- Academic Editor
†These authors contributed equally.
Background: Ribosome inactivating proteins (RIPs) are N-glycosylases found in various plants that are able to specifically and irreversibly inhibit protein translation, thereby leading to cell death. Their cytotoxic properties have attracted attention in the medical field in the context of developing new anticancer therapies. Quinoin is a novel toxic enzyme obtained from quinoa seeds and classified as a type 1 RIP (Chenopodium quinoa Willd.). Recently, quinoin was found to be cytotoxic to normal fibroblasts and keratinocytes in vitro, as well as to several tumor cell lines. Methods: The aim of this study was to evaluate the in vitro and in vivo genotoxicity of quinoin in a zebrafish model. We evaluated its ability to induce DNA fragmentation, genomic instability, and reactive oxygen species (ROS) generation by means of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) reaction, randomly amplified polymorphic DNA (RAPD) Polymerase Chain Reaction (PCR) technique, and dichlorofluorescine (DCF) assay, respectively. Results: Quinoin was found to cause genomic damage in zebrafish, as shown by DNA fragmentation, polymorphic variations leading to genomic instability, and oxidative stress. Interestingly, longer quinoin treatment caused less damage than shorter treatments. Conclusions: This study demonstrated ROS-mediated genotoxicity of quinoin toward the zebrafish genome. The reduced damage observed after longer quinoin treatment could indicate the activation of detoxification mechanisms, activation of repair mechanisms, or the loss of protein activity due to enzymatic digestion. In order to clarify the genotoxic actions of quinoin, further investigations of the response pathways to DNA damage are needed. Overall, the ability of quinoin to cause breaks and instability in DNA, together with its clear cytotoxicity, make it an interesting candidate for the development of new drugs for cancer treatment.
Edible parts of plants, such as seeds, fruits, roots and leaves, are important
for human nutrition because they contain metabolites and macromolecules that are
essential for growth and health, including carbohydrates, sugars, vitamins and
proteins [1]. However, different plant tissues also contain harmful or toxic
substances that are synthesized as defense mechanisms against biotic attack by
insects, fungi and viruses [2], and against abiotic environmental stresses such
as radiation and heat stress [3]. Most of these toxic substances belong to
several classes of chemical compounds, including phytates, tannins, alkaloids,
oxalates, saponins, cyanides and raffinose oligosaccharides, or to protein
families such as protease inhibitors, lectins,
Other known plant toxic proteins are ribosome inactivating proteins (RIPs) found in angiosperms. RIPs are enzymes (EC: 3.2.2.22) endowed with rRNA N-glycosylases activity. They are necessary for the removal of a specific adenine (A4324 in rats) in the Sarcin Ricin Loop (SRL) of 28S rRNA, which is involved in the interaction between ribosomes and elongation factors. This irreversible rRNA damage leads to the inhibition of translation and cell death by the apoptotic pathway [6, 7].
Interest in RIPs has increased in recent years due to their use in experimental studies as the toxic moiety of immunoconjugates that target various human pathologies, including cancer [8]. The anti-tumor potential of RIPs has been demonstrated in clinical studies with immunotoxins [9]. For example, saporin-S6 type 1 RIP, isolated from the seeds of Saponaria officinalis L., has been used to construct conjugates against numerous targets and shows good efficacy in hematological cancer models [10]. Stenodactylin, purified from the caudex of Adenia stenodactyla Harms, is a potent type 2 RIP that induces multiple cell death pathways, mainly involving apoptosis, but also necroptosis and free radical production in a neuroblastoma cell line [11]. Kirkiin, purified from the caudex of Adenia kirkii (Mast.) Engl., is a type 2 RIP that can completely inhibit protein synthesis and induce cell death by apoptosis at very low doses in a human neuroblastoma cell line [12].
Recently, a novel type 1 RIP called quinoin was purified from quinoa seeds and extensively characterized [13]. The quinoa plant (Chenopodium quinoa Willd.) is native to the Andes of South America [14]. Quinoa seeds, known as the ‘Golden Grain’, are rich in raw proteins and contain all of the essential amino acids, dietary fibers, B vitamins, and dietary minerals. The consumption of quinoa seeds has therefore continued to increase, together with the production of its derivatives such as bread, pasta, biscuits and cakes [15].
Nevertheless, quinoa seeds contain ANFs including saponins, phytic acid, oxalates, tannins, and trypsin inhibitors [16, 17]. In addition to cooking, quinoa seeds are subjected to other food processing methods such as extrusion, roasting, or mechanical abrasion, all of which remove ANFs [17, 18]. Aside from its health benefits, this pseudo-cereal could also be used in the medical field for cancer treatment, given the presence of quinoin.
The type 1 RIP quinoin (~29-kDa) can inhibit protein synthesis in vitro, shows remarkable thermostability (Tm ~68.2 °C), and is partially resistant to in vitro digestion systems [13]. Furthermore, quinoin is cytotoxic towards BJ-5ta (human fibroblasts), HaCaT (human keratinocytes), and several tumor cell lines in a dose- and time-dependent manner [13, 19]. As with most RIPs, quinoin can also depurinate DNA to cause genotoxic damage and subsequent apoptosis [20]. Quinoin is thus a novel toxin enzyme found in quinoa seeds (~3.0 mg/100 g of seeds) and also in smaller amounts in the sprouts [13].
The aim of the present study was to evaluate the in vitro and in vivo acute genotoxicity of quinoin, thereby improving our understanding of the mechanism of action of RIPs. For this purpose, we used the zebrafish model. This represents a valid genotoxicity model to replace mammalian models, since it shares approximately 75% of its genome with humans. Moreover, zebrafish show rapid development, allowing the toxic effects of a given substance to be monitored over a short exposure time [21, 22].
DNA fragmentation and Genomic Template Stability (GTS) were assessed in zebrafish blood cells exposed in vitro to different amounts of quinoin for different times, as well as in blood cells taken from zebrafish after intraperitoneal administration of different amounts of quinoin for several days. DNA damage is reported to be secondary to oxidative stress, and reactive oxygen species (ROS) have been identified as the main mediators of RIP-induced cell death [23]. We therefore estimated the level of ROS in exposed blood cells in order to correlate oxidative stress with DNA damage caused by quinoin.
This research provides new evidence for the harmful actions of quinoin in causing genomic and subsequent cellular damage, thus highlighting its potential incorporation into immunotoxins for the therapeutic targeting of cancer.
Chemicals for chromatography were obtained as previously reported [13], while standard reagents for protein purification and analysis were purchased from Sigma-Aldrich Solutions (Merck Life Science, Milan, Italy).
The purification of quinoin for in vitro and in vivo biological assays was performed as previously reported [13]. Briefly, a crude extract of quinoa seeds was acidified with glacial acetic acid and the soluble protein was fractionated according to differences in protein size (gel filtration chromatography) and charge characteristics (ion exchange chromatography). The protein obtained was first dialyzed against deionized water, then freeze-dried and stored at –20 °C. All general methods used for the analytical characterization of quinoin, such as Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE), Reverse Phase High Performance Liquid Chromatography (RP-HPLC), and measurement of the protein concentration by colorimetric assay (BCA assay), were carried out as previously described [24, 25].
