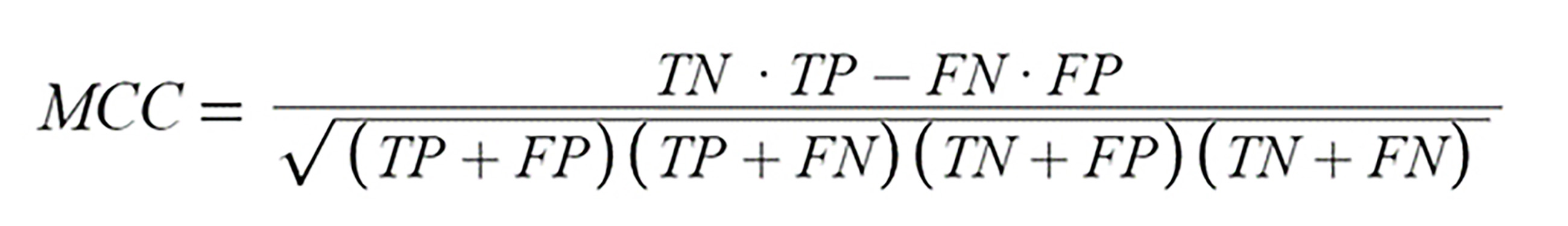1 REHS Program, San Diego Supercomputer Center, UC San Diego, La Jolla, CA 92093, USA
2 San Diego Supercomputer Center, UC San Diego, La Jolla, CA 92093, USA
3 BiAna, La Jolla, CA 92038, USA
4 CureScience Institute, San Diego, CA 92121, USA
5 Pacific Neuroscience Institute, Santa Monica, CA 90404, USA
6 Department of Neurosciences, UC San Diego, La Jolla, CA 92093, USA
Abstract
Background: The current standard for Parkinson’s disease (PD) diagnosis is often imprecise and expensive. However, the dysregulation patterns of microRNA (miRNA) hold potential as a reliable and effective non-invasive diagnosis of PD. Methods: We use data mining to elucidate new miRNA biomarkers and then develop a machine-learning (ML) model to diagnose PD based on these biomarkers. Results: The best-performing ML model, trained on filtered miRNA dysregulated in PD, was able to identify miRNA biomarkers with 95.65% accuracy. Through analysis of miRNA implicated in PD, thousands of descriptors reliant on gene targets were created that can be used to identify novel biomarkers and strengthen PD diagnosis. Conclusions: The developed ML model based on miRNAs and their genomic pathway descriptors achieved high accuracies for the prediction of PD.
Keywords
- machine learning
- Parkinson's disease
- miRNA biomarkers
- neural networks
- deep learning
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, trailing only slightly behind Alzheimer’s disease in prevalence [1]. With nearly a million people affected in the United States alone, PD is projected to impact 1.2 million individuals by 2030 and is expected to double in prevalence by 2040 [2, 3]. Despite its increasing prevalence, the current diagnostic methods for PD remain suboptimal.
The most popular diagnosis of PD is based on clinical criteria, which include the presence of motor symptoms such as bradykinesia (slowness of movement), rest tremor, and rigidity [4]. However, these methods are fraught with limitations. For instance, by the time motor symptoms manifest, significant neuronal loss has already occurred, closing the window for early therapeutic intervention. An earlier diagnosis may provide a therapeutic window to slow or prevent the progression of PD prior to the onset of motor impairments [3]. Another problem with the current diagnosis of PD is that a number of disorders can cause symptoms similar to those of PD, leading to potential misdiagnosis. People with Parkinson’s-like symptoms that result from other causes, such as multiple system atrophy and dementia with Lewy bodies, can be misdiagnosed to have Parkinson’s [4]. Moreover, the clinical diagnostic accuracy remains suboptimal, even when the condition is clinically fully manifest. The identification of prodromal disease is an even greater unmet need given that future disease-modifying therapies will have their greatest chance for success at this stage [5].
In light of these challenges, recent advances in biomarker research advocate for a multidimensional approach to PD diagnosis. A comprehensive review by He and coauthors discussed the limitations of existing biochemical markers and calls for the development of more reliable, early-stage markers [6]. This is precisely where our research comes into play. We leverage machine-learning (ML) algorithms to identify novel microRNA (miRNA) biomarkers for a more accurate diagnosis for PD. Our innovative approach aims to fill the existing gaps in biomarker research, offering a more comprehensive, accurate, and personalized diagnostic model for PD.
Current efforts in PD research are not only focused on understanding the disease mechanisms but also on the identification of reliable biomarkers. As outlined by the American Parkinson Disease Association, finding a biomarker for PD is crucial for early diagnosis, accurate differentiation from other neurological conditions, and effective monitoring of disease progression [7]. Our ML-based approach aligns with these objectives, aiming to provide a definitive, low-cost, and easily accessible means of diagnosing and monitoring PD.
miRNAs are small non-coding RNA molecules that play a crucial role in post-transcriptional regulation of gene expression. They are involved in various biological processes, including cell differentiation, proliferation, and apoptosis. Due to their stability in body fluids and tissue-specific expression patterns, miRNAs have emerged as promising biomarkers for various diseases, including neurodegenerative disorders like PD.
Recent studies have suggested that certain miRNAs may serve as potential biomarkers for Parkinson’s disease. For instance, a study by Gui and colleagues found that the levels of miR-4639-5p were significantly decreased in the serum of PD patients compared to healthy controls. Another study found that certain miRNAs, specifically miR-146a, miR-335-3p, and miR-335-5p, were downregulated in both idiopathic Parkinson’s Disease (iPD) and LRRK2-PD patients compared to healthy controls [8]. Additionally, miR-155 was found to be upregulated in LRRK2-PD compared to iPD patients. These findings suggest that these miRNAs could serve as potential biomarkers for PD potentially improving disease diagnosis efficiency and accuracy [9].
An exhaustive analysis in the field of PD biomarkers brings to light the
complexities and limitations of current diagnostic methods. The analysis sorts
biomarkers into clinical, imaging, and biofluid categories, each with its unique
drawbacks. For example, clinical markers like non-motor symptoms are promising
but not yet reliable for early-stage detection [10]. Imaging markers, although
precise, are financially burdensome and not widely available. Biofluid markers,
such as
To further deepen our understanding of miRNAs’ role in PD, recent studies have explored their influence on the functioning of microglia within the central nervous system. These innate immune cells in the brain are pivotal in the disease mechanisms of PD. When overly activated, microglia can intensify neuroinflammation, leading to the further decline of dopaminergic neurons. Research indicates that miRNAs can control both the activation levels and polarization states of these microglia, thereby affecting the course of PD [11]. This discovery paves the way for new therapeutic strategies, offering the possibility of using miRNAs to regulate microglia activity, which could potentially arrest or even reverse the progression of the disease.
Though miRNAs offer a hopeful avenue for diagnosing PD, their clinical utility has hit some roadblocks. One study delved into the use of plasma-circulating miRNAs, employing algorithms like k-Top Scoring Pairs and significance analysis of microarrays to craft a wide-ranging set of PD-predictive indicators. While the research showed strong predictive capabilities in an initial sample set, it faltered when tested on a different clinical sample [12]. This underscores the need for robust machine learning techniques that can navigate the complexities of varying clinical samples, amplifying the importance of our own work in harnessing machine learning to pinpoint trustworthy miRNA markers.
A new study in the field of PD research in Neural Plasticity has identified exosomal miRNAs, specifically miR‑342‑3p, as promising circulating biomarkers for PD [13]. This research not only addresses the limitations of current diagnostic methods but also opens the door for ML algorithms to analyze these new types of biomarkers for more accurate and earlier diagnosis. The discovery of exosomal miRNAs like miR‑342‑3p could revolutionize the way clinicians approach PD, allowing for diagnosis at much earlier stages than currently possible.
Machine-learning techniques offer a promising avenue for enhancing PD diagnosis as discussed in a recent study by Kang and colleagues, the authors demonstrated the successful application of machine learning in classifying diseases, particularly cancers, based on miRNA data can also be applied to PD [14]. ML techniques can be used to analyze complex proteomic and genomic measurements, which are crucial in identifying potential biomarkers like miRNAs for PD. These techniques can help in the early detection of the disease.
In a similar vein, a study by Amy Xu and coauthors demonstrates the application of machine learning (ML) in diagnosing Alzheimer’s disease (AD), another neurodegenerative disorder [15]. The authors developed a ML model that includes miRNAs and their genomic and pathway descriptors for the diagnosis of AD. This new application of ML into the discovery of new pathway descriptors can also be applied to PD.
