- Academic Editor
†These authors contributed equally.
Background: One of the crucial processes for small RNA synthesis and plant disease resistance is RNA interference (RNAi). Dicer-like (DCL), RNA-dependent RNA polymerase (RDR), double-stranded RNA binding (DRB), and Argonaute are important proteins implicated in RNAi (AGO). Numerous significant woody plants belong to the Juglandaceae; walnut is one of the four groups of woody plants on earth and one of the four groups of dried fruits. Methods: In order to correlate walnuts and their homologues, this work integrated numerous web resources from structural analysis and transcriptome data collected from gene families in order to elucidate the evolution and functional differentiation of RNA-related proteins in the walnut (Juglans rega) genome. Results: 5 DCL genes, 13 RDR genes, 15 DRB genes, and 15 AGO genes are found in the walnut genome and encode conserved protein domains and motifs with similar subcellular distribution.There are three classes and seven subclasses of walnut AGO proteins. RDRS are primarily split into four categories, whereas DRBs can be divided into six. DCLs are separated into four groups. The walnut RDR1 copy number of 9 is the exception, with 7 of those copies being dispersed in clusters on chromosome 16. Proteins are susceptible to various levels of purification selection, but in walnut, purification selection drives gene creation. These findings also indicated some resemblance in other plants belonging to the walnut family. Under various tissues and stresses, many RNA-related genes in walnut produced abundant, selective expression. Conclusions: In this study, the genome of the Juglandaceae’s DCL, RDR, DRB, and AGO gene families were discovered and analysed for the first time. The evolution, structure, and expression characteristics of these families were also preliminary studied, offering a foundation for the development and breeding of the walnut RNAi pathway.
The RNA interference (RNAi) pathway is the main pathway of biological resistance to viral hazards and the central pathway of small RNA formation [1, 2]. Existing studies have shown that RNAi is involved in coping with various abiotic and biotic hazard coordination processes and has an important role in plant growth and development [3, 4, 5, 6]. Dicer-like (DCL), RNA-dependent RNA polymerase (RDR) and Argonaute (AGO) are three classes of proteins that are core components of RNAi. DCL is a protein that enables the conversion of dsRNA to primary siRNA, and secondary siRNA can facilitate the production of more siRNA by retranscribing primary siRNA to synthesize dsRNA, while AGO forms an RNA silencing complex (RIGC) with siRNA, which in turn is involved in the resistance response through RISC. RISC is involved in the resistance response [7]. Second, double-stranded RNA-binding (DRB) can participate in dsRNA cleavage instead of DCL and contribute to AGO protein binding and downstream small RNA formation; therefore, DRB can also be considered an important component in the RNAi mechanism [8]. However, the relatively complex structures of the AGO, DCL, DRB, and RDR proteomes and the abundance of gene copy phenomena have led to difficulties in obtaining homologous genes directly for research. For this reason, RNAi core proteins have been identified in several species, such as hairy poplar (Populus trichocarpa), grape (Vitis vinifera), citrus (Citrus sinensis), tomato (Solanum lycopersicum), and rice (Oryza sativa) [5, 9, 10, 11]. In general, DCL proteins (DEAD box, RNA decapping enzyme structural domain, DUF283, ds RBD, RNase III and PAZ) and AGO (including at least one each of PAZ and PIWI structures) proteins are multiconserved structural class proteins, while RDR has only one RdRP structural domain. PAZ and PIWI structures are associated with the formation of RNA binding pockets, while the ds RBD is a key region for DCL and RNA recognition and binding [12, 13]. In contrast, DRB proteins include one or more DSRM (double-stranded RNA binding motif) structures, a result related to their hydrolytic activity on dsRNA [8]. The expression of DCL, RDR, DRB and AGO genes in a variety of plants is affected by a variety of biotic or abiotic adversities that enhance their expression levels, such as virus infestation, hormonal stress, drought, and low temperature [9, 14, 15, 16]. In addition, multiple basic life activities are also associated with DCL, RDR, DRB and AGO genes, such as methylation [17], fruit drop [5], and male sterility [18].
The multicopy nature of the RDR family is caused by the proliferation of three
ancient copies of RDR
Among them, walnuts are known as one of the four dried fruits, are rich in nutrients, and play an important role in the development of the world’s agricultural economy. However, in the walnut cultivation industry, several adversities seriously affect its high quality and quantity production [23], such as cold damage, diseases, and insect pests. In recent years, a high-quality walnut genome (Juglans regia) has been completely sequenced [24], and the evolutionary study of its RNAi system based on feasible methods is an important part of facilitating the application of model plant research on walnut plants. Second, the process of testing preliminary walnut disease resistance using gene expression will also enhance our understanding of natural resistance to walnut diseases and the functional differentiation of genes [25, 26, 27]. To this end, the DCL, RDR, DRB, and AGO genes possessed by the walnut genome were comprehensively analysed and investigated their structure, evolution, and possible interactions. Second, the analysis of potential RNAi-related expression signatures in walnut using transcriptomic data will enhance our understanding of natural disease resistance and gene functional differentiation in walnut. To this end, the DCL, RDR, DRB, and AGO genes were comprehensively analysed that are present in the walnut genome and investigated their structure, evolution, and possible interactions. Data were used to analyse their gene expression differentiation for RNAi-related genes, and these findings are expected to lay the foundation for the study of walnut breeding.
AGO, DCL, DRB, and RDR genes were identified using the following: firstly, Arabidopsis and tomato homologous proteins by blastp search; secondly. HMMER 3.61 was used to search for marker domains (RDR “RdRP”, AGO “PAZ, PIWI”, DCL “PAZ, dsRBD”, DRB “DSRM”) [5, 28, 29]; and SMART (http://smart.embl-heidelberg.de/) [30] and PFAM (http://pfam.xfam.org/) [31] websites were used to determine the marker structure domain completeness of the marker structure. Finally, manual merging and deduplication were performed.
Conservative motifs, gene structure maps and chromosome localization maps were predicted or drawn using MEME (search motif number 10 for each family, length limit 6–100) and TBtools v.1082 software (Huazhong Agricultural University, Wuhan, China) [32, 33]. In addition, the walnut genome used in this paper was obtained from the Hardwood Genome Database (https://hardwoodgenomics.org/), Arabidopsis from TAIR (https://www.arabidopsis.org/), tomato genome from Solanaceae genome (https://solgenomics.net/), and other genomes from EnsembelPlant (http://plants.ensembl.org/). Other genomes not mentioned in the methods were obtained from 3 databases (http://gigadb.org/, https://ngdc.cncb.ac.cn/, http://www.juglandaceae.net/).
Protein isoelectric points and relative molecular masses were predicted using EXPASy’s Prot-Paramtool online program (http://web.expasy.org/cgi-bin/protparam/protparam) [34]; protein subcellular localization was predicted using Softberry (http://www.softberry.com/).
