments through the direct discharge of wastewater treatment plants into surface or ground water; through leachate from landfills and leakages from sewer pipes, manure storage tanks, or lagoons; and through stormwater from manure-fertilized farmland [128]. The presence in the environment of medical antibiotics is mainly due to the discharge of treated wastewater from wastewater treatment plants, because conventional wastewater treatment is insufficiently effective at removing these compounds [132]. The entry of antibiotics into aquatic environments is also a result of their use in aquaculture [133].
High resistance to antibiotics used to treat common bacterial infections such as urinary tract infections, sepsis, sexually transmitted diseases, and some forms of diarrhea has been observed worldwide, indicating that the current arsenal of effective antibiotics is depleted. For example, in countries reporting data to the WHO Global Antimicrobial Resistance and Use Surveillance System (GLASS), the incidence of resistance to ciprofloxacin, an antibiotic commonly used to treat urinary tract infections, is between 8.4% and 92.9% for E. coli and between 4.1% and 79.4% for K. pneumoniae. Three major circumstances currently threaten the treatment of bacterial infections:
Among the various global challenges facing humanity, the greatest concern is the growing bacterial AMR and the need to design accelerated methods for its evaluation. Traditional test systems for analyzing the AMS of bacteria are considered the “gold standard”, because they are fairly proven and reliable and because they have been used for decades. But these methods have important drawbacks: they are based on recording changes in bacterial metabolism and require pure clinical isolates. Because of these drawbacks, they are time-consuming. Since a large number of antibiotics are currently in use, it is not rational to evaluate the AMS of isolates to all antimicrobials used in practice. The choice of the antibiotic being analyzed depends on the isolated microorganism, the focus and type of infection, the presence of concomitant diseases, and so on. Interpretation of the test results is based on the most commonly used CLSI and EUCAST standards [134]. A decisive point in the choice of antibiotics is the accurate identification of the bacteria. Although many brilliant reviews have been published on the evaluation of antibacterial sensitivity [134, 135, 136, 137, 138, 139], research continues to develop and validate new methods, including screening methods, for the analysis of AMS. In recent decades, several innovative approaches to the determination of AMS have been developed, including MALDI-TOF, flow cytometry, and isothermal microcalorimetry [134, 140]. Separate mention should be made of molecular genetic and “omics” methods of AMS analysis [141, 142, 143, 144, 145, 146, 147, 148]. The main focus of future research in this area will be on the standardization of these methods and the simultaneous testing of the sensitivity of microbes to various classes of antimicrobials.
In summary, most studies are shifting toward the design of rapid AMS evaluation systems, which do not require pure clinical isolates. These new systems can be used either directly or through simple sample pretreatment for the direct determination of AMS [72]. One promising area in the development of rapid AMS evaluation methods is biosensor technologies [81, 149, 150]. Unfortunately, there is still no single standard for the use of sensor technologies in AMS analysis. Nevertheless, through the efforts of many research groups, systems have already been developed that potentially can compete with traditional methods. But in terms of application of sensors in AMS analysis, a limiting point is the validation of these systems and repeated testing with clinical isolates.
The development of methods for accelerated AMS testing in bacteria, especially in their biofilms, is an urgent area for further research. Particular attention should be paid to sensor-based analysis systems that allow research to be conducted without sample pretreatment and results to be obtained within a short time.
OIG – Conceptualization and design, Project administration, Writing – original draft, Writing – review & editing. SSE – Formal Analysis, Software, Writing – original draft. OAK – Resources, Visualization, Writing – review & editing. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
Not applicable.
This work was funded by the Ministry of Science and Higher Education of the Russian Federation as a state assignment for the Saratov Scientific Centre of the Russian Academy of Sciences (research topic No. 121032300311-5).
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.