The integration of ML in the analysis of biomarker-based diagnostics can potentially revolutionize the way we approach PD, moving towards a more personalized, predictive model of medicine. This could lead to improved patient outcomes, as treatments could be administered earlier, slowing the development of the disease.
Fig. 1 presents a schematic representation of the study’s methodology. Dysregulated miRNAs in Parkinson’s Disease (PD) were identified and validated from previously published studies. Subsequently, the gene targets from the miRNAs were extracted from the miRpathDB database (v. 2.0, Saarland University, Saarbrücken, Saarland, Germany). These extracted features processed by pandas software (v. 2.1.3, NumFOCUS, Inc., Austin, TX, USA) were then inputted into the Waikato Environment for Knowledge Analysis (WEKA, v. 3.8.6; University of Waikato, Hamilton, New Zealand) and Keras (v. 2.13.1, Google LLC, Mountain View, CA, USA) platforms to construct ML models, aiming at PD classification. Attribute filtering techniques were employed to minimize the dimensionality of the initial dataset. Finally, the performance of various classification models was assessed and compared in terms of their accuracy.
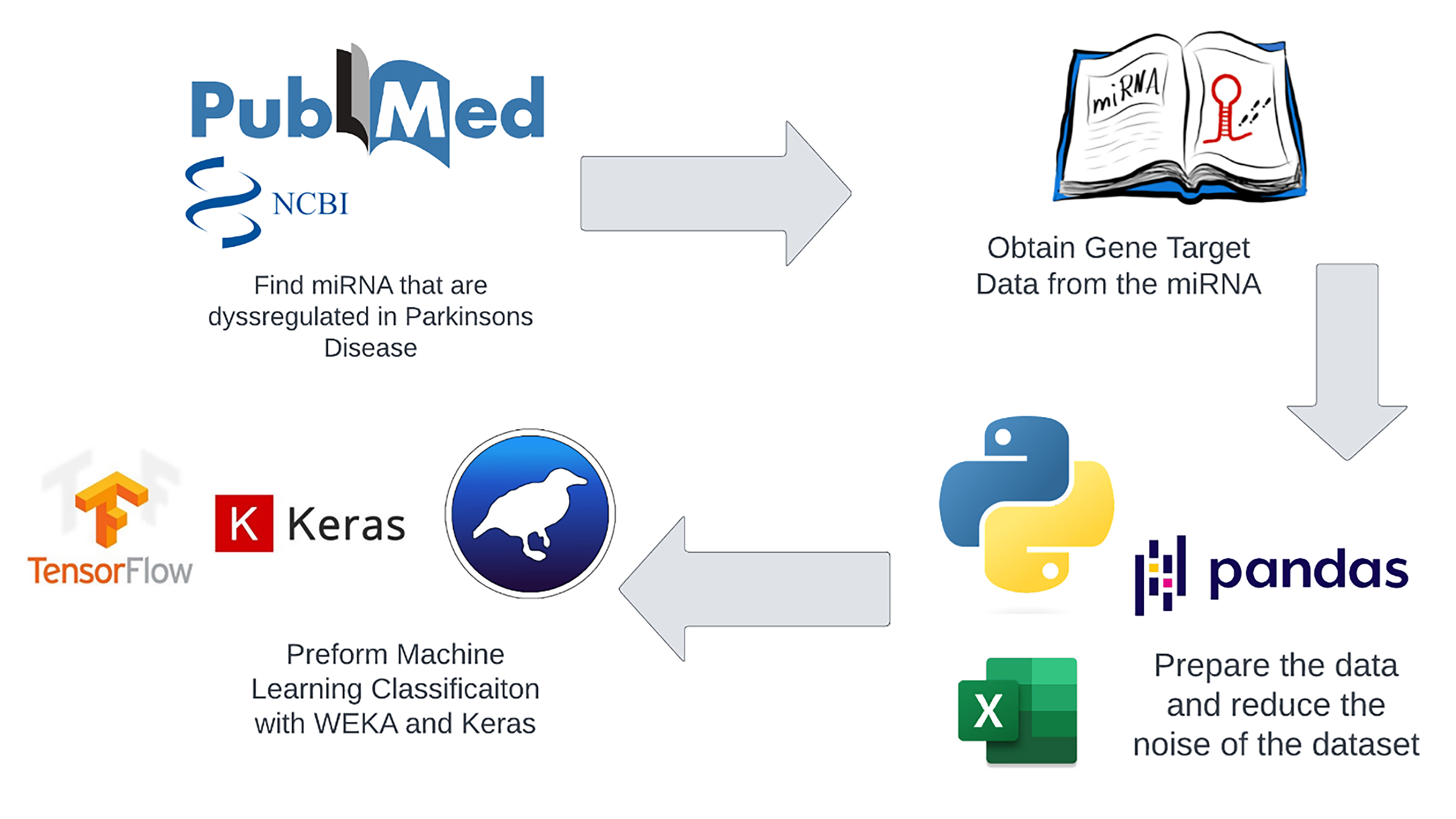 Fig. 1.
Fig. 1.Flowchart of methods. We used the published data to identify dysregulated microRNAs (miRNAs) in Parkinson’s disease (PD) and using miRPathDB database we extracted their gene targets. Then using gene targets as descriptors, we constructed machine-learning models with Waikato Environment for Knowledge Analysis (WEKA) and Keras platforms for PD diagnostics. We reduced the dataset dimensionality through attribute filtering. Then we cross-validated the created classification model and checked it performance on independent datasets.
Firstly, the study began with selecting dysregulated miRNAs significantly related to the development and pathogenesis of PD. Only circulating miRNAs (CSF, serum, plasma, peripheral blood mononuclear cells (PBMCs), and saliva) are of PD patients were extracted from a study done by Nies and coworkers [16]. Along with this list of miRNAs that are dysregulated in PD, a set of controls was collected randomly from the miRPathDB database. The inclusion criteria for the PD set involved selecting miRNAs that have been identified as significantly dysregulated in peer-reviewed studies focused on PD and are implicated in pathways known to be involved in PD pathogenesis. The exclusion criteria for the PD set ruled out miRNAs with conflicting evidence across multiple studies and those that are also significantly dysregulated in other neurodegenerative diseases. For the control set (not related to PD), the inclusion criteria consisted of miRNAs that have not been implicated in any neurodegenerative diseases and are considered to be stably expressed across multiple tissue types. The exclusion criteria for the control set eliminated miRNAs with known roles in other neurological disorders and those showing significant variability in expression across different tissue types. All included miRNAs are presented in Table 1.
| PD Set | Control |
| hsa-miR-138-5p, hsa-miR-338-3p, hsa-miR-431-5p, hsa-miR-30c-2-3p, hsa-miR-10394-5p, hsa-miR-1208, hsa-miR-1256, hsa-miR-1271-5p, hsa-miR-1286, hsa-miR-142-3p, hsa-miR-183-5p, hsa-miR-20a-3p, hsa-miR-2276-3p, hsa-miR-2278, hsa-miR-27b-5p, hsa-miR-299-3p, hsa-miR-3158-5p, hsa-miR-342-3p, hsa-miR-3606-3p, hsa-miR-3622a-3p, hsa-miR-365a-3p, hsa-miR-423-5p, hsa-miR-4266, hsa-miR-4272, hsa-miR-449c-3p, hsa-miR-452-3p, hsa-miR-4660, hsa-miR-4712-5p, hsa-miR-4714-5p, hsa-miR-4740-3p, hsa-miR-4762-5p, hsa-miR-4769-3p, hsa-miR-496, hsa-miR-5009-5p, hsa-miR-526b-5p, hsa-miR-548a-3p, hsa-miR-574-3p, hsa-miR-590-3p, hsa-miR-6082, hsa-miR-6128, hsa-miR-627-5p, hsa-miR-631, hsa-miR-647, hsa-miR-6719-3p, hsa-miR-6727-5p, hsa-miR-6757-3p, hsa-miR-6779-5p, hsa-miR-6792-3p, hsa-miR-6796-5p, hsa-miR-6811-3p, hsa-miR-6848-3p, hsa-miR-6873-3p, hsa-miR-6874-3p, hsa-miR-6882-5p, hsa-miR-6889-3p, hsa-miR-7162-5p | hsa-miR-103a-3p, hsa-miR-105-5p, hsa-miR-124, hsa-miR-126, hsa-miR-132-3p, hsa-miR-137-3p, hsa-miR-142-3p, hsa-miR-144, hsa-miR-146b-5p, hsa-miR-151, hsa-miR-153-3p, hsa-miR-16-2-3p, hsa-miR-181a-5p, hsa-miR-185-5p, hsa-miR-193a-3p, hsa-miR-195-5p, hsa-miR-19b-3p, hsa-miR-200a-3p, hsa-miR-214-3p, hsa-miR-22-3p, hsa-miR-221-3p, hsa-miR-222-3p, hsa-miR-24-3p, hsa-miR-27a-3p, hsa-miR-27a-3p, hsa-miR-28-5p, hsa-miR-29a-3p, hsa-miR-29c-3p, hsa-miR-301a, hsa-miR-30c-5p, hsa-miR-373-5p, hsa-miR-374a-5p, hsa-miR-1227-5p, hsa-miR-5586-3p, hsa-miR-4763-5p, hsa-miR-4308, hsa-miR-6759-5p, hsa-miR-3613-3p, hsa-miR-5682, hsa-miR-4781-3p, hsa-miR-1251-5p, hsa-miR-4431, hsa-miR-1914-5p, hsa-miR-4470, hsa-miR-33a-5p, hsa-miR-217-5p, hsa-miR-491-3p, hsa-miR-6508-5p, hsa-miR-16-1-3p, hsa-miR-190a-5p, hsa-miR-6870-5p, hsa-miR-6515-5p, hsa-miR-182-3p, hsa-miR-223-5p, hsa-miR-361-3p, hsa-miR-489-5p |
The choice to exclusively use miRPathDB 2.0 as our sole database for control miRNAs is due to its focus on high-quality, experimentally verified data. This aligned with our goal of ensuring data reliability. Primarily, miRPathDB is acknowledged for its robust data quality, as it curates miRNA targets from a variety of sources including peer-reviewed publications, thus ensuring a high level of data reliability. For our analysis, we included all predicted target genes for our selected miRNAs, using both intersection and union prediction methods.
