1 Center for Drug Safety Evaluation, Hangzhou Medical College, 310013 Hangzhou, Zhejiang, China
2 Medical School of Jinhua Polytechnic, 321016 Jinhua, Zhejiang, China
3 Department of Hepatology and Infection, Sir Run Run Shaw Hospital, Affiliated with School of Medicine, Zhejiang University, 310020 Hangzhou, Zhejiang, China
4 High Level Bio-safety Laboratory, Hangzhou Medical College, 310013 Hangzhou, Zhejiang, China
Abstract
Background: Non-alcoholic fatty liver disease (NAFLD) is a popular
chronic liver disorder with high morbidity and with no approved therapeutic
drugs. Fibrosis is a crucial drug efficacy indicator for NAFLD. Thus,
investigating the mechanisms of NAFLD-associated fibrosis and exploring effective
therapeutic targets is imperative. Methods: Gerbil NAFLD-associated
fibrosis model was constructed by feeding a high-fat and high-cholesterol diet.
The hematoxylin and eosin staining and the alanine transaminase (ALT) and
aspartate transaminase (AST) assays were used to determine liver tissue injury.
Masson staining and hydroxyproline (Hyp) level determination were used to assess
liver fibrosis. High-throughput mRNA sequencing was used to screen differentially
expressed genes in the NAFLD-associated fibrosis model. Cell Counting Kit-8 was
utilized to test cell viability. Results: Liver injury and fibrosis were
observed in the gerbil NAFLD-associated fibrosis model with increased ALT, AST,
and Hyp levels. The screened differentially expressed genes were mainly enriched
in “negative regulation of hemopoiesis”, “response to interleukin-1”, and
“granulocyte migration”. Zinc Finger and BTB Domain Containing
14 (Zbtb14) was upregulated in liver tissues of the gerbil NAFLD-associated
fibrosis model, patients with liver fibrosis, and hepatic stellate cells (HSCs).
Additionally, Zbtb14 regulated primary HSCs activation via the
Keywords
- non-alcoholic fatty liver disease
- fibrosis
- hepatic stellate cells
- Zbtb14
- β-catenin
Non-alcoholic fatty liver disease (NAFLD) is a chronic liver disorder with global prevalence [1]. Globally, 25% adults and 90% obese populations suffer from NAFLD [1, 2, 3]. Therefore, NAFLD is a huge threat to public health. NAFLD is a metabolic disease that presents as lipid deposition in liver cells but without alcohol abuse [4]. Currently, lifestyle change, including proper diet plans and exercises, is the most basic therapy strategy for patients with NAFLD [5]. However, most patients cannot complete the designed plan, resulting in less than satisfactory treatment outcomes [5]. At present, NAFLD has no approved pharmacological treatments [6]. NAFLD will develop into steatohepatitis, cirrhosis, and even cancer if patients with NAFLD delay treatment [5, 7]. Hence, investigating the potential drugs is important to prevent NAFLD.
Liver fibrosis, which is characterized by extracellular matrix deposits, results from advanced liver damage and is intimately linked to cirrhosis and liver cancer [8]. One-third of patients with NAFLD develop liver fibrosis in 4–5 years [9]. Liver fibrosis deterioration is an important NAFLD outcome indicator [10, 11]. Thus, liver fibrosis amelioration is an important indicator of the efficacy of drugs for NAFLD therapy. Hence, research into molecular mechanisms and the search for targets and drugs to alleviate liver fibrosis is important for treating NAFLD.
Hepatic stellate cells (HSCs) are the main matrix-secreting cells and exert
critical function in liver fibrosis development [12, 13]. HSCs are activated
after liver damage, making the cells acquire proliferative and contractile
characteristics, thereby expressing alpha-smooth muscle actin (
In this study, we constructed the gerbil NAFLD-associated fibrosis model, screened differentially expressed genes in this model, and investigated the role of a significantly upregulated gene in NAFLD-associated fibrosis and its possible mechanism.
The Experimental Animal Center of Zhejiang Academy of Medical Sciences (Hangzhou, China) offered 24 male 90-day-old Mongolian gerbils (Merionesunguiculatus). The animal experiments were approved by the local Ethics Committee of the Zhejiang Academy of Medical Sciences (approved number 2019-056) and were conducted the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Gerbils were allocated into the normal (n = 6) and model groups (n = 18). The normal group received a normal diet for 16 weeks. The model group received the HFD for 8, 12, and 16 weeks (n = 6 per group). The HFD recipes for inducing NAFLD and fibrosis models are referred to in our previously reported paper [17]. The HFD consists of 80.3% of ordinary feed, 10% of egg yolk powder, 7% lard oil, 2.5% cholesterol, and 0.2% cholate.