The websites STRING (https://string-db.org/), plant.map
(http://plants.proteincomplexes.org/), and Cytoscape 3.71 were used to predict
the AGO, RDR, DRB, and DCL interactions between proteins (species primarily
referred to as Arabidopsis, with a screening restriction of score
Tree building analysis was calculated using MEGA 7.0 based on AGO, DCL, DRB and
RDR protein sequences (Clustal W method alignment, posterior value bootstrap =
1000, and neighbour-joining method for tree building, Supplementary Table
1) [6, 38]. The evolutionary analysis based on repetition type was calculated
using MCScan X [39]. The screening principles for tandem duplicate gene pairs are
based on (1) the length of coverage on the pair exceeds 75% and the homology is
75%, and (2) the gene distribution should be close in physical distribution
[40]. Ka/Ks (synonymous substitution/nonsynonymous substitution) was calculated
using TBtools v.1082 (Bell Laboratories, New York City, USA), and differentiation time was calculated using T = Ks/2r (r
= 1.5
Raw sequencing data (SRP034866, SRP290361, SRP359475, SRP343997) in fastq format from SRA (https://www.ncbi.nlm.nih.gov/sra) was used to examine the expression levels of RNAi-associated genes in various walnut tissues and under various stressors [43]. The walnut genome from the Hardwood Genome Database was then compared to high-quality sequenced samples using HISAT2 [44]. Using featureCount [45] and a customized Perl script, raw counts for unique mapped reads were pooled, and transcript per million (TPM) values were generated for each gene. The sorghum upstream sequence (1000 bp before the 5th end of the coding sequence) was selected as the promoter sequence and submitted to the PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [46], and the results were then visualized using TBtools.
Juglandaceae homologous proteins of AGO, DCL and RDR was constructed three phylogenetic trees, it was found that walnut has 15 AGO genes, 5 DCL genes, 15 DRB genes, and 13 RDR genes. The lengths of the AGO and DCL proteins ranged from 900–1100 aa and 1400–2000 aa, respectively, while the lengths of the RDR proteins, except for JrRDR1e (386 aa) and JrRDR1f (630 aa), were approximately 1000 bp. While the DRB protein sizes are mostly distributed at 400–600 aa, some of them are near 200 aa (JrDRB6.1, JrDRB6.2, JrDRB7.2, JrDRB7.3) or near 1000 aa (JrDRB7.1).
In addition, the molecular mass of most of this walnut AGO, RDR, and DCL proteins was 100–200 kDa, with the largest protein being DCL1 (224 kDa) and the smallest being JrRDR1e (44 kDa). In contrast, the molecular weights of walnut DRB proteins are mainly located at 40–60 kDa, with the smallest protein being much smaller than other RNA-related proteins (JrDRB7.3, 18 kDa). The isoelectric point of the walnut AGO protein was between 8 and 9, and the subcellular localization was in the cytoplasm or nucleus, except for JrAGO2a, which was in the extracellular (secretory) region. The isoelectric point of the walnut DCL protein was between 6 and 8, and the subcellular localization was in the nucleus, while the isoelectric point of the walnut RDR protein was larger at between 5 and 9, and the subcellular localization was in the extracellular (secretory) region. The isoelectric points of walnut DRB proteins are mainly between 8 and 10, with a few between 5 and 6 (JrDRB4A, JrDRB7.2, JrDRB7.1), and the subcellular localization is dominated by the nucleus and chloroplasts. Interestingly, the subcellular localization of JrDRB7.4 showed the highest probability of being in the vesicles (Supplementary Table 2).
The evolutionary relationships between walnut and rice, poplar mullein, citrus, grape, tomato, Arabidopsis AGO, RDR, DRB, and DCL proteins are depicted in Fig. 1. To characterize the more likely true protein homology relationships, phylogenetic tree positions were used to name the walnut genes. As shown in Fig. 1A, AGO is mainly divided into three classes (I, II, and III), and walnut AGO proteins are distributed in seven subclasses among these three classes (I: AGO1, AGO5, AGO10; II: AGO2/3, AGO7; III: AGO6, AGO4). The copy number of AGO1, AGO4, and AGO10 proteins is 3, while the copy number of AGO2 and AGO5 proteins is 2, and the copy number of AGO6 and AGO7 proteins is 1. In addition, walnut AGO proteins have the highest homology with grape or citrus AGO proteins among most members. However, specifically, the three proteins of walnut AGO10, JrAGO10a and JrAGO10c formed a direct homologous pair with CsAGO10, while JrAGO10b formed a direct homologous pair with VvAGO10b. Walnut JrAGO6 formed a direct homologous pair with PtAGO16 but not with grape or citrus AGO proteins.
 Fig. 1.
Fig. 1.Phylogenetic tree analysis of AGO, RDR, DRB and DCL proteins. (A) JrAGO protein’s phylogenetic tree. (B) JrDCL protein’s phylogenetic tree. (C) JrRDR protein’s phylogenetic tree. (D) JrDRB protein’s phylogenetic tree. Note: Oryza sativa (Os); Arabidopsis thaliana (At); Solanum lycopersicum (Sl); Vitis vinifera (Vv); Populus trichocarpa (Pt); Citrus sinensis (Cs); Juglans regia (Jr).
Fig. 1B shows that DCLs are divided into four branches, DCL1, DCL2, DCL3, and DCL4, and walnut contains 1, 2, 1, and 1 gene in each of the four types of DCLs. jrDCL2s and jrDCL4 have the highest homology with citrus, followed by grape. JrDCL1 and JrDCL3 had the highest homology with grape, followed by citrus and mullein. Fig. 1C shows that RDR is mainly divided into four branches, RDR1, RDR2, RDR3, and RDR6. Walnut RDR2 and RDR3 are single copies, while the RDR6 copy number is 2, and the RDR1 copy number is 9. Walnut RDR3 has the highest homology with grape RDR3, while walnut RDR1, RDR2, and RDR6 have the highest homology with citrus or burkegonia. Fig. 1D shows that DRB can be clustered in the six published DRB branches (DRB1, DRB2, DRB3-5, DRB4, DRB6, and DRB7) and that DRBs are multicopy in all classes except DRB4, which is a single copy and has copy numbers of 2, 2, 4, 2, and 4, respectively. Walnut and P. trichocarpa and C. sinensis had the highest homology, and proteins from these three species tended to cluster on the same branch. In addition, in some categories, walnut DRB proteins showed a clear genetic gap between two (e.g., JrDRB7.1, JrDRB7.2, and JrDRB7.3 clustered on one node each and did not cluster together).
Among 15 AGO genes, there are 7 structural domains, 5 of which are
shared, while the remaining ArgoMid is dispersed among the class I proteins of
walnut AGO, according to the examination of walnut gene structures. The
gene structure showed that the number of introns of class III genes among the
three classes of AGO was only 3, while the intron structure of class I
and II genes was complex (
 Fig. 2.
Fig. 2.Gene structural analysis of JrAGO, JrDCL, JrRDR and JrDRB from Juglandaceae. (A) JrAGO gene family gene structure. (B) JrDCL gene family gene structure. (C) JrRDR gene family gene structure. (D) JrDRB gene family gene structure.