Gene-target data was collected for each miRNA associated with miRPathDB [17], as well as the miRNAs in the control group. miRPathDB is a comprehensive database that consolidates miRNA-target interactions and pathway annotations from multiple pathway databases, providing valuable insights into miRNA regulation and signaling pathways. It utilizes various sources such as TargetScan, miRTarBase, and DIANA-TarBase to compile miRNA–target interactions.
To systematize each miRNA’s unique features, we implemented an algorithm that employed a singular gene list (derived from the aggregate of each miRNA’s target genes), assigning a binary value (0 or 1) to each miRNA for each gene and pathway. Consequently, we built a dataset column-by-column, each representing a gene target attribute (e.g., TP53), and rows representing individual miRNAs with their binary descriptors. A “1” signifies the miRNA holding the gene target or pathway in the column, whereas a “0” denotes the absence of that specific genomic target or pathway.
While we acknowledge the binary coding system simplifies the intricacies of miRNA–target links, such as site binding or conservation factors, it serves the purpose of facilitating a general first-level analysis. Future work could delve into these finer details for a more nuanced understanding.
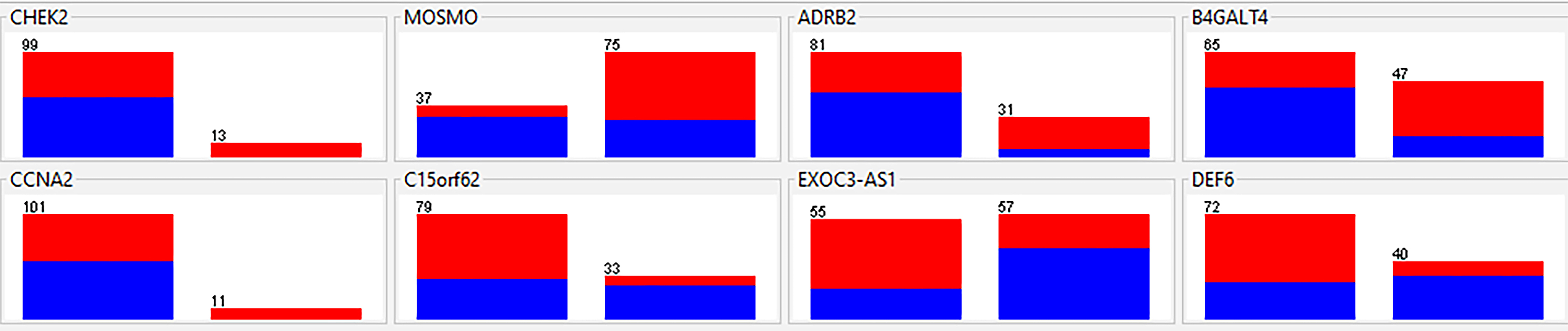
After the preparation, the dataset included 16,299 attributes (descriptors) for 56 associated miRNA and 56 control miRNA (Fig. 2). The process of attribute selection involved utilizing WEKA’s InfoGainAttributeEval module that ranks descriptors according to their capacity to differentiate between classes in a classification problem. By calculating the mutual information between each feature and the class variable, this algorithm quantifies the extent to which the feature provides information about the class [18].
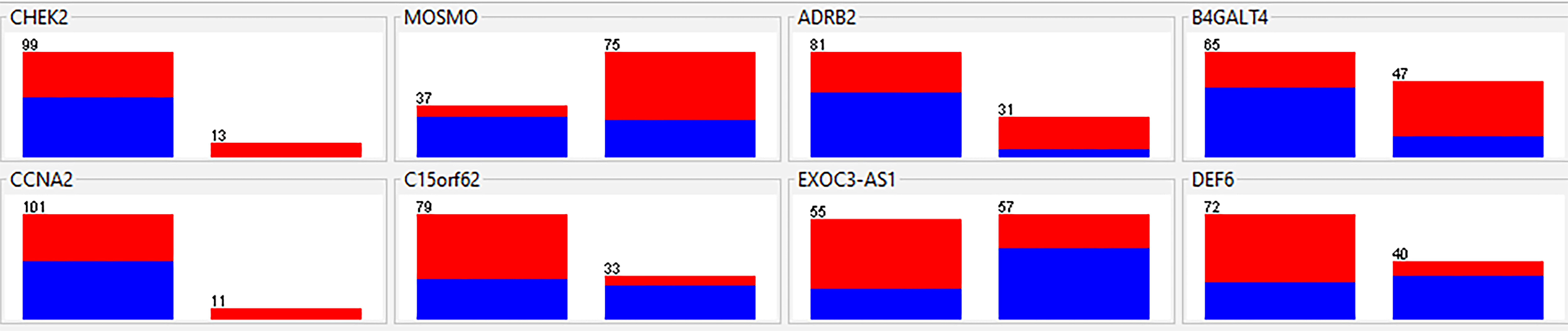 Fig. 2.
Fig. 2.Highest ranked descriptors by the InfoGainAttributeEval algorithm from WEKA program. The chart depicts the dataset’s attributes, presenting a visual representation of each classes’ gene targets. By utilizing the InfoGainAttributeEval algorithm, which measures the information gain provided by each attribute in relation to the class variable, the chart showcases the attributes that contribute the most relevant and discriminative information for distinguishing between different classes within the dataset. The classes are represented by colors: red for miRNA dysregulated in Parkinson’s Disease, and blue for the control group. The Y-axis shows the quantity of miRNA with a specific gene target. The left column shows miRNA without this gene target, while the right bar represents miRNA that have the specific gene target. Each gene target corresponds to a unique biological function or implication. For instance, ‘CHEK2’, also known as Checkpoint Kinase 2, is a gene that encodes for the protein CHK2. This protein plays a pivotal role in the cell’s response to DNA damage, thereby highlighting the significance of the gene target in the context of the miRNA dataset.
After employing WEKA’s InfoGainAttributeEval module, the number of descriptors was substantially reduced. We validated the efficacy of this reduced attribute set through additional analyses. Specifically, the reduced set showed higher classification accuracy of 93.3% on the independent testing set after reducing the number of descriptors, confirming that it retains sufficient statistical power for reliable classification.
On the miRNA training and testing datasets, ML analysis was performed using the WEKA program environment. This open-source workbench includes several tools for data cleaning and filtering, classification and pattern recognition [18]. In addition to the ML analysis performed using WEKA, the TensorFlow Python library Keras was used to generate a neural network for making predictions from the data. Keras is a high-level neural networks application programming interface (API) that provides an interface to build, train, and evaluate neural networks. It is built on top of lower-level libraries such as TensorFlow (v. 2.10, Google LLC, Mountain View, CA, USA), which handle the computations underlying the neural network [19]. The neural network utilizes a Sequential model with 5 layers. The first layer is a 32-neuron Dense layer with Rectified Linear Unit (ReLU) activation, which use a threshold function to introduce non-linearity. The second layer is also a 32-neuron Dense layer using ReLU activation but includes Dropout, which randomly deactivates 20% of neurons during training to prevent overfitting. This is followed by two hidden Dense layers, also using ReLU activation, of 128 and 64 neurons, respectively, allowing for complex pattern recognition. The final layer is a single-neuron Dense output layer with sigmoid activation. Sigmoid activation transforms the output into a probability value between 0 and 1, representing the likelihood of the binary classification. The architecture is illustrated in Fig. 3.
