- Academic Editor
-
-
-
†These authors contributed equally.
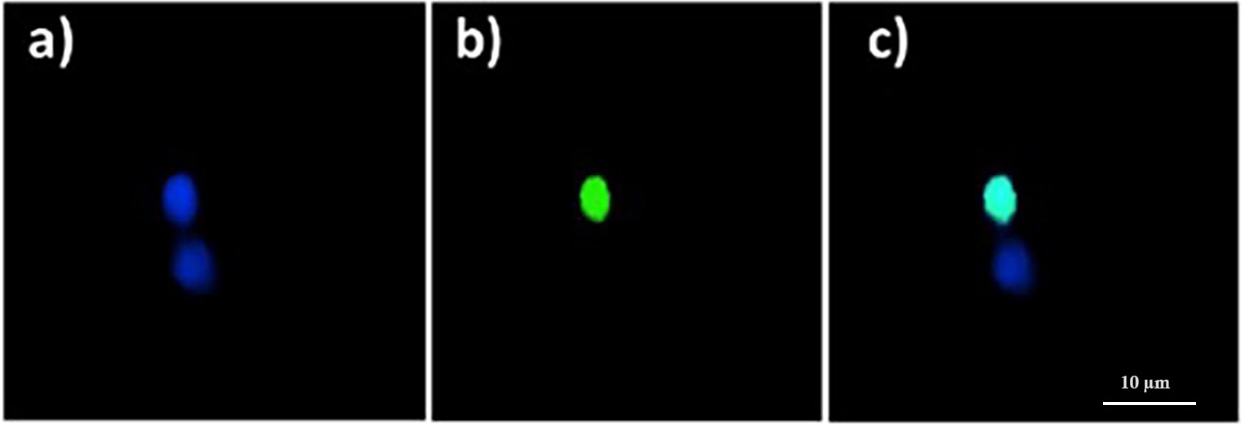
Background: Ribosome inactivating proteins (RIPs) are N-glycosylases found in various plants that are able to specifically and irreversibly inhibit protein translation, thereby leading to cell death. Their cytotoxic properties have attracted attention in the medical field in the context of developing new anticancer therapies. Quinoin is a novel toxic enzyme obtained from quinoa seeds and classified as a type 1 RIP (Chenopodium quinoa Willd.). Recently, quinoin was found to be cytotoxic to normal fibroblasts and keratinocytes in vitro, as well as to several tumor cell lines. Methods: The aim of this study was to evaluate the in vitro and in vivo genotoxicity of quinoin in a zebrafish model. We evaluated its ability to induce DNA fragmentation, genomic instability, and reactive oxygen species (ROS) generation by means of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) reaction, randomly amplified polymorphic DNA (RAPD) Polymerase Chain Reaction (PCR) technique, and dichlorofluorescine (DCF) assay, respectively. Results: Quinoin was found to cause genomic damage in zebrafish, as shown by DNA fragmentation, polymorphic variations leading to genomic instability, and oxidative stress. Interestingly, longer quinoin treatment caused less damage than shorter treatments. Conclusions: This study demonstrated ROS-mediated genotoxicity of quinoin toward the zebrafish genome. The reduced damage observed after longer quinoin treatment could indicate the activation of detoxification mechanisms, activation of repair mechanisms, or the loss of protein activity due to enzymatic digestion. In order to clarify the genotoxic actions of quinoin, further investigations of the response pathways to DNA damage are needed. Overall, the ability of quinoin to cause breaks and instability in DNA, together with its clear cytotoxicity, make it an interesting candidate for the development of new drugs for cancer treatment.