The model group was sacrificed for dynamic mechanical studies at 8, 12, and 16
weeks. The normal group was spared at week 16. Abdominal aorta blood was
harvested [18], and serums were prepared using centrifugation (3500
Hematoxylin and eosin, Masson’s trichrome, and Sirius red staining were performed on liver tissues for the histopathological study using the microscope (Leica DM2500, Wetzlar, Germany). A single-blinded pathologist evaluated liver damage, inflammation, and fibrosis, following the scoring criteria described in a previous study [19]. Ten random areas were obtained from each liver slice.
The serum contents of ALT (Cat No. BC1555), AST (Cat No. BC1565; all from Beijing Solarbio Science & Technology Co., Ltd, Beijing, China), and Hyp (Cat No. A030-1; Jiancheng Biotech. Sci. Inc., Nanjing, China) were determined by Commercial kits following the provider’s instructions.
High-throughput mRNA sequencing was accomplished in LC-BIO Technologies (Hangzhou, China) Co., Ltd. Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used to isolate RNA samples. A 2100 Bioanalyzer was used for quantification, and RNA 6000 Nano LabChip Kit (Agilent, Santa Clara, CA, USA) was used for purification. Afterward, short fragments were prepared from the purified mRNA. Then, the mRNA-Seq sample preparation kit (Illumina, San Diego, CA, USA) was applied to construct the cDNA library. Sequencing was conducted on the Hiseq4000 platform by the 150PE strategy. The cleaned reads were assembled de novo by Trinity version 2.4.0 (Broad Institute, Cambridge, MA, USA). The transcripts were clustered, and the unigenes were searched against the Gene Ontology (GO) database.
The DEGs between the normal and model groups at 16 weeks were screened using
Limma (3.32.5) with the criteria, p
Primary HSCs were harvested from gerbil livers treated with or without the HFD
for 16 weeks following previously reported protocols [20]. The livers were
digested with 1 mg/mL of pronase, 0.4 mg/mL of collagenase IV, and 0.2 mg/mL of
DNase I at 37 ℃ for 30 min. Then, the tissues were made into pieces and digested
again for 15 min. Subsequently, Dulbecco’s Modified Eagle Medium (DMEM) with 10%
fetal bovine serum (FBS) (HyClone, Logan, UT, USA) was used to finish the
digestion. The digestive tissues were filtered, and the filtrate was centrifuged
and removed from the supernatant. Afterward, cells were resuspended in DMEM, and
39.5% of percoll solution (Solarbio, Beijing, China) was mixed into the cells to
separate HSCs. The HSCs were cultured using DMEM plus 10% FBS with or without 10
µM of XAV-939 (Selleck Chemicals, Shanghai, China), which is a
Wnt/
Small interfering RNAs (siRNAs) were used for gene silencing. Two Zbtb14 siRNAs
(siZbtb14-1: 5
Primary HSCs were maintained in a 96-well plate and treated with 10 µL of Cell Counting Kit-8 (CP002; Signalway Antibody, Greenbelt, MA, USA) for 60 min. OD450 nm was observed.
The contents of types I (Cat No. CSB E08083m) and III collagen (Cat No. CSB E07925m) in cell supernatant were tested by Commercial kits (CUSABIO, Houston, TX, USA) following the stated protocols by the supplier.