Adult zebrafish (Danio rerio) were maintained at the Department of Biology, University of Naples Federico II. The handling of animals and the experimental procedures were in accordance with the local regulation and international guidelines for the use and care of laboratory animals [26] and approved by the Italian Ministry of Health (Permit Number: 529/2019-PR). Efforts were made to minimize animal suffering and the number of specimens used. The zebrafish were reared in an aquarium tank at a temperature of between 24 and 29 °C, with a photoperiod of 10 hours dark and 14 hours light, and a pH of ~7.2 to mimic the conditions of their natural habitat [27].
Each organism was anesthetized with ethyl 3-aminobenzoate methane sulfonate (MS-222, Sigma Aldrich, Darmstadt, Germany) to minimize suffering. Blood was collected from the gills of 10 zebrafish using a heparinized syringe. In order to obtain a sufficient number of cells for all tests, a pool of blood cells from different specimens was subdivided into aliquots, with each containing different final concentrations of quinoin (1, 5 and 10 µg/µL). Cells were exposed to quinoin for 30, 60 and 90 minutes, after which the genotoxicity tests were performed.
After 2 weeks of acclimation, 45 zebrafish were sorted into 5 L tanks as follows: 12 specimens for each group were divided into three tanks. The remaining specimens were sorted into one other tank and injected with 1 µL of sterile water to serve as a negative control. Following zebrafish anesthesia as reported above, inoculation with 1, 5 and 10 µg quinoin was performed by intraperitoneal injection into the abdominal cavity posterior to the pelvic girdle using a 10 µL 35G blunt steel Hamilton-type micro needle (SYR 10 µL – 1701 RN, WO: 1900029358). After administration of the quinoin, zebrafish specimens were transferred to suitably prepared tanks and their swimming movements were monitored to ensure they were not stressed. At 15 and 30 days after injection, approximately 30 µL of blood was collected from the gills of each fish with a heparinized syringe and the tests were performed.
DNA fragmentation was determined using an in situ cell death detection
kit (Roche Diagnostics, Basel, Switzerland) as previously reported [28]. Briefly,
blood cells were mixed with phosphate-buffered saline (1
DNA was isolated from zebrafish blood cells using the High Pure PCR Template
Preparation Kit (ROCHE Diagnostics). Genomic DNA amplification was performed
using primer 6 (5
In order to calculate the intracellular percentage of ROS, 13 µM of
2
The percentages of DNA fragmentation, genome stability and intracellular ROS
were expressed as the mean
All zebrafish specimens survived after quinoin injection. No signs of suffering or changes in physical condition or movement were observed during the 30 days following injection of the three quinoin doses.
Cellular DNA fragmentation as observed with the TUNEL technique is shown in Fig. 1. DNA fragmentation in zebrafish blood cells increased significantly following
in vitro exposure to quinoin (Fig. 2). This was observed for all
treatment times, regardless of the quinoin concentration used. However, DNA
fragmentation decreased with increasing exposure time, with values ranging from
19.25
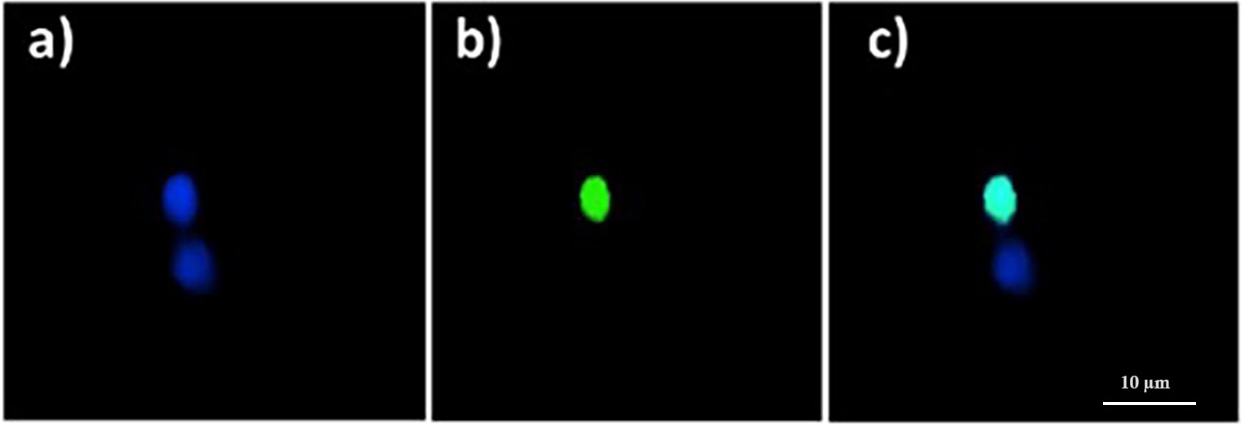 Fig. 1.
Fig. 1.Quinoin-treated zebrafish erythrocytes showing fragmented DNA
(green nuclei), and no DNA fragmentation (blue nuclei), as observed with the
Terminal deoxynucleotidyl transferase (TdT) 2
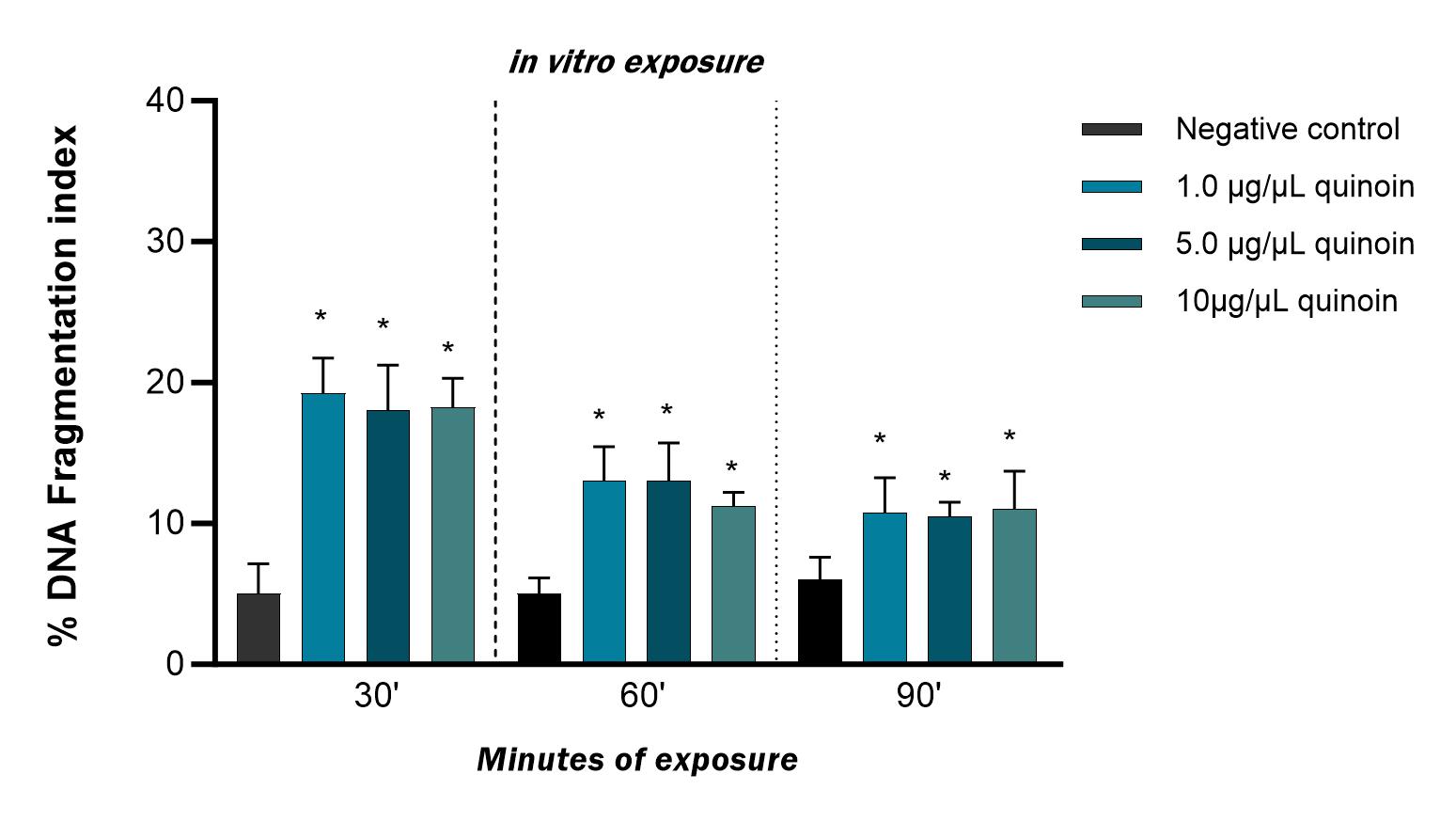 Fig. 2.