Collinearity analysis was done to look at the evolutionary history of the AGO, RDR, and DCL genes in walnut. The distribution of the five DCL genes in walnut on chromosomes 5, 6, 7, 8, and 11 is depicted in Fig. 3A. Thirteen RDR genes were distributed on chromosomes 1, 2, 3, 11, and 16, with the exception of RDR1b (Jr01) and RDR1c (Jr02), and nine clusters of RDR genes were arranged in the middle and upper parts of chromosome 16. Fifteen AGO genes were distributed on chromosomes 1, 3, 4, 5, 6, 7, 10, 12, 13, 14, and 15, forming five collinearity pairs. The 14 DRB genes were distributed on chromosomes 1, 3, 4, 5, 6, 8, 9, 10, 11, and 14, while JrDRB6.2 was located on the fragment that could not be loaded onto the chromosome. Two DCL2 genes, 4 DRB genes, and 9 AGO genes formed duplicated gene pairs resulting from the duplication of the fragment. In addition, all these collinearity pairs generated by fragment duplication were formed by strong purifying selection (Fig. 3A and Fig. 4, Supplementary Tables 3,4).
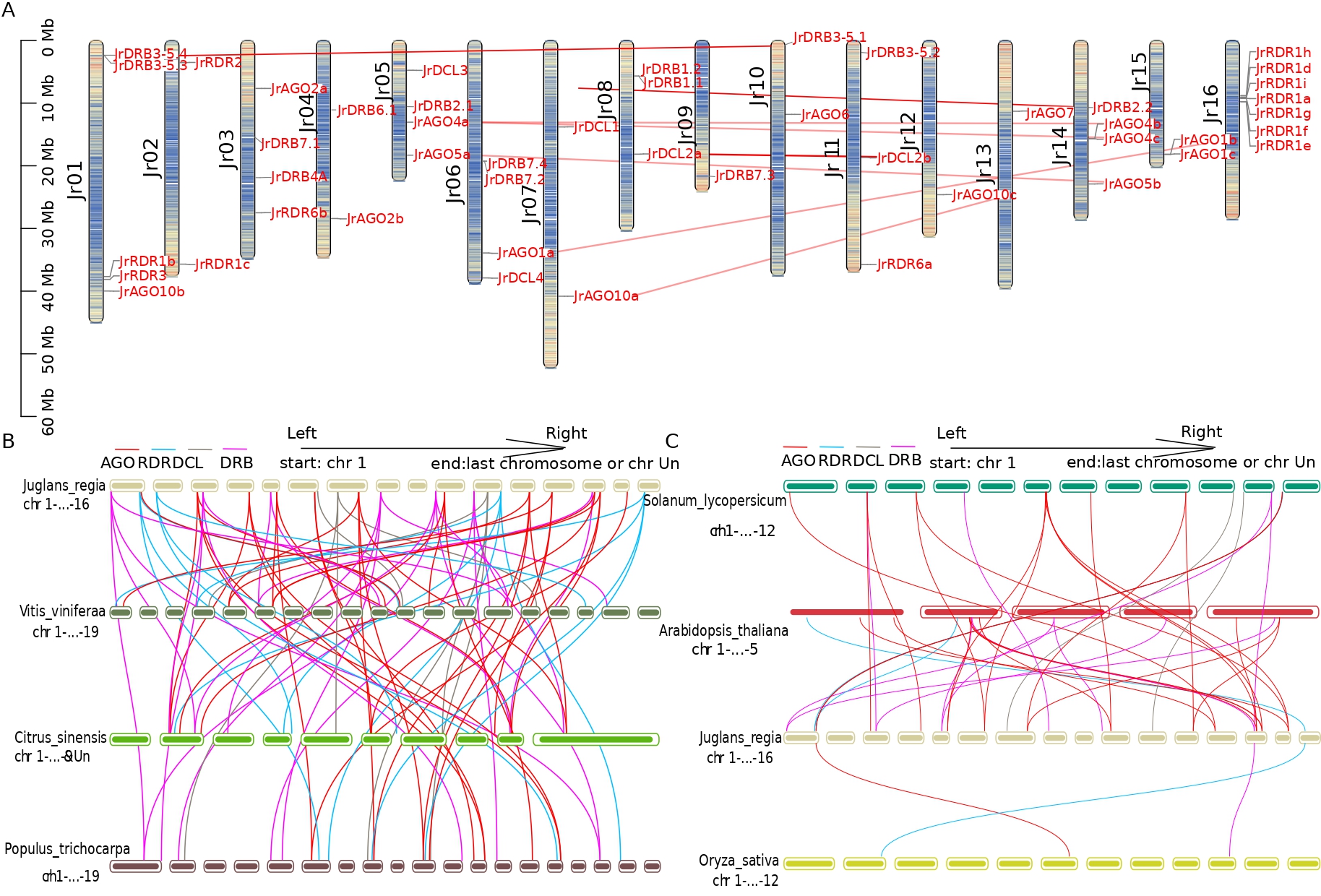 Fig. 3.
Fig. 3.Chromosomal localization and collinearity analysis of walnut JrAGO, JrDCL, JrRDR and JrDRB with different species. (A) The position and collinearity of JrAGO, JrDCL, JrRDR and JrDRB gene families on the chromosome (the red line indicates the composition of the gene pair relationship). (B) Collinearity of JrAGO, JrDCL, JrRDR and JrDRB in walnut, grape, kiwi and poplar (red line indicates AGO gene family, blue line indicates RDR gene family, brown line indicates DCL gene family, purple line indicates DRB gene family). (C) Collinearity of JrAGO, JrDCL, JrRDR and JrDRB in walnut, tomato, Arabidopsis and rice (red line indicates AGO gene family, blue line indicates RDR gene family, brown line indicates DCL gene family, purple the connecting line indicates the DRB gene family). The colour depth of the chromosome in (A) represents the gene distribution density within 10,000 bp. The darker the colour is, the higher the gene density.