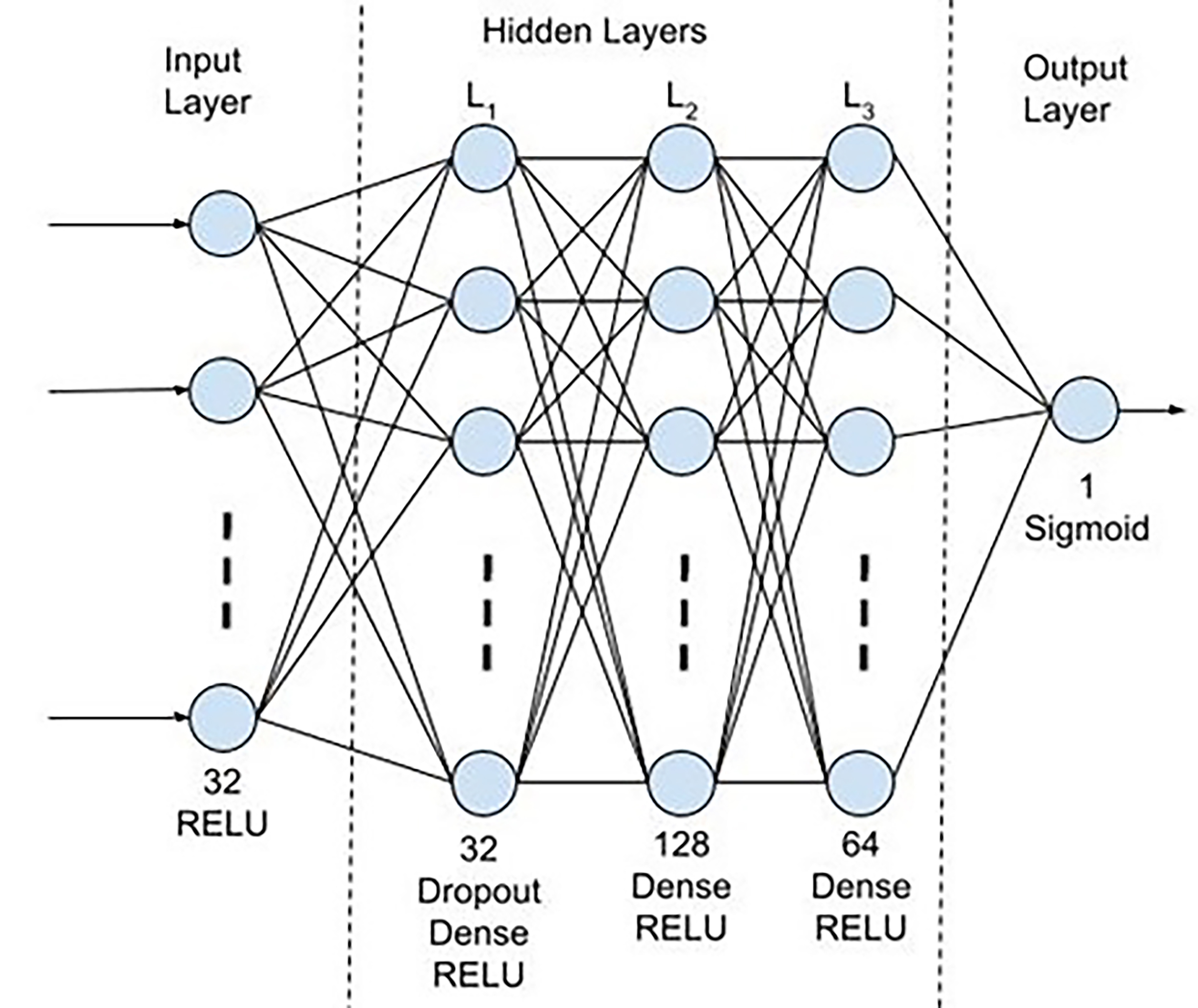 Fig. 3.
Fig. 3.Schematic of Sequential model architecture. The schematic diagram visually represents a Sequential neural network model. It consists of five layers: an input layer, three hidden layers including a dropout layer, and an output layer. RELU, Rectified Linear Unit.
The objective of our model was to identify patterns of prospective miRNA blood-based biomarkers specific to PD for diagnostic applications. Each discrete dataset underwent individual analysis through various ML classification algorithms. We subsequently evaluated these models’ validity by applying them to test data, ensuring no overlap with the respective training datasets. At the end of this stage the ML models were developed and cross-validated.
In a second—validation phase, we introduced the model to an independent and distinct set of miRNAs significantly associated with PD, as well as another set tied to a different pathology, specifically breast cancer. The model’s effectiveness was gauged by its accuracy, which is defined as the proportion of correctly classified instances to the total instances.
The cross-validation test sets yielded above 90% accuracy for the Hoeffding Tree algorithm, the Naïve Bayes, Multilayer Perceptron, and the Sequential Model that was trained using Keras. The training sets for the model contained 112 miRNAs (with 56 miRNAs being PD biomarkers and 56 not), which was then filtered from the dataset reducing the number of attributes from 16,299 to 61. The main parameters of models prepared with different algorithms are presented in Table 2. The highest accuracy model was the Sequential Model, which was then used to make predictions on independent data (Fig. 4).
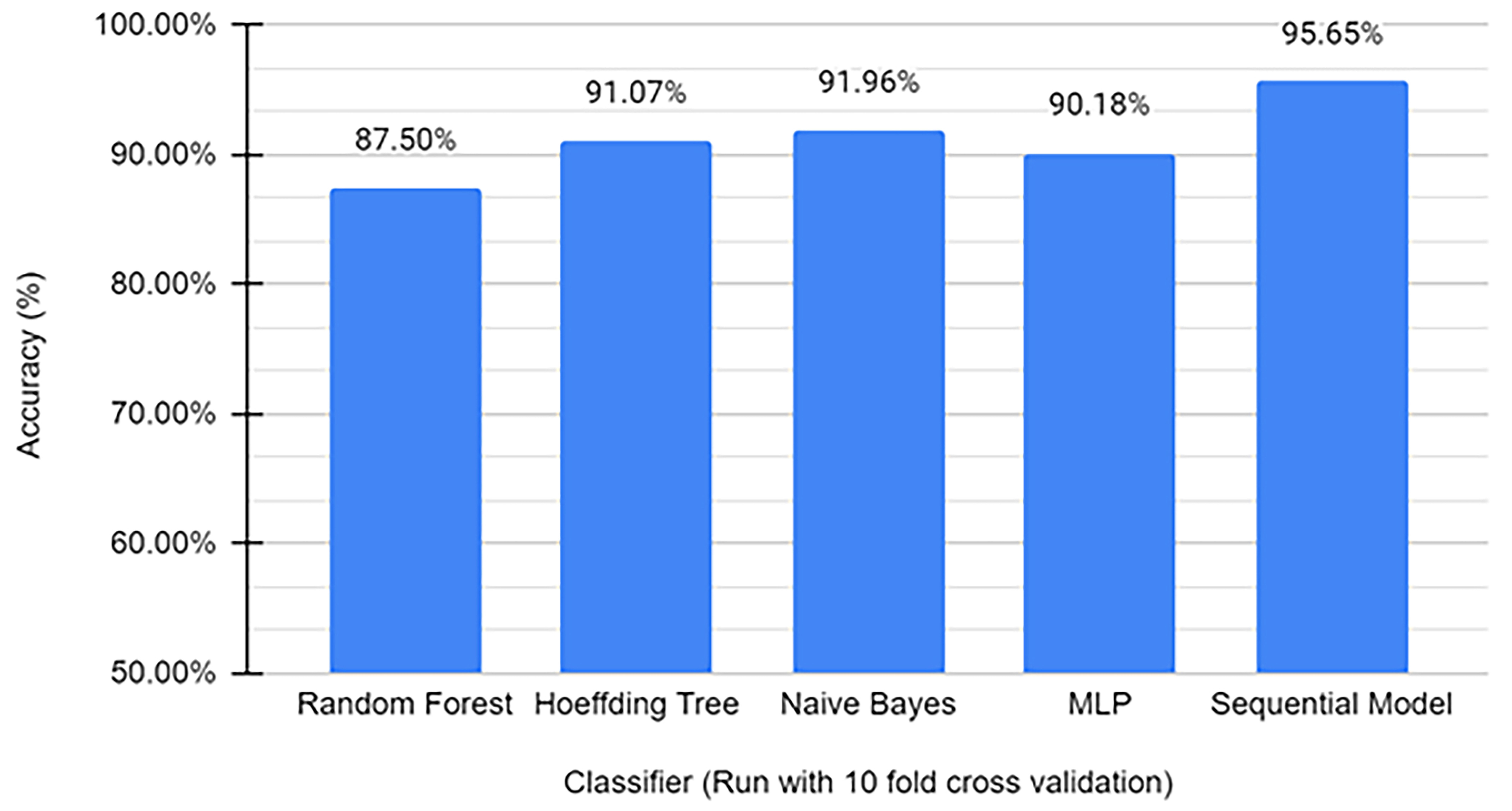 Fig. 4.