Fig. 2.Percentage of DNA fragmentation observed in zebrafish blood
cells after 30, 60 and 90 minutes of in vitro exposure to different
quinoin concentrations. * p
A time-dependent decrease in the percent DFI was also observed after in
vivo treatment with quinoin (Fig. 3). Administration of 1, 5 and 10
µg quinoin resulted in DFI percentages of 39.75
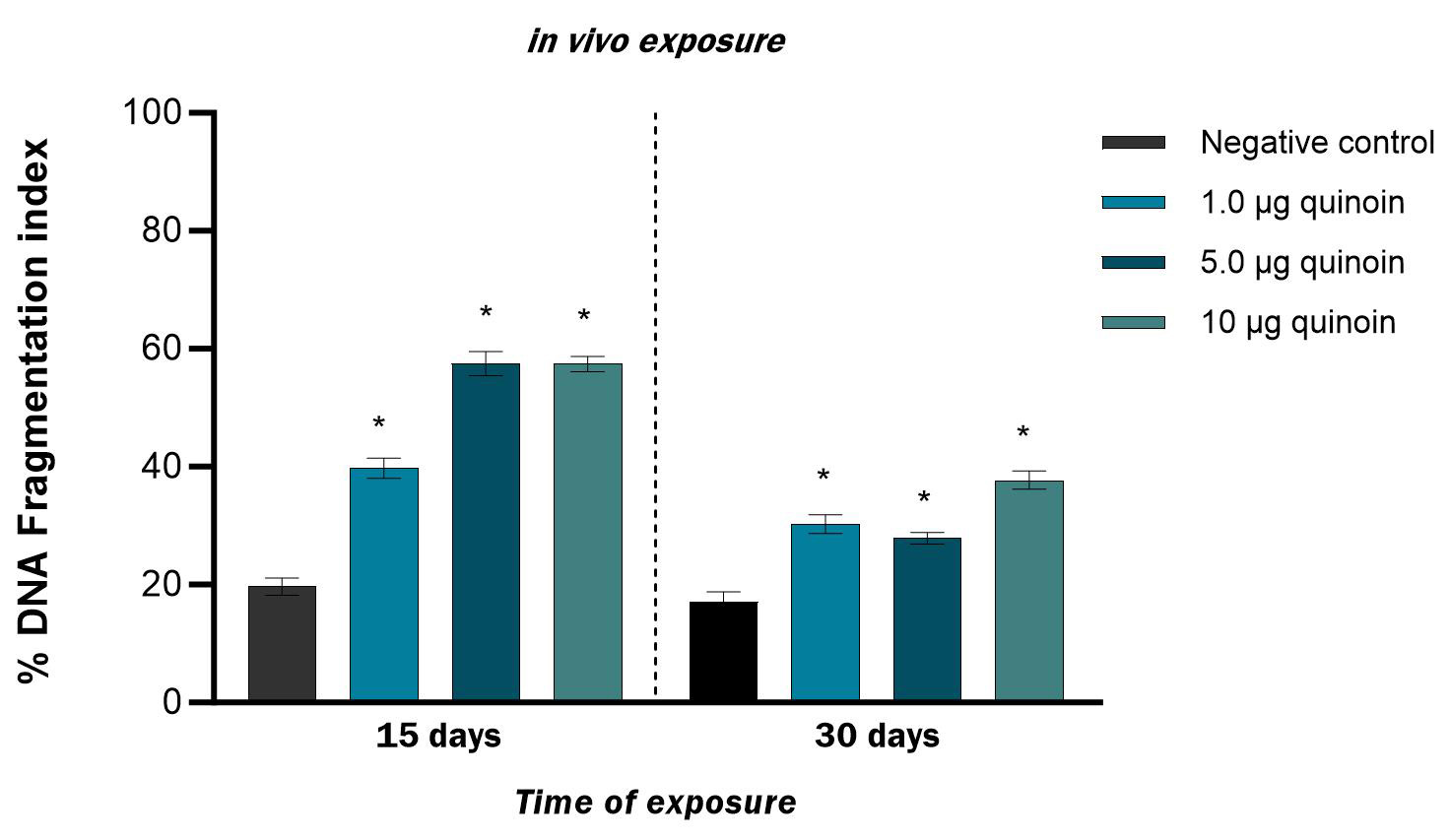 Fig. 3.
Fig. 3.Percentage of DNA fragmentation observed in zebrafish blood
cells after 15 and 30 days of in vivo injection of different amounts of
quinoin. * p
A quinoin concentration of 1 µg/µL induced a single polymorphic variation (lost band) after 30, 60 and 90 minutes of in vitro treatment (Table 1, Fig. 4). At 5 µg/µL quinoin, three variations were observed after 30 minutes of treatment, and one variation at each of 60 and 90 minutes exposure. At the highest concentration of quinoin (10 µg/µL), three variations were observed after 30 minutes of treatment, and one variant band at each of 60 and 90 minutes.
| Quinoin concentration | Minutes of exposure | Lost Bands | Gained Bands |
| 1 µg/µL | 30 | 510 bp | - |
| 60 | 510 bp | - | |
| 90 | - | 350 bp | |
| 5 µg/µL | 30 | 510 bp | 350 bp |
| 780 bp | |||
| 60 | - | 350 bp | |
| 90 | - | 780 bp | |
| 10 µg/µL | 30 | 510 bp | 350 bp |
| 820 bp | |||
| 60 | - | 420 bp | |
| 90 | - | 420 bp |
Control bands: 200 bp, 210 bp, 300 bp, 400 bp, 500 bp, 510 bp, 600 bp, 800 bp, 1000 bp.