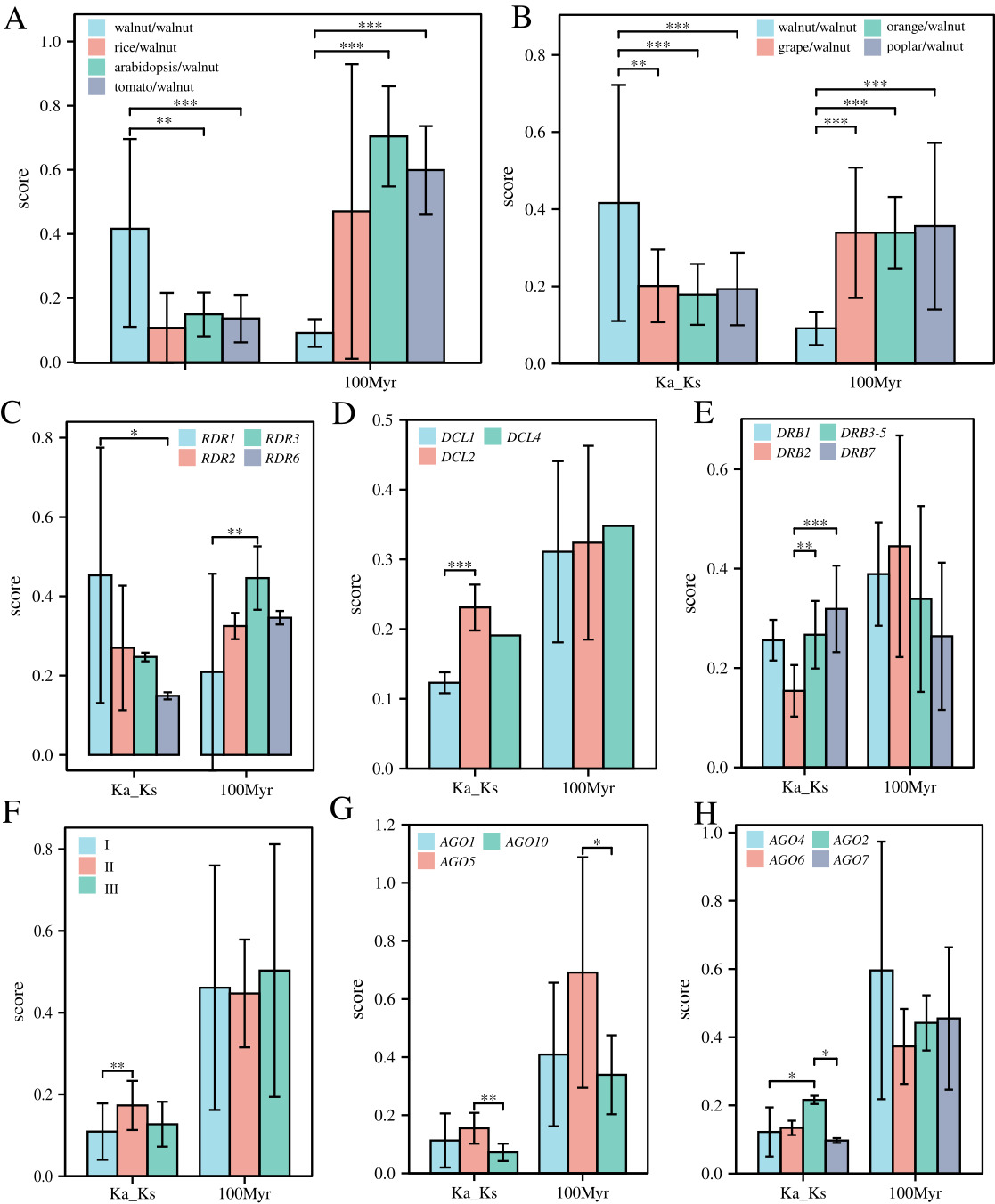 Fig. 4.
Fig. 4.Evolution rate (Ks/Ks) and differentiation time analysis of
collinear gene pairs between AGO, RDR, DRB,
DCL and different species in walnut. Evolution rate (Ks/Ks) and
differentiation time analysis in (A) non-woody plants, (B) non-woody plants, (C)
different RDR, (D) different DCL, (E) different DRB,
(F) different AGO type, (G) different AGO-I type protein, (H)
different AGO-II and III type protein. The significance marker was NS,
p
Colineages were relatively abundant in woody plants (23 pairs in grapes, 24 pairs in citrus, and 32 pairs in burgundy poplar). Grapes are a vine woody plant that formed 23 collinearity pairs with 23 genes of walnuts (including 9, 3, 7, and 4 collinearity pairs of AGO, DCL, DRB, and RDR genes); notably, among them, collinearity pairs of DRB4, DRB6, DCL3, and AGO1 were not found at this stage. Citrus aurantium is a domesticated and important woody fruit tree that forms 24 collinearity pairs with 24 walnut genes (including 11, 2, 7, and 4 collinearity pairs for AGO, DCL, DRB, and RDR genes). Burkholderia is an old woody plant that forms a total of 32 collinearity pairs with 20 walnut genes (including 13, 3, 10, and 6 collinearity pairs for AGO, DCL, DRB, and RDR genes); interestingly, collinearity pairs for AGO1 are accessible in citrus and burkholderia, while collinearity pairs for DRB4, DRB6, and DCL3 are similarly lacking, and covariance with walnut DCL4 could not be found in citrus and woolly poplar (Fig. 3B).
Only three pairs of collinearity (involving three genes, JrRDR1d, JrAGO10b, JrDRB2.2) were found between monocotyledonous rice and dicotyledonous walnut; relatively few collinearity were also found between herbaceous dicotyledonous plants and walnut; 18 pairs of collinearity involved 15 walnut genes (involving AGO10, AGO1, AGO4, AGO5, AGO6, JrAGO7, DRB2, DRB3-5, DRB7, RDR1, RDR3) in Arabidopsis; and 22 pairs of collinearity involved 19 walnut genes (involving AGO10a, AGO1, AGO2, AGO4, AGO5, AGO6, AGO7, DCL1, DCL2, DRB1, DRB2, DRB7, RDR3) in tomato (Fig. 3C).
In addition, all collinearity pairs between species were formed by purifying selection, but the degree of purifying selection differed among walnut DCL, AGO, DRB, and RDR genes. Little difference was noted in the degree of purifying selection and divergence times among the six species, but the internal collinearity pairs of walnut were subject to weaker purifying selection pressure (~2-fold weaker) and diverged much later (~10 Myr) than the divergence of other genes among the species (30–70 Myr) (Fig. 4A,B). RDR1 (~20 Myr) diverged later than RDR3 (~45 Myr). DCL2s (~0.27) were subjected to much less purifying selection pressure than DCL1 (~0.14) genes, while DRB2 (~0.139) was subjected to much more purifying selection pressure than DRB3-5 (~0.257) and DRB7 (~0.307) genes and much higher selection pressure for purification (Fig. 4C–E). For the AGO gene family, AGO-I (~0.079) was subjected to much higher purification selection pressure than the AGO-II class (~0.205); specifically, AGO genes AGO2s were subjected to much lower purification selection pressure than AGO4s and AGO7s, while AGO5s were subjected to much lower purification selection pressure than AGO10s and diverged earlier (Fig. 4F–H).
As the Fig. 5 shown, tandem repeats have an important role in the copy duplication of RNAi-related families in walnut, where it was found 21 possessing possible tandem repeat relationships, which consisted of 1 JrRBD7.4-JrRBD7.2 relationship and 2 AGO1, AGO4, DRB1, DRB3-5 and 6 RDR1 (Fig. 5A, Supplementary Tables 3,4). Moreover, the relationship is composed of genes on 6 chromosomes, independent of the other 10 chromosomes. It is particularly interesting that all 2 AGO genes of chromosome 15 form tandem repeats first. Further analysis revealed that these gene pairs included 3 pairs of genes with very short evolutionary times (Ka/Ks = 0, JrDRB3-5.4-JrDRB3-5.3, JrDRB1.2-JrDRB1.1, JrAGO4b-JrAGO4c) and 1 pair of positively selected genes (JrRDR1f- JrRDR1e), and the selection pressure of the remaining genes was purified selection. In addition, the distribution time of AGO1 is approximately 7.6 Myr (JrAGO1b-JrAGO1c), while the differentiation time of DRB7 is approximately 0.9 Myr (JrDRB7.4-JrDRB7.2) and that of RDR1 is approximately 1.5–11 Myr. Uniquely, the divergence time of the JrRDR1f- JrRDR1e gene pair subjected to positive selection pressure was extremely close at only approximately 0.13 Myr (Fig. 5B,C).