Fig. 4.Machine-learning accuracies on identifying PD from miRNA biomarkers. The Y-axis represents the accuracy of each model, and different models are represented by individual bars on the graph. This visual representation enables a straightforward comparison of the performance strengths of each model, providing a scientific basis for evaluating and selecting the most effective model for PD classification based on its accuracy.
| Model | Kappa statistic | Mean absolute error | Root mean squared error | Relative absolute error | ROC | MCC |
| Random Forest | 0.75 | 0.2876 | 0.3483 | 0.574770 | 0.9160 | 0.754 |
| Hoeffding Tree | 0.8214 | 0.0961 | 0.2721 | 0.192052 | 0.9660 | 0.822 |
| Naïve Bayes | 0.8393 | 0.09 | 0.2611 | 0.179895 | 0.9692 | 0.840 |
| MLP | 0.8036 | 0.1064 | 0.2719 | 0.212583 | 0.9676 | 0.805 |
MLP, Multilayer perceptron; ROC, Receiver operating characteristic; MCC, Matthews correlation coefficient.
The Hoeffding Tree model outperformed Random Forest in our analysis, a result that may seem counterintuitive given Random Forest’s ensemble approach. This outcome is likely due to unique dataset characteristics that favor the Hoeffding Tree’s online learning method. It also suggests that our Random Forest model may have been overfitting as its the parameters was not optimized.
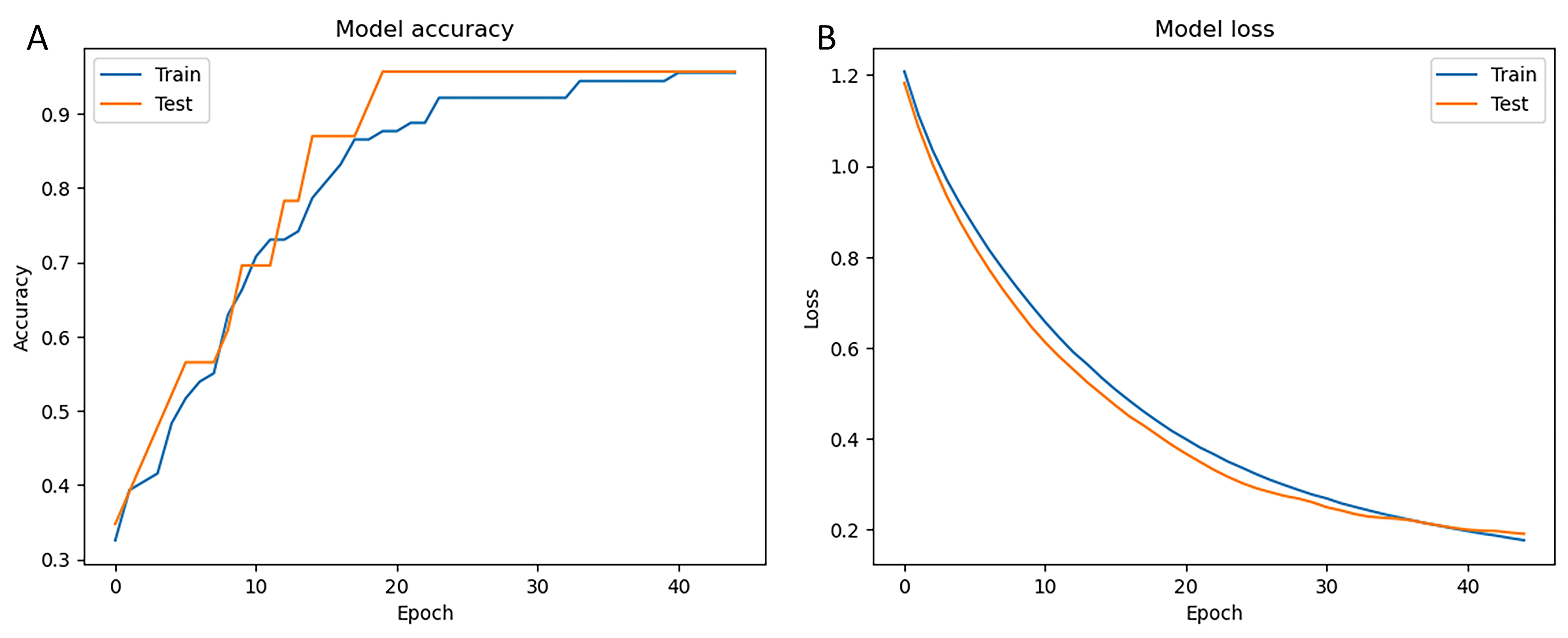
The Sequential Model that was created using Keras was able to reach accuracies of 95.65% (Fig. 5). To further eliminate potential bias and prevent overfitting, we created an independent test set comprising miRNA dysregulated in PD (Table 3). The independent test set used for the additional was extracted from a study done by Ming-Che Kuo and coauthors [20]. We selected eight miRNAs from this source that have been significantly associated with PD. There was no overlap between these miRNAs and those utilized in the training set. This independent set allowed us to test the model’s capacity to accurately classify new, unseen data. Upon analysis of this independent test set, the Sequential Model continued to show strong performance, achieving an accuracy of 93.3% (Fig. 6).
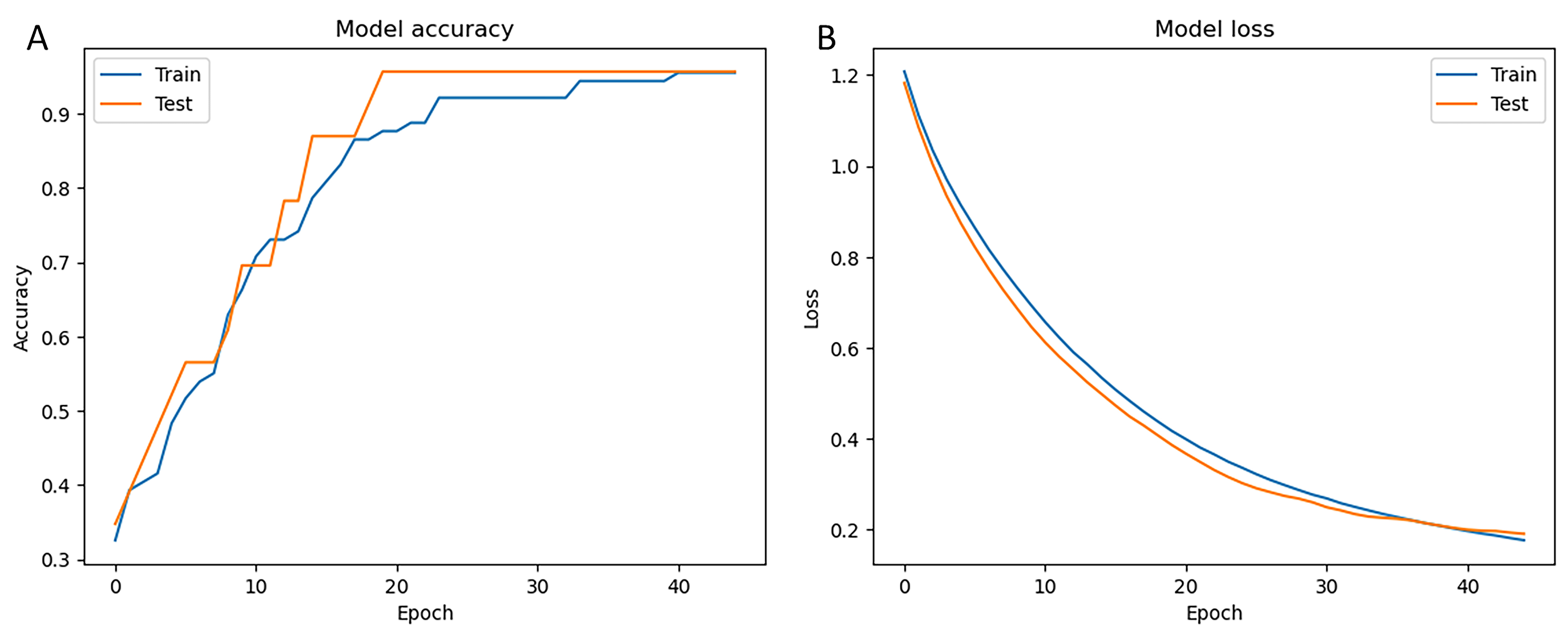 Fig. 5.