A total of 5 µg of RNA samples were harvested by TRIzol and used for
producing cDNAs utilizing the Hifair® Ⅱ 1st Strand cDNA Synthesis
SuperMix for qPCR (gDNA digester plus) (Cat No. 11123ES60, Yesen, Shanghai,
China). The SYBR®Green reagent (Thermo Fisher Scientific, Waltham, MA, USA) was
applied to ABI PRISM 7300 RT-PCR system (Applied Biosystems, Hammonton, NJ, USA) to complete the qPCR
assay. The relative expression was obtained using the
2
RIPA reagent (Sigma-Aldrich, St. Louis, MO, USA) was used to collect protein samples, and the
bicinchoninic acid assay method was used to determine the concentration
measurement. Then, 25 µg of proteins were run on the SDS-PAGE, blotted to
PVDF membranes, and probed to primary and secondary antibodies. Antibodies
included anti-ZBTB14 (Santa Cruz, Santa Cruz, CA, USA; sc-514298), anti-
Liver tissue samples were obtained from 36 patients with liver fibrosis (Mild, n
= 18, 44.4% male; Severe, n = 18, 44.4% male) who received liver biopsies from
Sir Run Run Shaw Hospital, Affiliated with School of Medicine, Zhejiang
University. Healthy normal liver tissues were obtained from 12 volunteers (age,
38.5
Data are described as mean
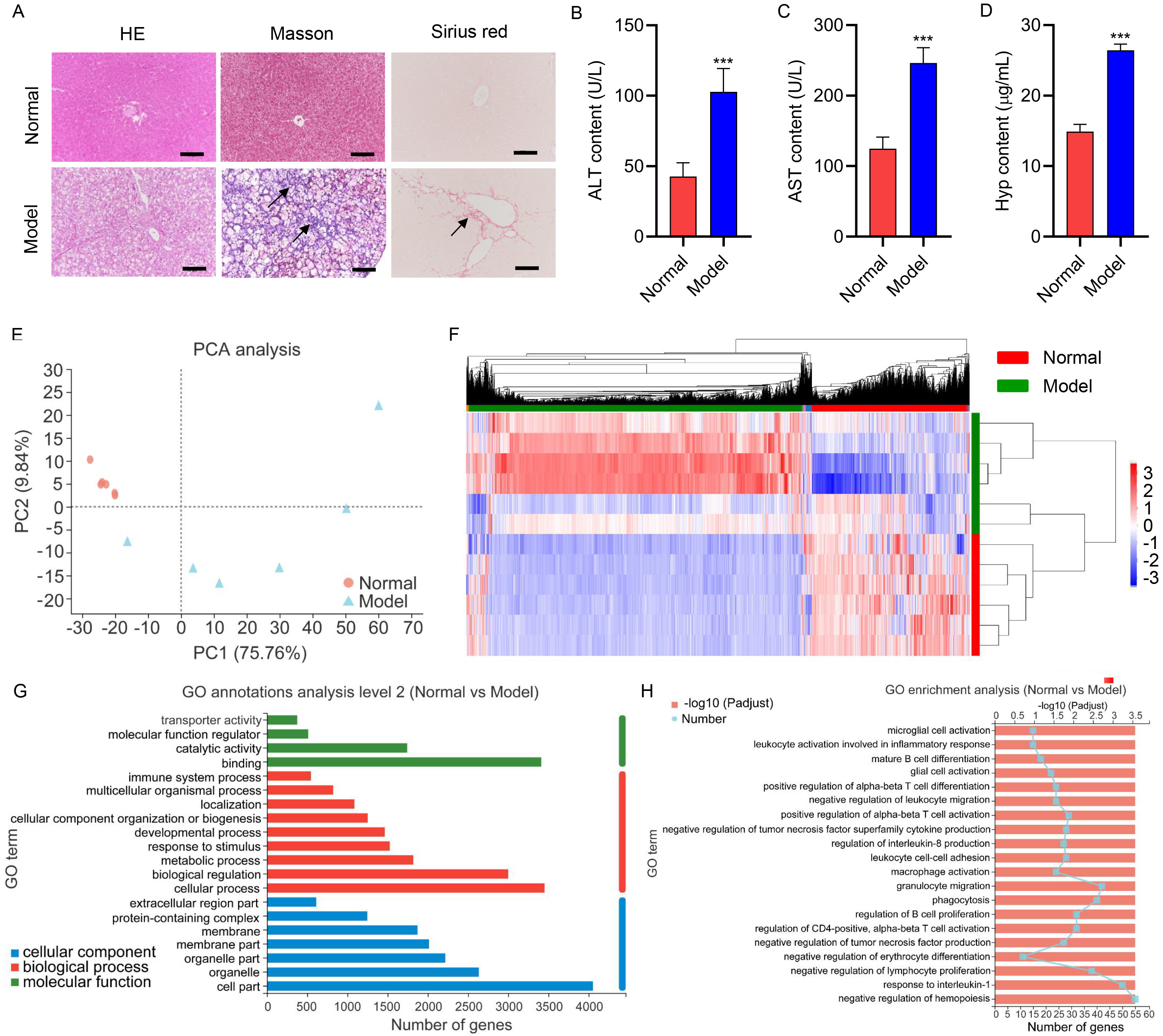
The gerbil model was constructed by giving an HFD to investigate the mechanism
of NAFLD-associated fibrosis development. Histological staining results indicated
no discernible histological alterations in the normal gerbils, which were
reflected by intact sinusoidal spaces, distinct nuclei, and central veins (Fig. 1A). However, the gerbils in the model group exhibited ballooning, denaturation,
hepatocyte necrosis, and obvious fibrosis (Fig. 1A). Additionally, the model
group presented elevated serum ALT, AST, and Hyp levels compared to the normal
group (p
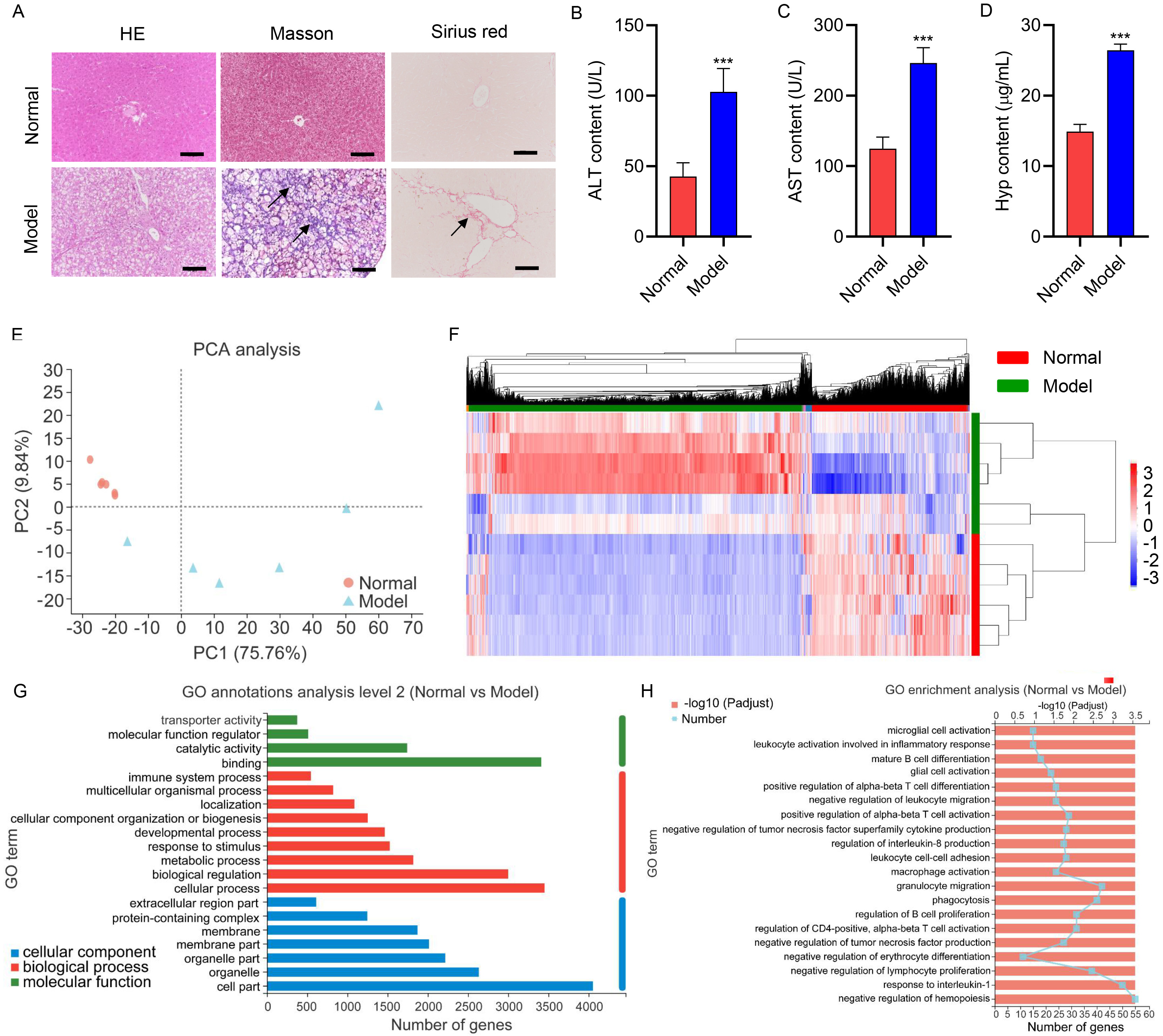 Fig. 1.
Fig. 1.Hepatic injury of gerbils during the high-fat and
high-cholesterol diet. Gerbils were treated with the HFD for 16 weeks (Model; n
= 6). Untreated gerbils were used as the normal control group (Normal; n = 6).
(A) Representative HE, Masson, and Sirius red stained section of liver tissues
(scale bar, 100 µm). Arrows indicated the fibrotic lesion localization.