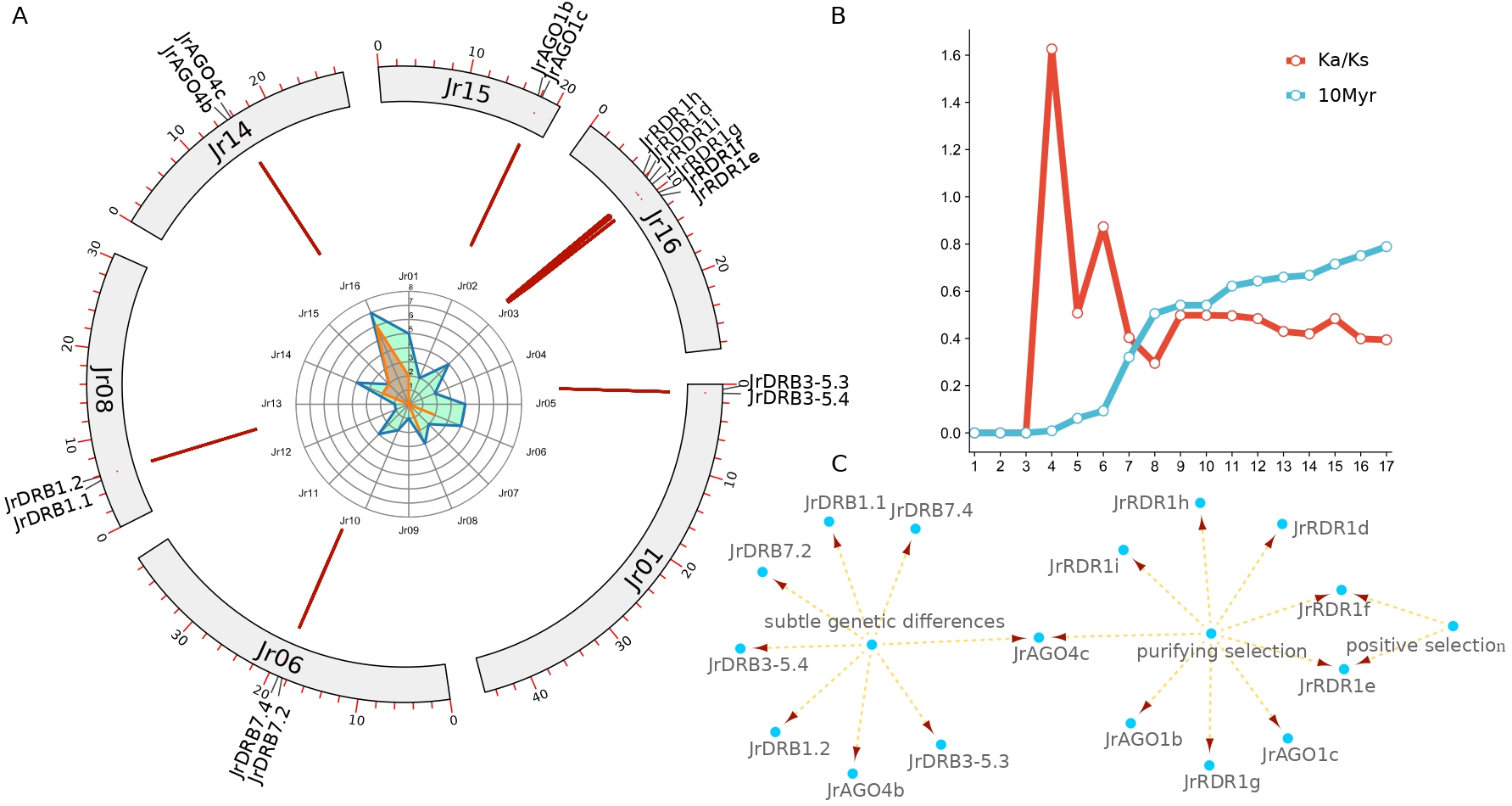 Fig. 5.
Fig. 5.Detection of tandem repeats in walnut AGO, RDR, DRB and DCL. (A) Distribution characteristics of tandem repeat gene pairs. (B) Evolutionary rate (Ks/Ks) and differentiation time distribution. (C) Selection pressure analysis of different genes.
The collinearity between walnut RNAi-related families and two other important dried fruits, and 24 pairs of collinearity were found between walnut and two different dried fruits (Fig. 6). The collinearity between walnut and filbert involved 22 walnut genes, while the collinearity with apricot involved 24 different walnut genes (Fig. 6). Interestingly, 17 genes could be detected in both collinearities, including AGO, DCL, DRB, and RDR genes in the numbers 9, 1, 4, and 3 (Fig. 6A). After careful examination, it was found that the genomes of apricot and walnut had less AGO2 and DRB1 and more DCL2-4 than filbert, while DRB4 and DRB6 did not show collinearity in either dried fruit. In addition, the purifying selection pressure of walnut internal genes was less than the interspecific purifying selection pressure of different dried fruit plants, while the purifying selection pressure of AGO genes was also greater than that of DRB and RDR; additionally, there was no significant difference in selection pressure of single subtle different gene classes (Fig. 6B–D). In addition, gene pairs within walnut diverged later than those within apricot and even later than those within filbert; however, there was no significant difference in divergence time between gene classes (Fig. 6B–D).
 Fig. 6.
Fig. 6.Intraspecific collinearity of AGO, RDR, DRB and DCL in Prunus dulcis. (A) JrAGO, JrDCL, JrRDR, and JrDRB collinearity between Juglans regia and Corylus avellana and Prunus dulcis (red line indicates AGO gene family, blue line indicates RDR gene family, brown line indicates DCL gene family, purple line Indicates the DEB gene family). (B) The syntenic gene pairs Ka/Ks and differentiation time of AGO, JrDCL, JrRDR and JrDRB gene families in different species (light blue in the violin diagram indicates walnut, pink indicates apricot, and light green indicates hazelnut; “*” indicates that there is a difference, “**” means significant difference, “***” means extremely significant difference). (C) Collinear gene pairs Ka/Ks and differentiation time of JrAGO, JrDCL, JrRDR and JrDRB gene families in the species (light blue in the violin plot indicates AGO collinear genes, pink indicates DCL collinear genes, light green indicates RDR colinear genes Linear genes, purple means DRB collinear genes; “**” means significant difference, “***” means extremely significant difference). (D) Collinear gene pair Ka/Ks and differentiation time among the three AGOs in the species type (light blue in the violin plot indicates type I AGO, pink indicates type II AGO, and light green indicates type III AGO).