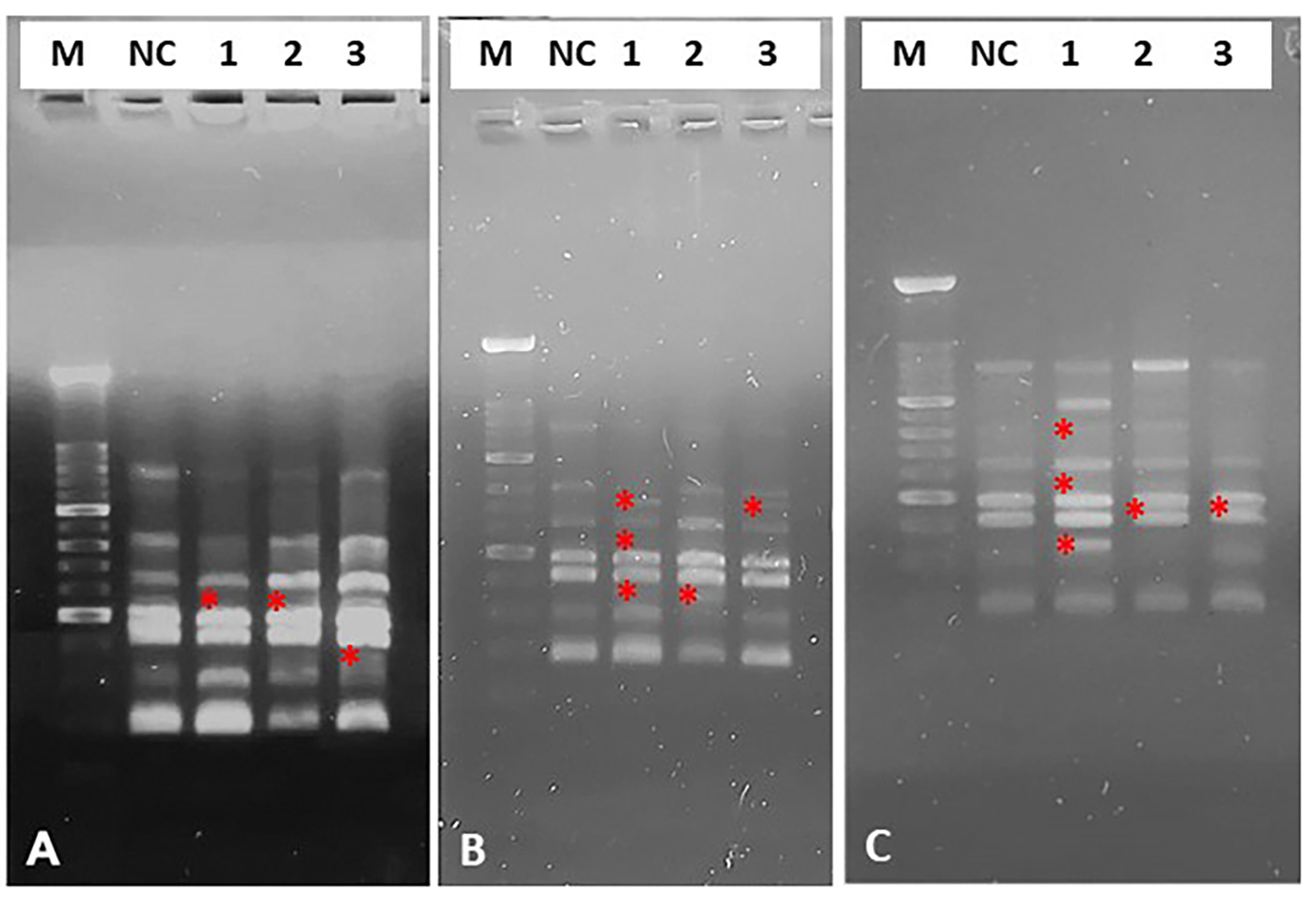 Fig. 4.
Fig. 4.Representative image of electrophoretic profiles of zebrafish DNA following 30 (A), 60 (B) and 90 (C) minutes of in vitro treatment with different quinoin. M, 100 bp DNA Marker; NC, negative control; 1, 1 µg/µL quinoin; 2, 5 µg/µL quinoin; 3, 10 µg/µL quinoin. * polymorphic bands.
The electrophoretic profile of zebrafish DNA 15 days after 1 µg quinoin injection revealed three variations, whereas 30 days after administration of 5 and 10 µg quinoin only one variation was found, regardless of the amount of quinoin administered (Table 2; Fig. 5).
| Quinoin | Days of exposure | Lost Bands | Gained Bands |
| 1 µg | 15 | - | 280, 320, 550 bp |
| 30 | 520 bp | - | |
| 5 µg | 15 | - | 280 bp |
| 30 | 520 bp | - | |
| 10 µg | 15 | - | 280bp |
| 30 | 520bp | - |
Control bands: 200 bp, 220 bp, 300 bp, 400 bp, 520 bp.
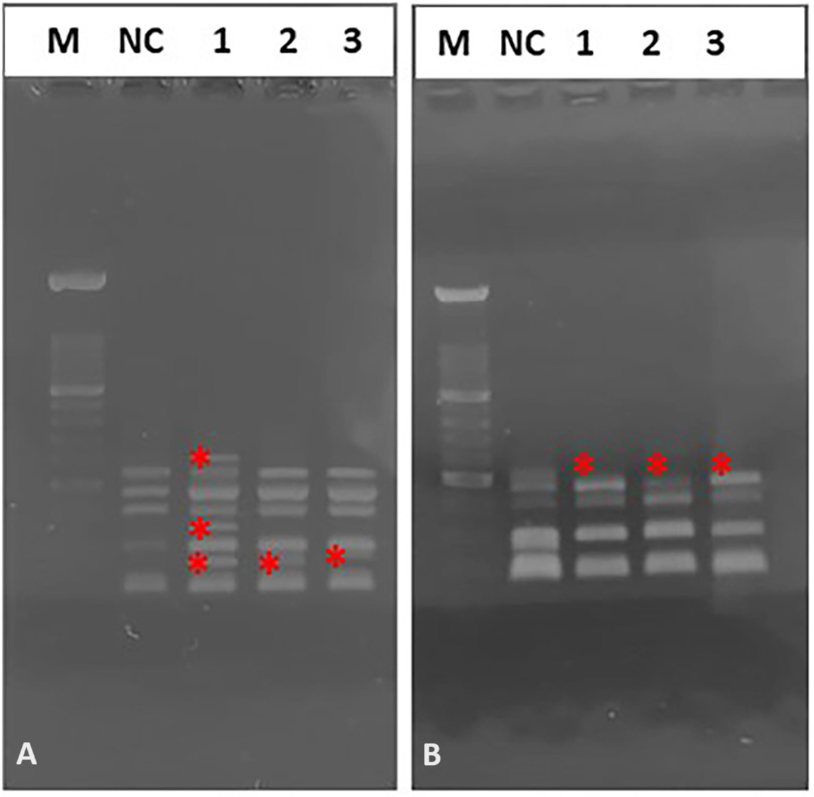 Fig. 5.