Fig. 5.Model accuracy and Model loss of Sequential Model. (A) The graph represents the model’s accuracy and loss over multiple epochs, with each epoch being one complete pass through the entire training dataset. Higher accuracy values indicate better performance in correctly classifying the classes. The accuracy is illustrated over a series of epochs, showing how the model’s performance grows as it loops over the training dataset. (B) The graph illustrates the model loss, which indicates the difference between the predicted and actual values during the training process. Lower values suggest a better fit between the model’s predictions and the actual values.
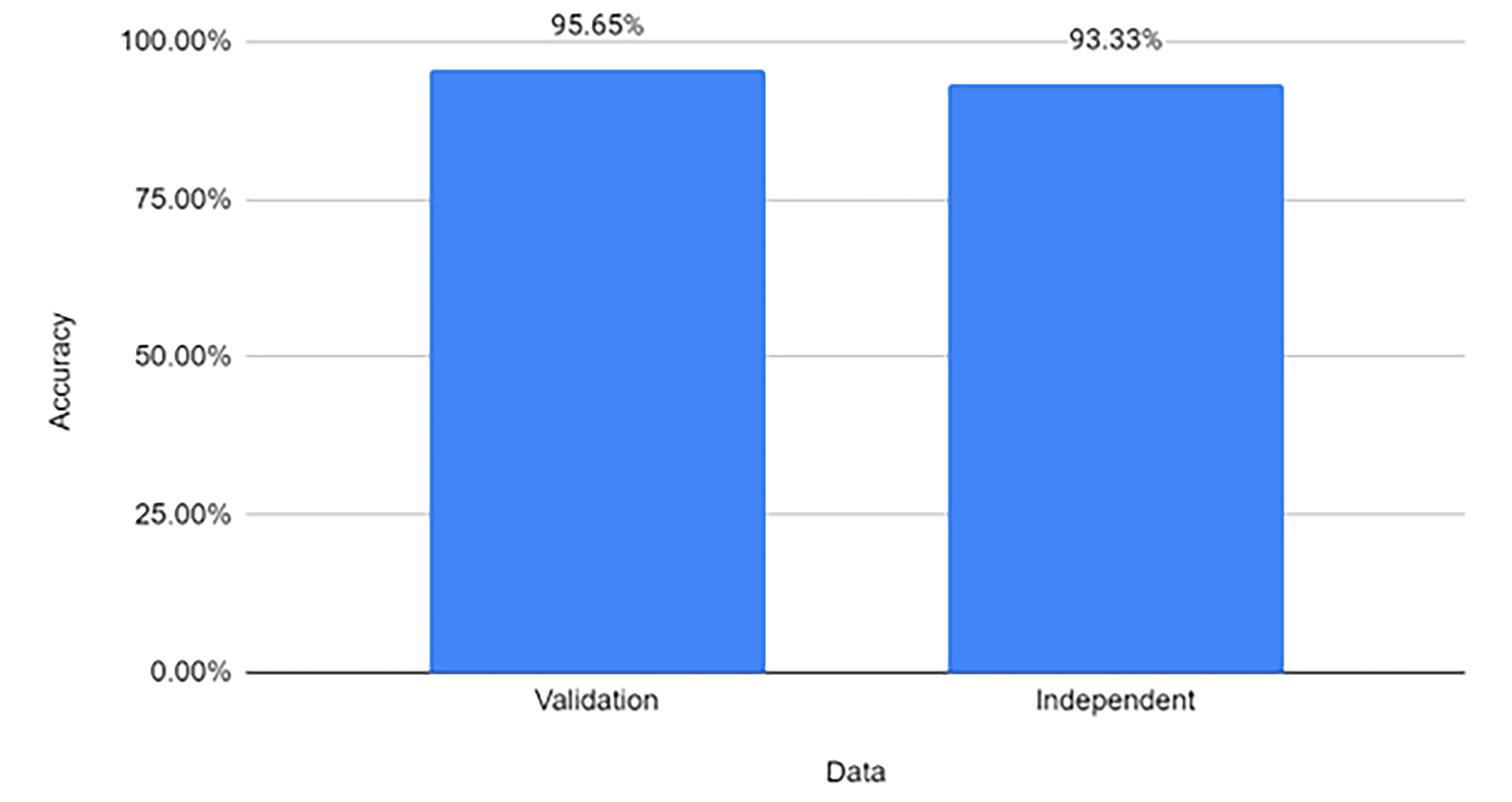 Fig. 6.
Fig. 6.Accuracy of Model on validation data versus independent PD data.
| Independent PD testing set |
| hsa-miR-7-5p, hsa-miR-139-5p, hsa-miR-330-5p, hsa-miR-495-3p, hsa-miR-154-5p, hsa-miR-501-3p, hsa-miR-874-3p, hsa-miR-145-3p |
Following the validation of our independent set, we initiated additional testing to ascertain that our models are selective for PD. We aimed to confirm that the models could specifically detect PD and not misclassify other conditions as PD. To verify this specificity, we challenged our classifier with miRNA datasets of a different disease—Breast Cancer (BC, Table 4). The BC miRNA dataset was drawn from the comprehensive research conducted by van Schooneveld and coworkers [21]. Our classifier model, trained on PD-specific miRNA data, was then run against the BC miRNA dataset. The expectation was a low number of false positives, indicating the model’s capability to discern between the miRNA profiles of different diseases.
| Breast Cancer miRNA validation set |
| miR-205, miR-375, miR-30a, miR-342-5p, miR-497, miR-122, miR-27b-3p, miR-21, miR-210, miR-9 |
The results were in line with our expectations. While the model achieved an accuracy of 95.65% on the PD-specific miRNA test set, its accuracy was significantly lower at 40% on the BC miRNA test set (Fig. 7). Although these findings suggest a degree of specificity for PD, further studies with multiple control groups and additional statistical tests are required for robust confirmation.
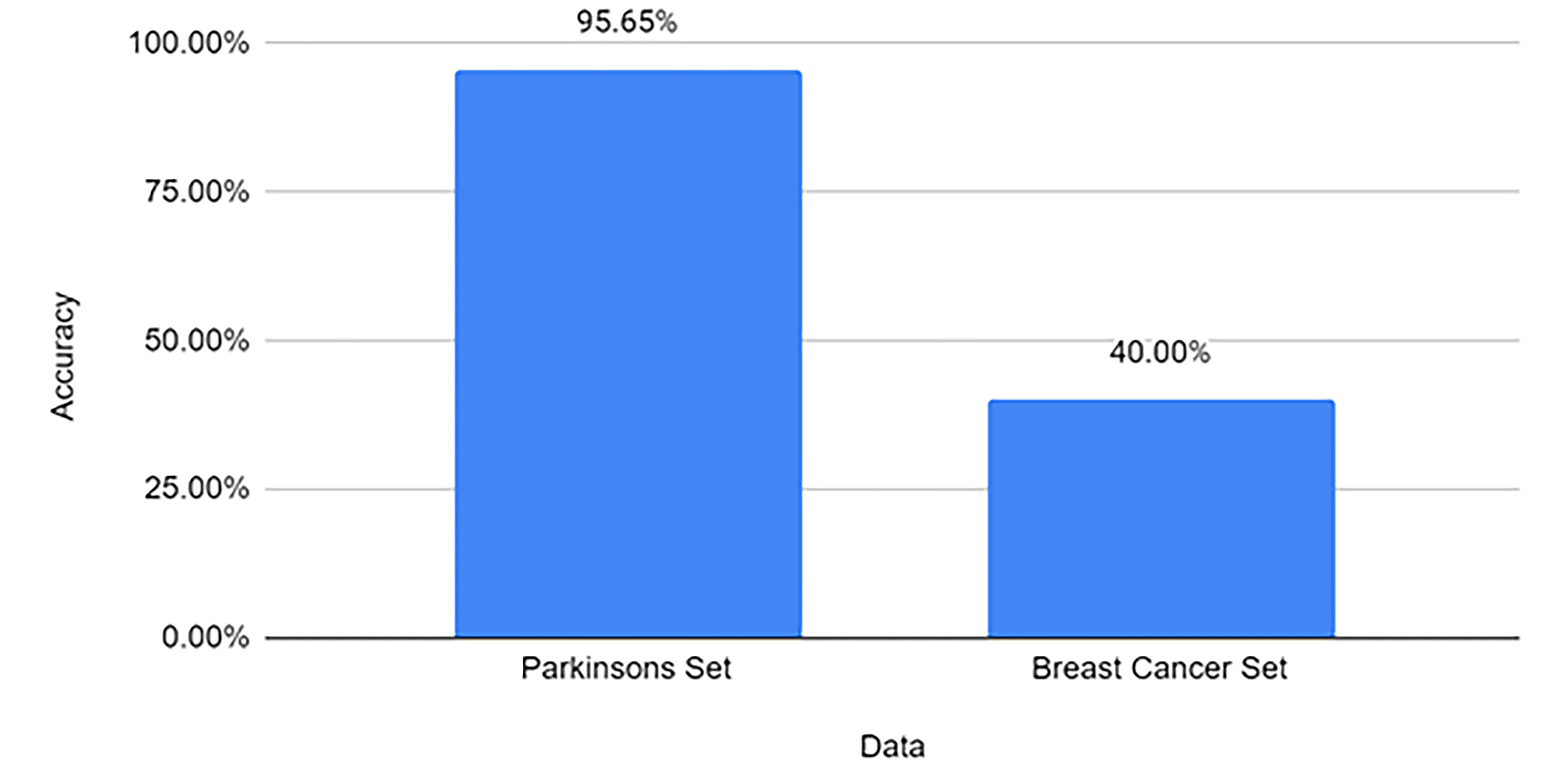 Fig. 7.