In this section, the potential evolution of RNA-associated proteins was parsed in Juglandaceae. It was first analysed the three accessible species of the proximate genus (Pterocarya stenoptera, Carya illinoinensis, C. cathayensis), six congeneric species (J. mandshurica, J. cathayensis, J. hindsii, J. microcarpa, J. nigra, J. sigillata) and one hybrid (J. microcarpa x regia) to identify RNA-associated proteins in their genomes. The results showed that we identified 181, 40, 105, and 67 possible coding genes for RDR, DCL, AGO, and DRB. Our preliminary analysis of their protein physicochemical properties showed that RDR, DCL, AGO, and DRB of all Juglandaceae maintained large molecular weights overall, with isoelectric points mainly distributed approximately at 5–8 (Supplementary Table 5).
When these possible proteins were examined, the presence of 2 RdRP domains was noted in a small number of RDR proteins, while the presence of DSRM structural domains in DRB proteins was widely distributed in different numbers (one, two, three, four). In contrast, in DCL and AGO, abundant structural domains are present, while a considerable abundance of structural domains are missing. For example, 68 proteins possess the distribution characteristics of Piwi, PAZ, N-terminal, L1, L2, and Mid structural domains, while in 24 proteins, Mid structural domains are missing (Supplementary Fig. 1).
Furthermore, a phylogenetic tree of each RNAi-related protein class was constructed from 17 species. The results showed that, on the premise that the protein clusters of each subclass were correctly separated, for AGO, RDR, DRB, and DCL proteins, the proteins of the pecan family were individually separated from those of other plants to form a unique branch. Notably, the pecan family appears to be younger than the RNAi-associated proteins of rice, grape, Arabidopsis, and six other species, as their branches are all at the end of each class of proteins rather than between proteins of several species (Fig. 7A–D).
 Fig. 7.
Fig. 7.Phylogenetic analysis of the same species of JrAGO, JrDCL, JrRDR and JrDRB. (A) The phylogenetic relationship of RDR in Juglandaceae. (B) Phylogenetic relationship of DRB in Juglandaceae. (C) Phylogenetic relationship of DCL in Juglandaceae. (D) Phylogenetic relationship of AGO in Juglandaceae. (E) AGO, DCL, RDR and DRB in 10 species of Juglandaceae and grape, rice, poplar, tomato, Arabidopsis and kiwifruit.
To correctly interpret the evolution of RNAi-associated proteins in the peccary family, the quantitative differences among protein subclasses were compared and showed that all regular protein taxa could be found for almost all species (DCL: DCL1, DCL2, DCL3, DCL4; RDR: RDR1, RDR2, RDR3/4/5, RDR6; DRB: DRB1, DRB2, DRB3-5, DRB6, DRB7; AGO: AGO1, AGO2/3, AGO4, AGO5, AGO6, AGO7, AGO10). The genomes of some of these species appear to be amplified for many gene taxa, such as RDR1, AGO5, DRB3-5, RDR6, and DCL2. More typically, the Walnut family seems to have a consistent expansion for RDR1, with a large number of RDR1s found in various Walnut species (Fig. 7E).
Meanwhile, collinearity analysis in walnut species was performed at the complete
chromosome level and measured the selection pressure and divergence time of
collinearity pairs. The results showed that there were 42 and 46 collinearity
pairs in the genomes of walnut, iron walnut and Mandshurian walnut, which
involved 29 and 33 walnut genes, respectively. In addition, 47 collinearity pairs
involving 33 walnut genes existed between walnut and hybrid walnut (J.
microcarpa x regia). A closer examination revealed that there were significant
differences in the divergence times between the genes within walnut and the
collinearity pairs of J. microcarpa, and in addition, there were
significant differences in the relative divergence times of the relationships
between walnut in J. microcarpa and hybrid walnut. In addition, the
RDR gene had significantly less selection pressure for purification
between these different walnut species than the AGO gene, but the
difference in selection pressure between subclasses was not significant.
Differentiation times and selection pressures for all the same protein subclasses
with walnuts showed significant differences between species and genes in
different classes, where the differences in purification selection pressures
between gene classes were AGO
Using promoter elements as a starting point, a preliminary analysis was carried out to investigate the functional differentiation of walnut RNAi-associated genes. The findings demonstrated the presence of numerous tissue-specific and adversity-related cis-regulatory elements in RNAi-related genes, which included many tissue-specific elements (e.g., seed-specific regulation: RY-element; meristem expression: CAT-box; endosperm expression: GCN4_motif) and adversity-related elements (e.g., defense and stress responsiveness: TC-rich repeats; low-temperature (Supplementary Fig. 3 and Supplementary Table 6). This implies that genes connected to walnut RNAi might be involved in tissue-specific expression and stress response.
Furthermore, the expression was analysed from transcriptome data of walnut RNAi-related genes. The findings indicated that RNAi-related genes had varying degrees of tissue specificity in various tissues, but the changes in gene expression levels were more notable, with 21 genes having greater expression levels and 27 genes having lower expression levels. Among them, the same subclasses of proteins did not have similar expression profiles, such as AGO10 (AGO10c showed high expression in most tissues, while expression was significantly downregulated in mature leaf-jun08 and dehiscing hull, and AGO10a and AGO10b were mostly low and upregulated in some tissues) and RDR1 (RDR1a showed high expression in most tissues, while expression was significantly downregulated in mature embryo, and the other eight RDR1 genes, i.e., RDR1b-i, were mostly expressed at low levels) (Supplementary Table 7 and Fig. 8). In addition, their potential stress responses (including the N response and heat stress response) were examined. The expression of 16 RNAi-related genes was highest in the root fraction at different N supply concentrations, while the expression of 14 RNAi-related genes remained low and the expression levels of the remaining 18 genes were essentially in the middle. Notably, the expression levels of JrDRB7.1, JrDCL2b, and JrRDR1i increased with the change in N supply concentration; in particular, the expression levels of JrRDR2 and JrRDR1h seemed to be significantly correlated with an increase in N concentration. In addition, RNAi-related genes were mostly heat-treated, resulting in restricted expression. Among them, approximately 60% of RNAi-related genes were highly expressed at 0 h of heat treatment, while they were approximately 30% higher after 2 h and 6 h of heat treatment. Interestingly, the expression of AGO2 (JrAGO2a and JrAGO2b) proteins was induced by heat treatment (Supplementary Fig. 4).
 Fig. 8.
Fig. 8.Heatmap of tissue-specific expression and abiotic stress expression of walnut RNAi-related genes. Note: The colour bar represents the log2 expression level of each gene (TPM, transcripts per million). Colour bar annotation is included at the top of the image.