Fig. 5.Representative image of electrophoretic profiles of zebrafish DNA following 15 (A), and 30 (B) days after administration of different quinoin amount. M, 100 bp DNA Marker; NC, negative control; 1, 1 µg/µL quinoin; 2, 5 µg/µL quinoin; 3, 10 µg/µL quinoin. * polymorphic bands.
The polymorphic profiles obtained with the RAPD-PCR technique were used to calculate the genomic stability of the template. Genome stability was reduced after in vitro treatment with 1, 5 and 10 µg/µL quinoin for 30, 60 and 90 minutes, with a maximum reduction of 23% observed after 30 minutes of 10 µg/µL quinoin (Fig. 6).
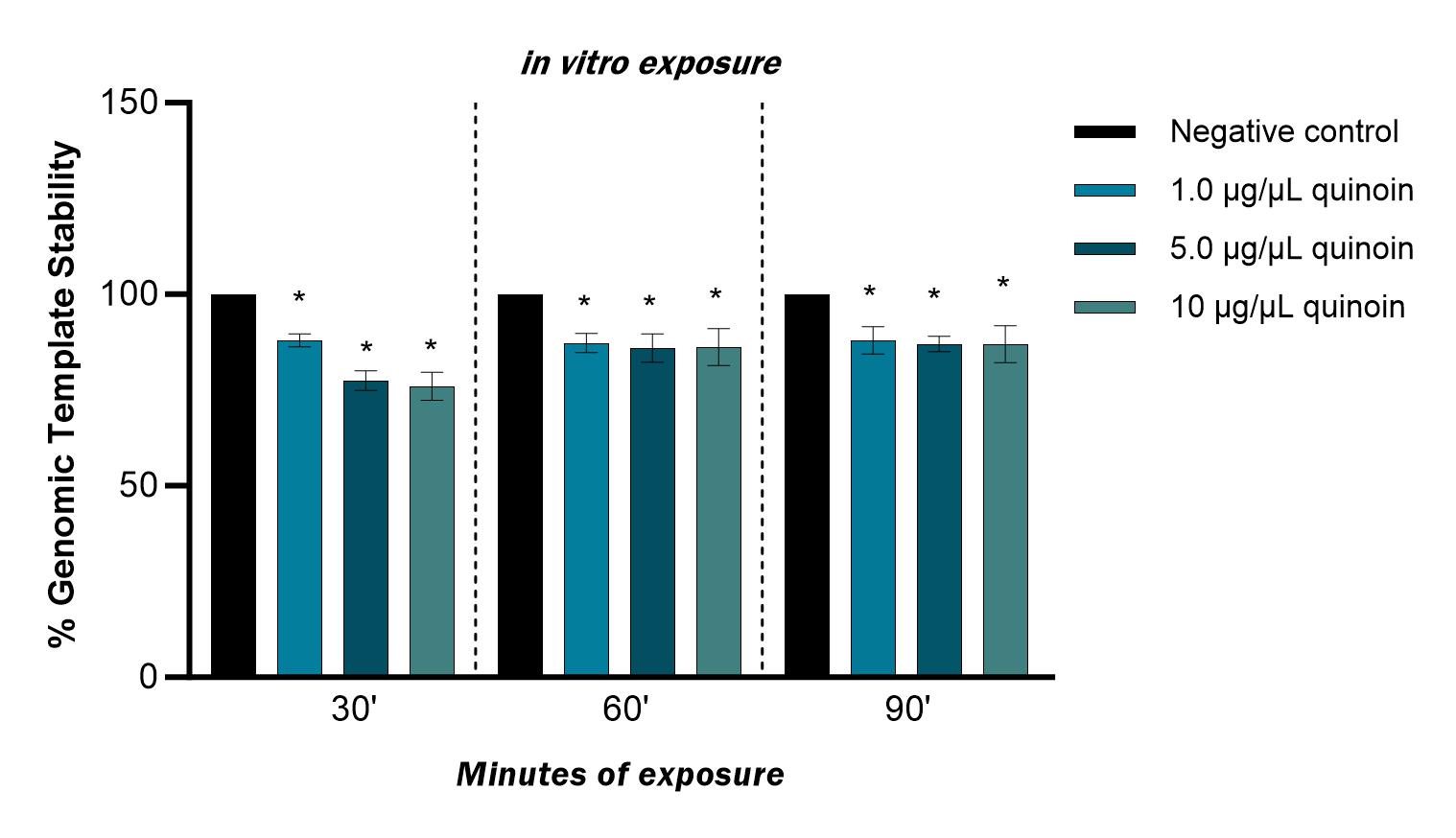 Fig. 6.
Fig. 6.Percentage of genomic template stability observed in zebrafish
blood cells after 30, 60 and 90 minutes of in vitro exposure to
different quinoin concentrations. * p
15 days after in vivo administration of 1 µg quinoin reduced genome stability by 60%. Administration of 1, 5 and 10 µg quinoin reduced genome stability 30 days after injection by an average of approximately 20% (Fig. 7).