Fig. 7.Model’s accuracy on PD testing set vs Breast Cancer (unrelated PD) set. The graph displays the accuracy of the model on the Y-axis and differentiates between the two datasets.
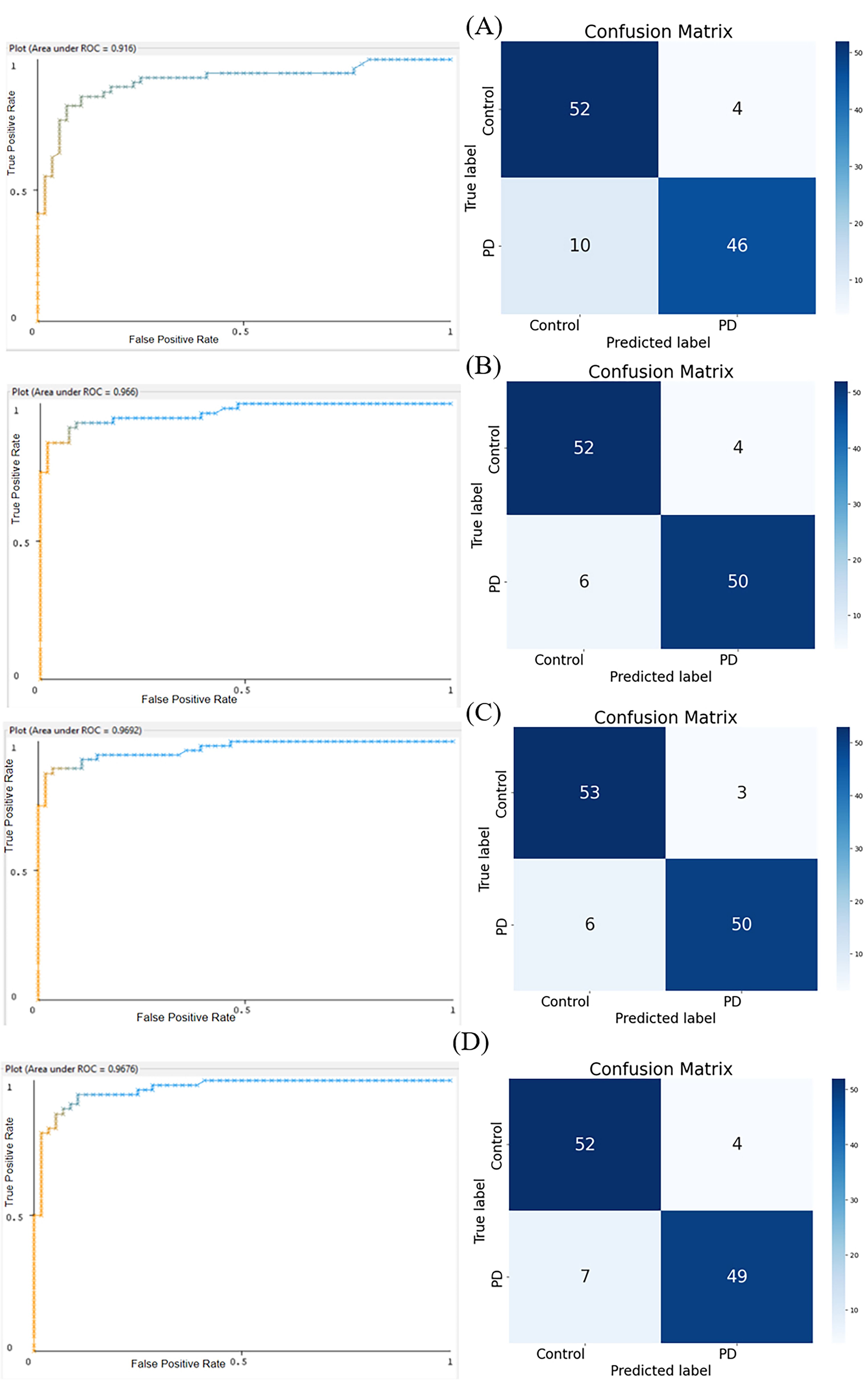
The Area Under the Receiver Operating Characteristic Curve (AUROC) values for each model are as follows: Random Forest scored 0.916, Hoeffding Tree scored 0.966, Naïve Bayes scored 0.9692, Multilayer Perceptron (MLP) scored 0.9676, and the Sequential model scored a 0.992. These values indicate high classifying power for all models, with Naïve Bayes, MLP, and the Sequential models showing the highest performance (Fig. 8).
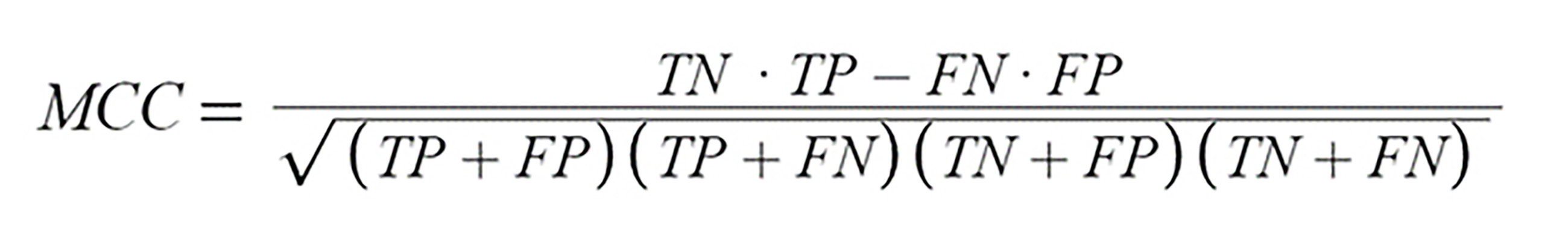 Fig. 8.
Fig. 8.Mathews Correlation Coefficient Formula. True Positives (TP) are instances where the model correctly identifies a positive case as positive. True Negatives (TN), on the other hand, are instances where the model accurately labels a negative case as negative. False Positives (FP) occur when the model mistakenly identifies a negative case as positive. Lastly, False Negatives (FN) are instances where the model incorrectly labels a positive case as negative.
From the confusion matrices of each model, the Matthews Correlation Coefficient (MCC) was computed. The MCC values, calculated using the formula from Fig. 8, for the Random Forest, Hoeffding Tree, Naïve Bayes, and MLP models, and the Sequential model were 0.754, 0.822, 0.840, and 0.805, 0.914, respectively. These values indicate that the Sequential model achieved the best balance between sensitivity and specificity among the tested models.