Due to the previous importance of RNAi-related genes for disease, it was
analysed transcriptome data based on walnut induced by Colletotrichum
gloeosporioides [27]. The results showed that most of the walnut RNAi-related
genes showed significant upregulation of expression compared to uninfested walnut
leaves (fold change
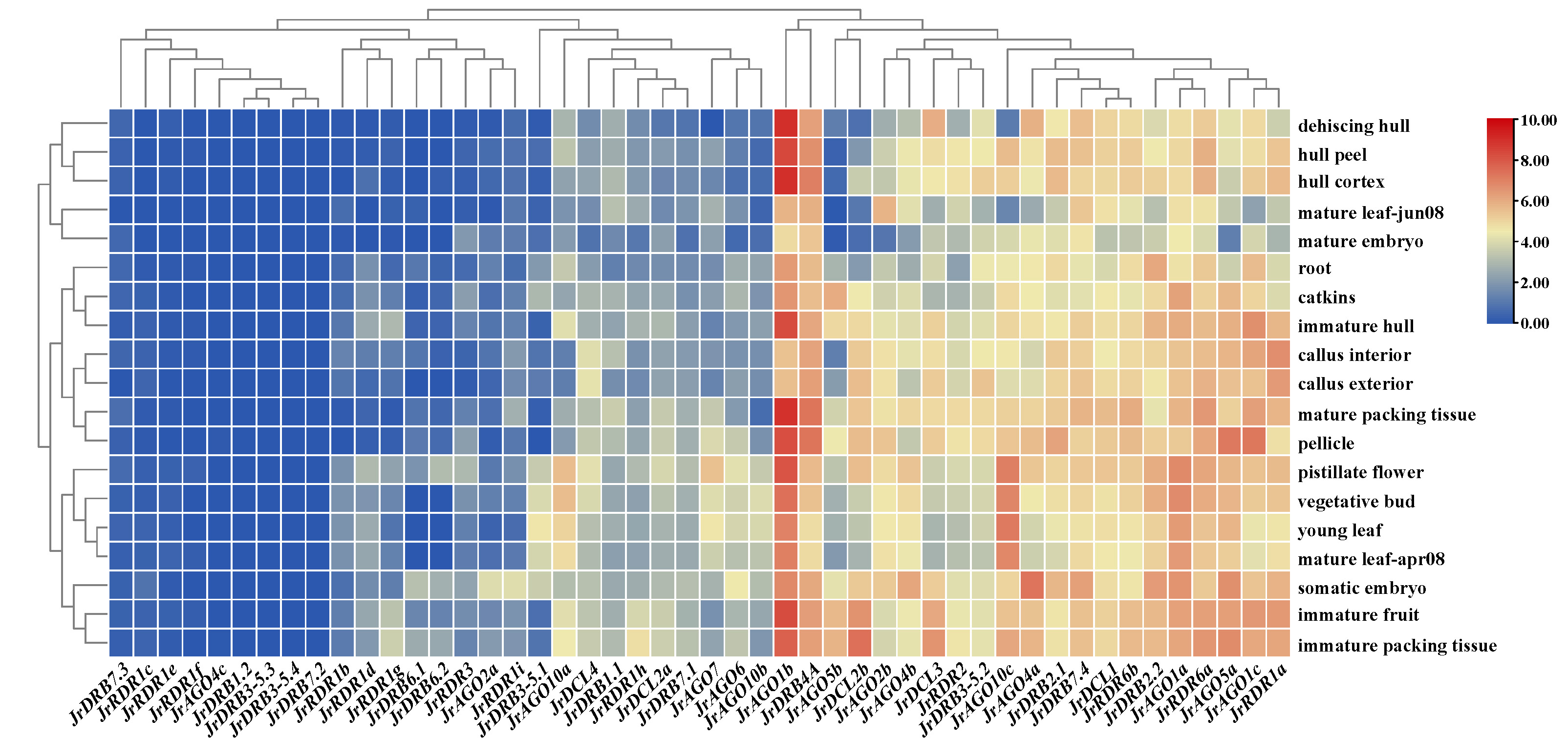 Fig. 9.
Fig. 9.Analysis of the expression pattern of JrAGO, JrDCL, JrRDR and JrDRB after infenction with Colletotrichum gloeosporioides. ‘4-23’ and ‘B73’ are plant material of Juglandaceae. The colour bar represents the log2 expression level of each gene (TPM, transcripts per million). Colour bar annotation is included at the top of the image. The heatmap is coloured according to expression values, with blue, yellow and red representing low, medium and high transcription abundance, respectively.
In recent years, sequencing of many genomes of Juglandaceae plants has been completed [47]. Evolutionary studies of their RNAi systems based on feasible methods are an important component to facilitate their production applications and help to preliminarily resolve natural disease resistance and gene functional differentiation in walnuts. For this purpose, the DCL, RDR, DRB, and AGO genes present in the walnut genome were comprehensively analysed and studied their structure, evolution.
DCL genes can generally be classified as DCL1, DCL2, DCL3, and DCL4, and the copy number is generally no more than three [5, 14, 48, 49, 50, 51, 52, 53]. The walnut DCL gene also includes the above features, and only DCL2 produces two copies of the walnut DCL gene. In fact, DCL2 and DCL3 may be copied in the DCL family, including multicopy DCL2 (DCL2a and DCL2b) reported from poplar, rice and maize, and cotton plants [53]. Meanwhile, we also have this multicopy feature in the walnut family, even up to four copies in Juglans nigra. In addition, DCL2 duplication may be associated with independent evolution of monocotyledons and true dicotyledons after differentiation [54]; in the collinearity feature section of the text, the collinearity of the JrDCL2b gene was found only in non-herbaceous plants in dicotyledons, suggesting that the evolutionary process of this gene may depend on the evolution of plant stem organs. Second, previous results suggest that DCL3 replication may be a unique event in eudicots [53], but we suggest that DCL3 replication may be heavily dependent on the formation of some species-specific branches rather than a universal replication event in eudicots, since the Juglandaceae explored here are all prepared with only a single DCL3.
A total of six RDR genes have been identified in Arabidopsis, but in more studies, it has been shown that RDR4 and RDR5 may be copies of RDR3, thus delineating the four categories of RDR1, RDR2, RDR3, and RDR6. Walnut RDR genes contain four classes of RDR1, RDR2, RDR3, and RDR6 with a total of 13, of which RDR6 copy number is 2, while RDR2 and RDR3 are single copies, which is similar to the structure in most studies. However, the RDR1 copy number of 9 is unique, seven of which are arranged in clusters on chromosome 16, which is much more than the one to three in the rest of the species [5, 14, 25, 28, 29, 55]. Interestingly, aberrant expansions of RDR have long been noted in polyploid species and wild species, but it appears that there are clear differences in the evolutionary patterns of RDR in different taxa of species [56]. For example, 3 and 5 copies of RDR3/4/5 are present in Arabidopsis and European oilseed rape, while the massive expansion of RDR1 in the walnut genome does not seem to be reflected in others, and the massive expansion of RDR1 is present in a variety of Juglandaceae species.
The AGO gene family is a typical multicopy family classified into three classes and 11 subfamilies (I: AGO1, AGO5, AGO10; II: AGO2, AGO3, AGO7; III: OsAGO15, AGO9, AGO8, AGO6, AGO4). Walnut AGO genes were distributed in seven subclasses of the three classes (I: AGO1, AGO5, AGO10; II: AGO2/3, AGO7; III: AGO6, AGO4) with copy numbers of 1 to 3. These results were similar to most species [5, 6, 31, 39, 57, 58, 59, 60]. Interestingly, while DCL and RDR have produced a large number of expansions unique to Juglandaceae, there has not been a large expansion of genes in AGO, where the number of walnuts is 15 and the number of other Juglandaceae plants is mostly also in the range of 12–17.