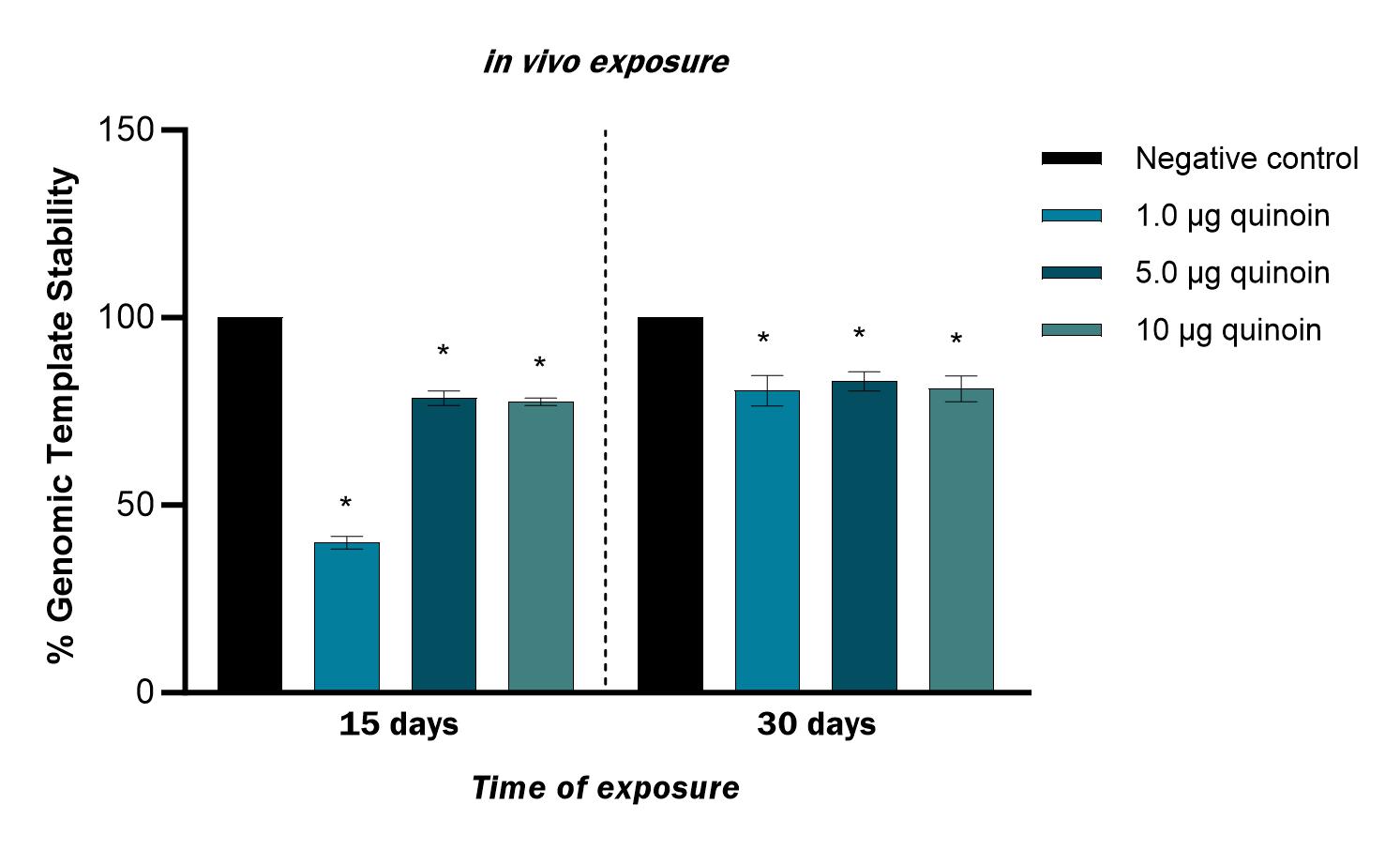 Fig. 7.
Fig. 7.Percentage of genomic template stability observed in zebrafish
blood cells 15 and 30 days after in vivo injection of different amounts
of quinoin. * p
Fig. 8 shows intracellular ROS, as seen with the 2
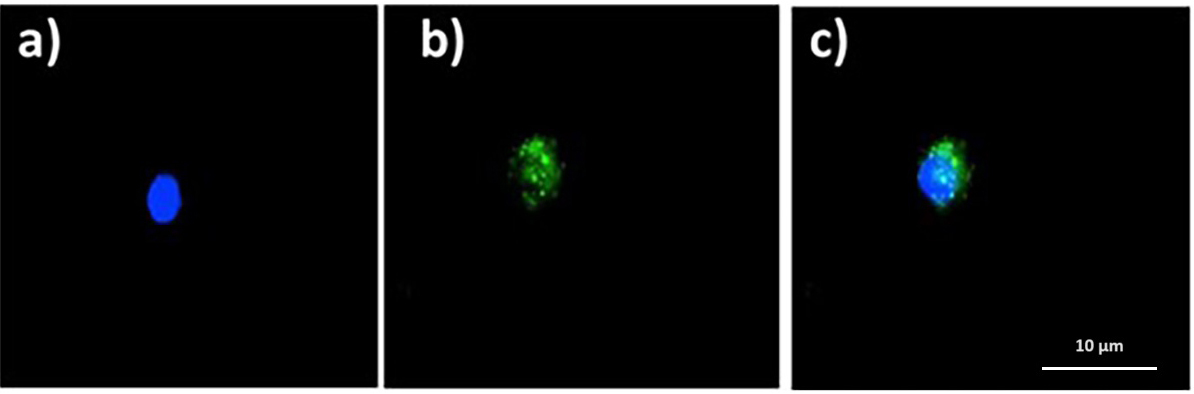 Fig. 8.
Fig. 8.Quinoin-treated zebrafish erythrocytes with intracellular ROS
(green fluorescence) after DCF assay. (a) DAPI. (b) FITC. (c) Merge. ROS,
reactive oxygen species; DCF, 2
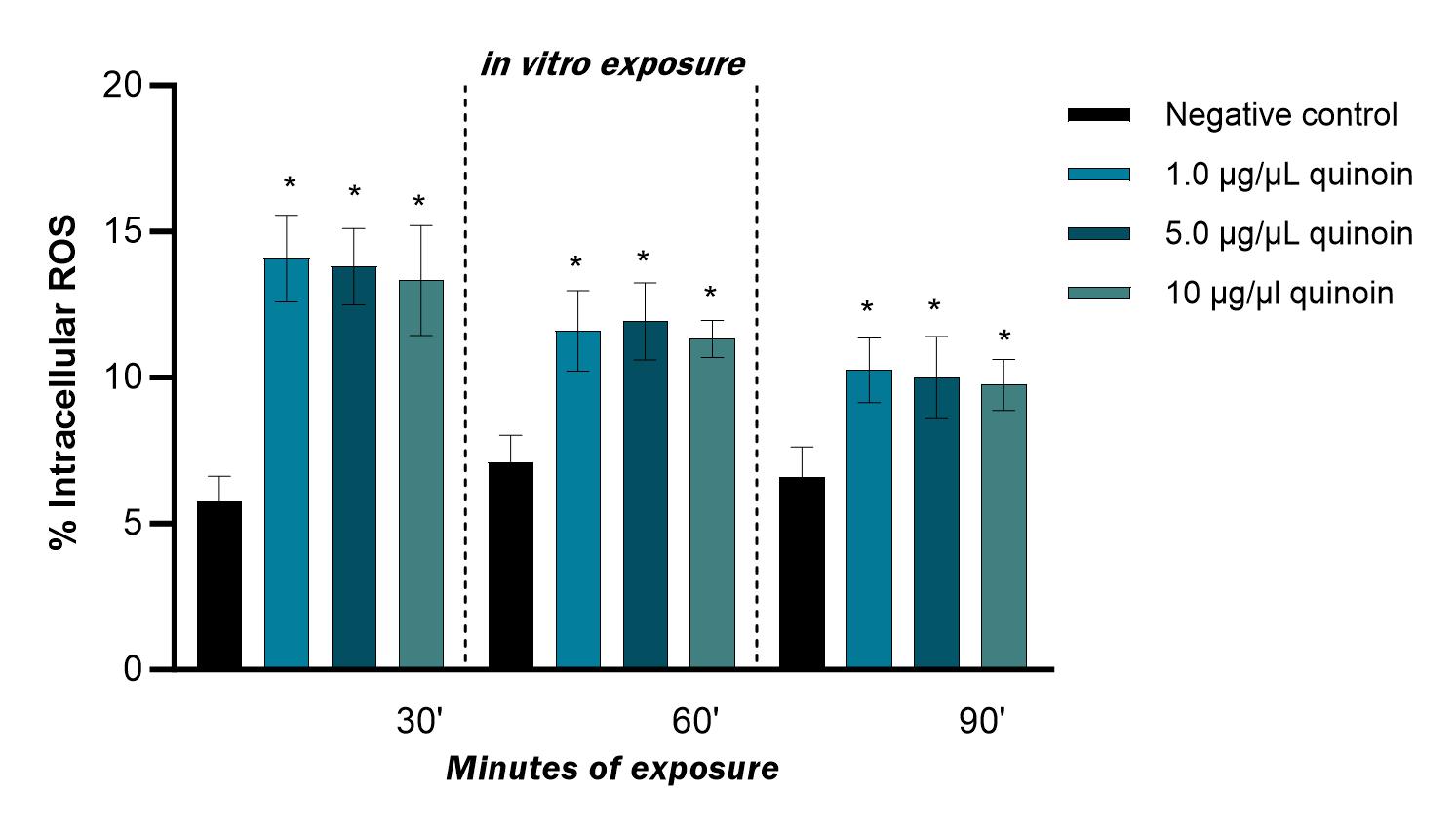 Fig. 9.