For the Random Forest confusion matrix as shown in Fig. 9A, the top left cell, representing True Positives, contains 52 instances, indicating that the model correctly identified these cases as positive. The bottom right cell, representing True Negatives, contains 46 instances, showing that these cases were correctly identified as negative. However, the model was not perfect. The top right cell contains 4 instances, representing False Positives, where the model incorrectly classified negative cases as positive. Similarly, the bottom left cell contains 10 instances, representing False Negatives, where the model incorrectly classified positive cases as negative.
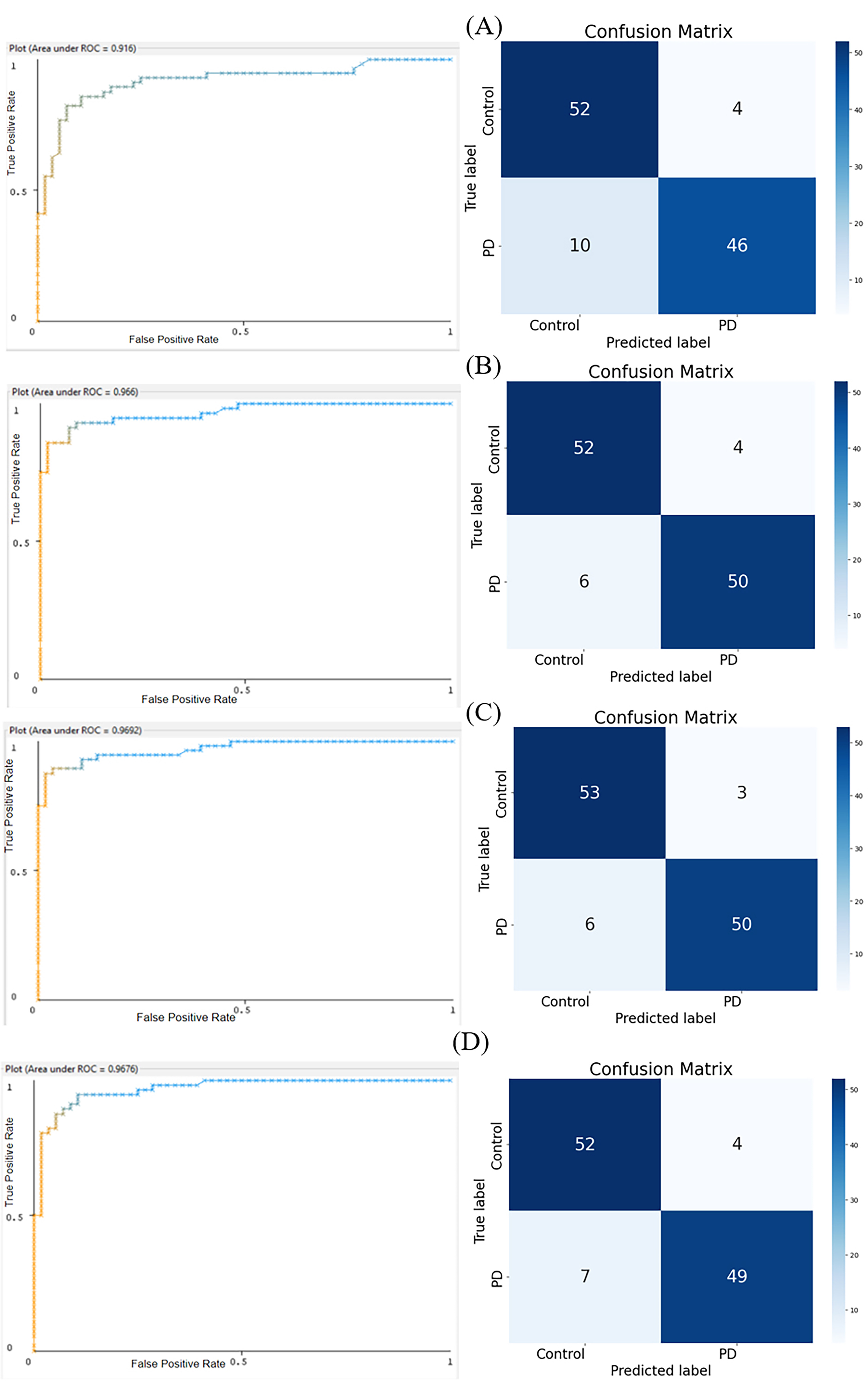 Fig. 9.
Fig. 9.Areas Under the Receiver Operating Characteristic Curves (AUROC) Plots (left panels) and Confusion Matrices (right panels). (A) Random Forest: AUROC = 0.9160. (B) Hoeffding Tree: AUROC = 0.9660. (C) Naïve Bayes: AUROC = 0.9692. (D) Multilayer Perceptron Algorithm: AUROC = 0.9676.
The confusion matrix for the Hoeffding Tree model (Fig. 9B) shows a slightly different accuracy. The top left cell, representing True Positives, contains 52 instances, showing that these cases were correctly identified as positive. The bottom right cell, representing True Negatives, contains 50 instances, indicating that these cases were correctly identified as negative. However, the model did make some errors. The top right cell contains 4 instances, representing False Positives, where the model incorrectly classified negative cases as positive. The bottom left cell contains 6 instances, representing False Negatives, where the model incorrectly classified positive cases as negative.
The confusion matrix for the Naïve Bayes (Fig. 9C) model presents yet another pattern. The top left cell, representing True Positives, contains 53 instances, indicating that these cases were correctly identified as positive. The bottom right cell, representing True Negatives, contains 50 instances, showing that these cases were correctly identified as negative. However, the model was not without errors. The top right cell contains 3 instances, representing False Positives, where the model incorrectly classified negative cases as positive. The bottom left cell contains 6 instances, representing False Negatives, where the model incorrectly classified positive cases as negative.
The confusion matrix for the MLP (Fig. 9D) model shows another pattern. The top left cell, representing True Positives, contains 52 instances, indicating that these cases were correctly identified as positive. The bottom right cell, representing True Negatives, contains 49 instances, showing that these cases were correctly identified as negative. However, the model did make some mistakes. The top right cell contains 4 instances, representing False Positives, where the model incorrectly classified negative cases as positive. The bottom left cell contains 7 instances, representing False Negatives, where the model incorrectly classified positive cases as negative.
Our study’s results may be useful in early diagnosis of PD. It is known that a patient has already experienced a significant and widespread loss of brain cells and brain and autonomic nervous system functions by the time they display the classic motor symptoms of PD and are given a diagnosis. Therapeutic interventions intended to slow or stop the progression of PD are severely constrained by this late-stage diagnosis. Our study demonstrates a possibility for diagnostic early PD early on, before motor impairments start to manifest.
Using microRNAs (miRNAs) as biomarkers is a promising strategy for a PD early diagnosis. Our results suggest that a set of miRNAs, which are known to be dysregulated in PD, may be used as biomarkers for these purposes. By facilitating earlier therapeutic interventions, these miRNAs may increase the precision and efficacy of PD diagnosis. More investigation is needed to validate these results and pinpoint the most trustworthy miRNA biomarkers because the use of miRNAs as PD biomarkers is still a relatively new field. Our study investigated the use of machine-learning (ML) techniques with miRNA biomarkers for diagnostics of PD. The application of ML to the evaluation of biomarker-based diagnostics has the potential to transform how we approach PD and move medicine closer to a more individualized, predictive model. According to our study, the top-performing ML model, trained on miRNA dysregulated in PD, had a 95.65% accuracy rate for diagnostics of PD. Due to its high degree of accuracy, machine learning (ML) has the potential to be an effective tool for the early detection of PD, predicting how the disease will develop, and tailoring treatment plans.
To validate these results and improve these diagnostic tools, additional study is needed. The creation of efficient diagnostic tools will be essential in enhancing patient outcomes and slowing the progression of this crippling illness as the prevalence of PD rises.
Our research tackles early PD diagnosis using miRNAs and machine learning. The best model achieved 95.65% accuracy, showing promise for early PD detection. While our binary coding is a simplified approach, it’s a useful starting point for future, more nuanced studies. Tests against other diseases suggest the model is PD-specific, but more work is needed to confirm this. As PD rates rise, the urgency for reliable early diagnostic tools grows.
The software developed is available on reasonable request. The data utilized and/or examined in the present study can be obtained from the corresponding author upon a reasonable request.
VLK, SK, and IFT—conceptualization of the study, planning of the models’ development and testing; AK—data mining, development and testing of the models including Keras model. All authors made substantive intellectual contributions to this article. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity. All authors read and approved the final manuscript. All authors contributed to editorial changes in the manuscript.
Not applicable.
Not applicable.
This research received no external funding.
The authors declare there are no conflicts of interest. IFT and VLK are president and CEO of BiAna. There is no conflict of interest with the current study.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.