DRB mainly consists of four subclasses (DRB1, DRB2-3-5, DRB4, DRB6), where DRB2 and DRB7 are sometimes considered independent branches [22]. However, no matter how to slice the DRB, the DRB gene of walnut is twofold expanded among various subbranches compared to Arabidopsis. Meanwhile, substantial expansion of DRB1 and DRB3 remained in other walnut plants, but other branches were lost and selectively expanded.
Walnut has seven core and two variable structural domains, of which PAZ confers DCL binding ability to RNA, and RNase III (Ribonuclease_3), Helicase_C, DEAD is the endonuclease active domain of DCL enzymes, which enhance DCL’s ability to deconvolve double-stranded molecules [60, 61]. In contrast, the DSRM and DND1_DSRM structural domains are only distributed within the JrDCL1 and JrDCL4 proteins, which may lead to enhanced RNA binding capacity of DCL1 and DCL4, resulting in functional differences with DCL2s and DCL3. Walnut RDR enzymes exist only in an RdRP structural domain of variable length, which is the core region where RDR exercises its function [57, 62].
Walnut AGO proteins possess seven structural domains, five of which are shared, and the remaining ArgoMid is distributed in the class I proteins of walnut AGO. Gly-rich_Ago1 is only distributed in walnut AGO1s protein, and PAZ and PIWI are key to the formation of the core active region of AGO, and the simultaneous presence of ArgoN, ArgoMid, and PIWI would be more helpful for the formation of AGO functional regions. Thus, these specifically distributed structures may lead to functional differences in different nucleolar AGO proteins [14, 17, 18, 63]. DRB proteins include one or more DSRM (double-stranded RNA binding motif) structures, a result related to their hydrolytic activity on dsRNA [8]. Walnut DRB enzymes are present in one or more DSRM structural domains, which are the key regions that guarantee their hydrolytic activity on dsRNA [57, 62]. Among them, walnuts possess 27%, 66%, and 5% of the proteins possessing one, two, or three DSRM structures, respectively. Meanwhile, other walnut family plants possessing 1–4 DSRM structure proteins are abundantly present, but the proteins with 2 DSRM structures remain the most abundant. Second, the Methyltransf_11 structure was also found in the JrDRB7.1 protein, a result that may have a unique role.
A range of sRNAs, including miRNA, trans siRNA (ta-siRNA), natural antisense transcript-siRNA (nat-siRNA), and viral-derived siRNA (vsiRNA), are involved in specialized RNA silencing to control invasive nucleic acids from endogenous (mostly transposon) or external (primarily viral) sources [7]. These four main components interacted with one another often in the current study’s rich PPI network analysis, which may have mediated the many RNA silencing pathways in which they may have been engaged. Interestingly, walnut exhibits a large increase of some RNAi-related genes that interact favorably with members of other families.
The RNAi pathway effectively mediates plant adaptation to the outside world, and the small RNAs produced are used to further regulate plant resistance [48, 64]. Available evidence suggests that the RNAi pathway is mainly accomplished by three classes of proteins, DCL, RDR, and AGO, with DCL2, DCL4, RDR2, RDR6, AGO1, AGO2, AGO3, AGO4, AGO5, AGO7, AGO10, and AGO18 being the most important for plant RNAi resistance effects, while other DCL1 and DCL3 led by proteins appear to exist to mediate small RNA formation as well as enrich neofunctionalization [7, 50, 51, 65]. Second, DRB proteins can also effectively complement many aspects of the system [22].
Various gene families may often be somewhat selective for the type of tissue, as is the case for RNAi-associated gene families [5, 26, 56, 66]. At the same time, these genes that produce tissue specificity can often also produce important effects on specific tissue life activities or tissue resistance. Walnut RNAi-associated genes show unique tissue specificity for different tissues. Moreover, the same protein subclasses do not have similar expression profiles, such as AGO10 and RDR1. This finding greatly reflects the unique functional differentiation of RNAi and its potential resistance to pathogens (infected tissues may be richer in resistance genes).
Indeed, many different gene families in many species have produced rich functional differentiation, leading to a large functional expansion of enriched genes, e.g., MADS and cytochrome P450 [67, 68]. Due to the unique tissue expression of RNAi genes and the low tissue expression of a small number of genes, we speculate that stress may induce differential expression of these RNAi-related genes. In fact, heat stress and nitrogen stress are two completely different types of stresses (environmental factor forcing and nutritional stress). The results showed that the expression of 16 RNAi-related genes was highest in the root fraction, and the expression of JrDRB7.1, JrDCL2b, and JrRDR1i increased with N supply concentration. Most of the RNAi-related genes were heat-treated, resulting in restricted expression, although a small number of proteins, including AGO2 (JrAGO2a and JrAGO2b), could be upregulated by heat treatment-induced upregulated expression. This suggests that the RNAi-associated gene family produced significant functional differentiation, and in particular, some taxa of proteins may develop unique response characteristics to different tissues or stresses.
Plants are shielded from viral infection by RNA silencing [69]. This antiviral
immunity involves the creation of virus-derived tiny interfering RNA (viRNA),
which results in effector complexes that silence certain viruses.
Verticillium dahliae, Fusarium virguliforme,
Magnaporthe oryzae, Verticillium longisporum [70, 71, 72, 73, 74],
and Botrytis cinerea are examples of fungal infections that have been
demonstrated to differentially express plant miRNAs when inoculated into plants
[75]. Most significantly, two Verticillium species and other major RNA silencing
machinery mutants show altered sensitivity to fungal infections [70, 76].
Moreover, it has been noted that Botrytis cinerea suppresses plant defenses by
stealing the host’s RNA interference pathway and producing short RNAs [77]. To
better understand how these factors contribute to biotic stress, transcriptome
data from walnut exposed to C. gloeosporioides revealed that the
majority of these genes’ expression levels were dramatically increased (fold
change
In summary, walnut AGO, DCL, DRB and RDR gene families were identified for the first time, completed an analysis of their sequence characteristics, evolutionary features (within species, within genus, within family, between woody species, within dicotyledons, between different orders at multiple levels, respectively), and finally, characterized the AGO, DCL, DRB and RDR genes based on transcriptome. Finally, based on the transcriptome, the potential functional was also characterized from differentiation of AGO, DCL, DRB and RDR genes These findings are expected to lay the foundation for the study of breeding research in walnut and have reference value for the evolutionary study of plant resistance.
All data included in this study are available upon request by contact with the corresponding author.
YL and YF are the designers and executors of this paper, YF completed the data analysis and wrote the first draft of the paper; YF, JW and XH helped in analyses; XP design, discuss with us, read it and polish it; All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
Many thanks for help from Associate professor Zhibo Zhao, Wenzhuo Feng, Xiangru Chen, Rong Fan, Zhaifu Yang, Fenghua Tian, Kaihuai Li, Professor Xin Xie, Yong Wang.
This work was supported by Guizhou Provincial Science and Technology Projects (Qian Ke He Ji Chu-ZK [2022] General 071), the National Natural Science Foundation of China (32060673), Guizhou Provincial Key Technology R&D Program (No.qiankehezhicheng[2020]1Y131), and projects of Guizhou University (No. Gui Da Pei Yu[2019]52) co-funded.
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