Fig. 9.Percentage of intracellular ROS observed in zebrafish blood
cells after 30, 60 and 90 minutes of in vitro exposure to different
quinoin concentrations. * p
A significantly higher percentage of cells with intracellular ROS was found 15
days after administration of 1, 5 and 10 µg of quinoin, with a
maximum of 26.2
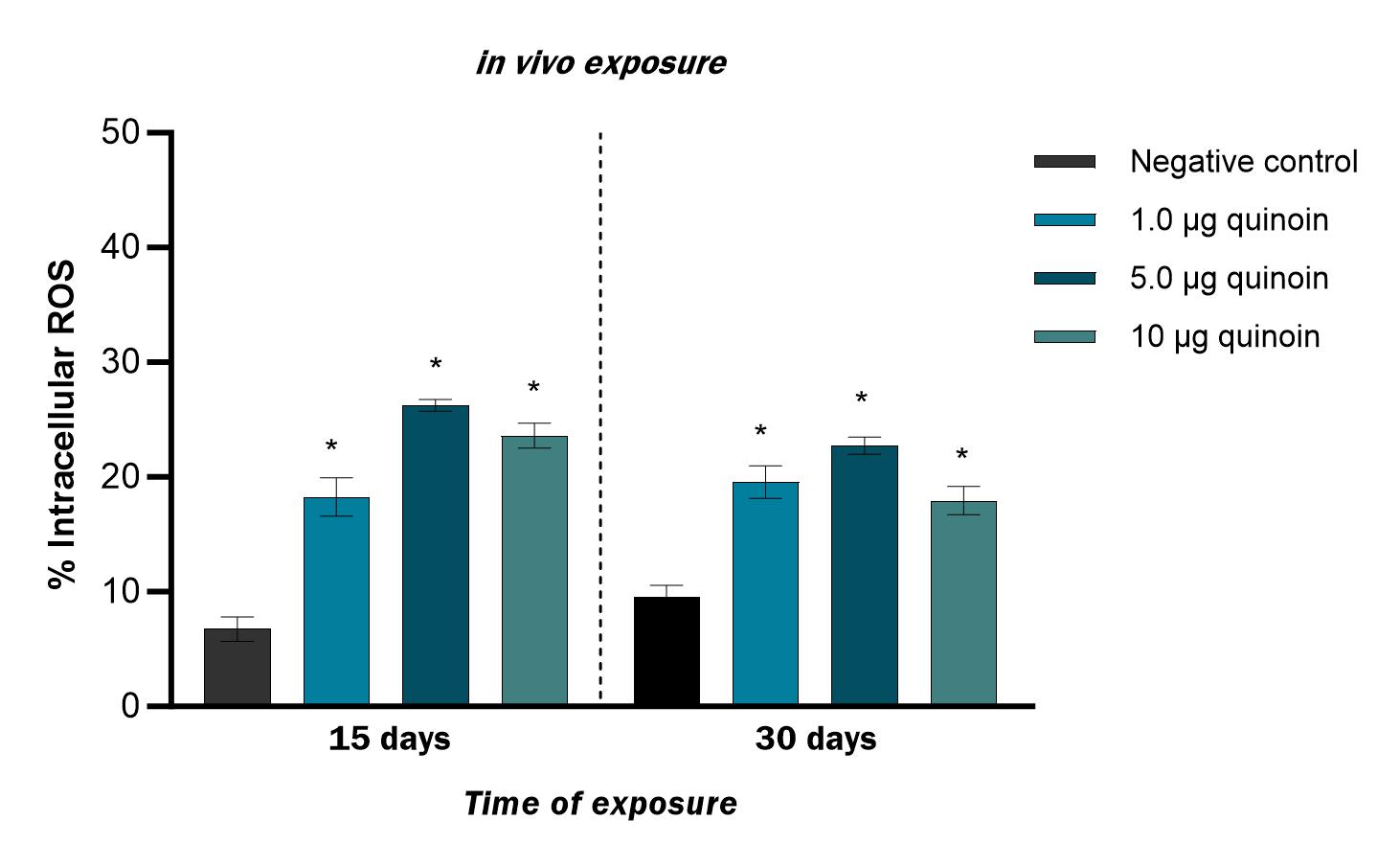 Fig. 10.
Fig. 10.Percentage of intracellular ROS observed in zebrafish blood
cells 15 and 30 days after in vivo administration of different amounts
of quinoin. * p
RIPs are toxic enzymes that can damage ribosomes and inhibit protein synthesis. They have attracted much attention in the medical and agriculture fields due to their antitumor, antifungal, antibacterial, antiviral and insecticidal activities [30]. RIPs are mainly found in the edible part of plants, such as seeds, leaves and roots. However, some RIPs are not harmful if the plant source is consumed after appropriate cooking. This is because they are denatured by heating, and hence their toxicity is reduced and/or eliminated [31]. Nevertheless, RIP activity has been detected in some plant tissues that are eaten raw, such as Allium cepa L., Spinacia oleracea L., Apium graveolens L., and Daucus carota L., or the seeds of Lycopersicon esculentum L. [32]. The low concentration of RIPs in these products is likely to make them safe for consumption.
Quinoa is one of the plant products that is eaten raw or cooked, and was recently included in the Mediterranean diet for its antioxidant and anti-inflammatory properties [33].
Quinoa is a pseudo-cereal belonging to the Chenopodiaceae family. It is an excellent source of nutrients and a valid alternative to commonly used cereals and carbohydrates, as it is rich in fiber, minerals, high quality proteins, vitamins (B, C and E group) and essential fatty acids. Quinoa is also an ideal food for coeliacs because it is gluten-free [17]. The consumption of quinoa seeds has been shown to increase intestinal permeability, improve the absorption of certain proteins, and produce anti-inflammatory effects [34]. However, quinoa also contains ANFs that can react with nutrients and interfere with their absorption [35]. It is particularly rich in saponins, which may reduce its potential benefits.
Quinoin is a novel type 1 RIP that was recently extracted from quinoa seeds. As with other RIPs, this enzyme acts by removing a specific adenine in 28S ribosomal RNA (rRNA), thereby blocking translocation and inducing cell death [13]. Its cytotoxicity has been demonstrated in both fibroblasts and glioblastoma tumor cell lines, which show clear changes in cell morphology and reduced cell growth following exposure to quinoin [19].
Although RIPs were previously thought to act only on rRNA, it is now known that many can also remove adenine bases from naked RNAs and from DNA. These genotoxins are capable of causing damage to nuclear DNA, leading to apoptosis [36].
In light of previous results showing a cytotoxic effect of quinoin in various experimental models, the aim of the present work was to evaluate the potential acute genotoxic effects of this enzyme in vitro and in vivo using the well-known zebrafish model system [21].
To this end, DNA fragmentation, genome stability interference and oxidative stress were assessed in zebrafish blood cells after in vitro exposure to different quinoin concentrations for different times and following different days after in vivo administration of several quinoin amounts.
The results obtained using the TUNEL reaction were consistent with a previous report in which quinoin was described as able to induce programmed cell death [19]. Indeed, DNA fragmentation is the main feature of apoptosis and is used as a specific marker of this process [37]. Compared to the negative control, the percent of DNA fragmentation was significantly higher in all samples treated with quinoin. Similarly, genomic template stability was reduced after quinoin treatment, as observed by the appearance of polymorphic bands. These results indicate that quinoin can induce damage at the genomic DNA level, either directly or indirectly [38], and also confirming that it can act on rRNA as well as DNA.
Many genotoxic substances are known to indirectly affect the cell nucleus through the production of free radicals [39]. Indeed, high percentages of intracellular ROS were detected here after both in vivo and in vitro exposure to quinoin, thus unequivocally indicating the presence of oxidative stress-mediated DNA damage.
Based on experimental evidence in the literature, it was hypothesized that quinoin acts on zebrafish cells by slowing down normal mitochondrial function. It has been reported that abrin, a RIP from Abrus precarious L., induces the shutdown of antioxidant protein-1 (AOP-1). AOP-1 normally activates antioxidant factors to help maintain the redox balance in cells. Blocking this system causes the release of cytochrome C and activation of the apoptotic pathway, with ROS identified as mediators of AOP-1-induced apoptosis [40]. Although the mechanism by which quinoin leads to oxidative stress has not yet been elucidated, its ability to cause ROS-mediated DNA damage has been demonstrated.
It is interesting to note that the percentage of damage was fairly consistent between the different experimental groups, except for the lowest amount of toxin used (1 µg). In the in vivo experiments, 1 µg of quinoin resulted in more instability of the genomic template compared to the higher amounts (5 µg and 10 µg). Alterations in RAPD profiles are due to induced molecular events such as genomic rearrangements, point mutations, deletions and insertions. These can be transient and can also be restored by DNA repair mechanisms, hence they do not always lead to apoptotic processes. Moreover, polymorphic variations do not always result in DNA fragmentation, as this can depend on the type of mutagenic substance, the cell type, and the type of induced polymorphism (appearance or disappearance) [29, 41, 42].
In support of the hypothesis for instant and transitory changes in RAPD profiles, both in vitro and in vivo experiments showed that longer quinoin exposure times resulted in less damage than fewer time of in vitro exposure, in the same way, the damage found 30 days after injection was reduced compared to 15 days after injection. The reduction in damage with prolonged in vitro treatment and many days after administration could indicate cellular adaptation and activation of endogenous repair mechanisms, but may also be due to a reduction in protein N-glycosylase activity.
Although the above hypothesis of cellular adaptation needs to be confirmed with further studies, the weak resistance of quinoin was recently demonstrated using an in vitro pepsin-trypsin (PT) enzyme system [13]. The results showed that most of the toxin was hydrolyzed in the presence of pepsin and trypsin. However, our study does not involve oral administration and therefore digestion in the gastro-intestinal tract cannot be assumed.
Considering, however, that from the aforementioned study it emerges that a small percentage of quinoin remained active even in the presence of hydrolytic enzymes, it is still advisable to treat quinoa prior to consumption in order to inactivate/reduce ANFs. However, our study does not support any obvious risk to health from quinoa, given that the relative quantities of quinoin administered to zebrafish were significantly higher than those found in 100 g of quinoa consumed by humans [13]. Furthermore, lower amounts of quinoin did not cause significant damage in the zebrafish genome (data not shown).
Although less damage was observed with prolonged in vitro treatment by quinoin and after many days from quinoin in vivo administration, genotoxicity remained evident at all times. The proven cytotoxicity of quinoin in various tumor lines in vitro could be the basis for its use in the development of new drugs for cancer therapy.
So far, however, the clinical translation of RIPs for cancer treatment is still a long-term goal. The main difficulty lies with gaining entry into the cell due to vascular, intra-tumoral and intracellular barriers. In order to solve these problems, various strategies are being developed which allow cytotoxic proteins to effectively penetrate cells and to block their activity during blood circulation, but are then activated near the target cells/tissues [43].
Quinoin isolated from quinoa seeds is a type 1 RIP that exhibits ROS-mediated genotoxic activity in the zebrafish model. Although this model organism appears to respond to treatment with quinoin by activating cellular defense mechanisms that reduce damage, significant in vitro and in vivo genotoxicity remained at all times. Further studies are needed to elucidate the prolonged genotoxicity of quinoin by effective DNA damage-response pathways, and to clarify the genotoxic mechanism of action by quinoin. The use of morpholinos to knockdown DNA damage response genes in zebrafish treated with quinoin could provide important insights in this area.
The acute genotoxic activity of quinoin opens up new possibilities for the use of this enzyme as a therapeutic tool in the development of new anticancer drugs.
However, non-specific cellular uptake of RIPs could potentially cause collateral toxicity [44]. As recently suggested by Asrorov et al. [45], these cytotoxic proteins could be tied to nanoparticles and to antibodies to improve the efficiency of tumor-targeted delivery.
The data presented in this study are available on request from the corresponding author.
All authors contributed to the study conception and design. Genotoxicity test were performed by FM, MC and RS. Quinoin purification and enzymatic assays were performed by SR and NL. The first draft of the manuscript was written by FM and LR, and all authors commented on previous versions of the manuscript. ADM and LR contributed with conceptualization, project administration, resources, funding acquisition, supervision, writing - review and editing. All authors read and approved the final manuscript and participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity. All authors contributed to editorial changes in the manuscript.
All used protocols were in accordance with the local regulation and international guidelines for the use and care of laboratory animals and approved by the Italian Ministry of Health (Permit Number: 529/2019-PR).
The authors would like to express their gratitude to Dr. Maria della Corte, Dr. Donatella Paciolla and Dr. Angela Barretta for their contribution in maintenance and treatment of zebrafish.
This work was supported by the research project PRR.AP026.014 - PNRR - IR0000009 - Title: ‘Potentiating the Italian Capacity for Structural Biology Services in Instruct-ERIC’. Acronym: ITACA.SB. Announcement: ‘MUR D.D. 0003264 del 28/12/2021’. In addition, this work was supported by the University of Campania ‘Luigi Vanvitelli’ and University Federico II.
The authors declare no conflict of interest. Given their role as Guest Editor Members, Filomena Mottola and Lucia Rocco had no involvement in the peer review of this article and had no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Gheyath K. Nasrallah.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