Serum (B) ALT, (C) AST, and (D) Hyp concentrations. (E) PCA analysis of genes
profiles of liver tissues isolated from gerbils. (F) Heatmap of the
differentially expressed genes. (G) GO annotations analyze the differentially
expressed genes in molecular function, cellular components, and biological
processes. (H) GO enrichment analysis of the differentially expressed genes.
***p
High-throughput mRNA sequencing was performed to screen DEGs to better study the pathogenesis of NAFLD-associated fibrosis. Principal Component Analysis (PCA) analysis of genes profiles of liver tissues isolated from gerbils revealed that the model samples appeared separated from normal samples (Fig. 1E), indicating the strongly differentiated expression profile of the model group from the normal group. The DEGs between the two groups were analyzed, and the heatmap of the DEGs was presented in Fig. 1F. The top 10 upregulated and downregulated genes in the model group were described in Tables 1,2, respectively. Additionally, GO annotations analysis of the DEGs was conducted in the aspects of molecular function, biological process, and cellular component. The molecular function includes “binding”, “catalytic activity”, and “molecular function regulator” as key functions involved by DEGs (Fig. 1G). DEGs were closely related to the “cellular process”, “biological regulation”, and “metabolic process” in the biological processes (Fig. 1G). DEGs were principally associated with the “cell part”, “organelle”, and “organelle part” in the cellular components (Fig. 1G). Furthermore, GO enrichment results demonstrated that DEGs were chiefly enriched in “negative regulation of hemopoiesis”, “response to interleukin-1”, and “granulocyte migration” (Fig. 1H).
| Gene name | Log |
p value | p adjust | Normal | Model |
| Mast3 | 1.935 | 1.38 × 10 |
1.25 × 10 |
0.752 | 6.650 |
| Bmf | 2.822 | 4.32 × 10 |
2.61 × 10 |
0.587 | 9.392 |
| Znf148 | 1.218 | 3.39 × 10 |
1.18 × 10 |
0.733 | 4.402 |
| Usp33 | 1.072 | 4.28 × 10 |
1.46 × 10 |
1.753 | 8.338 |
| Mmp12 | 5.760 | 2.19 × 10 |
5.74 × 10 |
1.887 | 261.7 |
| Tdp2 | 1.440 | 5.62 × 10 |
1.27 × 10 |
1.333 | 8.300 |
| Colec12 | 6.666 | 2.16 × 10 |
4.65 × 10 |
0.345 | 89.96 |
| Zbtb14 | 1.555 | 4.83 × 10 |
9.93 × 10 |
1.708 | 11.83 |
| Fbxo11 | 1.072 | 8.48 × 10 |
1.67 × 10 |
2.462 | 11.75 |
| Ppargc1b | 3.630 | 1.14 × 10 |
2.22 × 10 |
0.058 | 1.720 |
NAFLD, non-alcoholic fatty liver disease.
| Gene name | Log |
p value | p adjust | Normal | Model |
| Socs3 | –4.33249 | 6.33 × 10 |
1.15 × 10 |
100.7 | 10.84 |
| Csrnp1 | –3.57326 | 1.76 × 10 |
1.59 × 10 |
16.58 | 3.022 |
| Rasd1 | –7.8831 | 9.56 × 10 |
5.77 × 10 |
39.35 | 0.368 |
| Btg2 | –4.61946 | 2.03 × 10 |
9.18 × 10 |
94.39 | 8.588 |
| LOC110557684 | –3.12558 | 6.57 × 10 |
2.38 × 10 |
10.13 | 2.597 |
| Irs2 | –5.72892 | 3.83 × 10 |
1.16 × 10 |
66.78 | 2.782 |
| Gadd45g | –4.07069 | 5.33 × 10 |
1.38 × 10 |
90.33 | 11.52 |
| Midn | –1.99354 | 4.42 × 10 |
1.00 × 10 |
35.63 | 19.75 |
| Spidr | –1.54349 | 1.60 × 10 |
3.21 × 10 |
6.655 | 5.083 |
| Irf1 | –2.92794 | 6.33 × 10 |
1.15 × 10 |
73.13 | 20.33 |
Among DEGs, Zbtb14 was one of the top 10 upregulated genes in the
gerbil NAFLD-associated fibrosis model. However, the role of Zbtb14 in
NAFLD-associated fibrosis remained elusive. The Zbtb14 expression was first
verified in gerbils after being treated with the HFD for 8, 12, and 16 weeks to
explore its function in NAFLD-associated fibrosis. Results indicated increased
Zbtb14 in the liver of gerbils in the 8-, 12-, and 16-week groups versus the
normal group (p
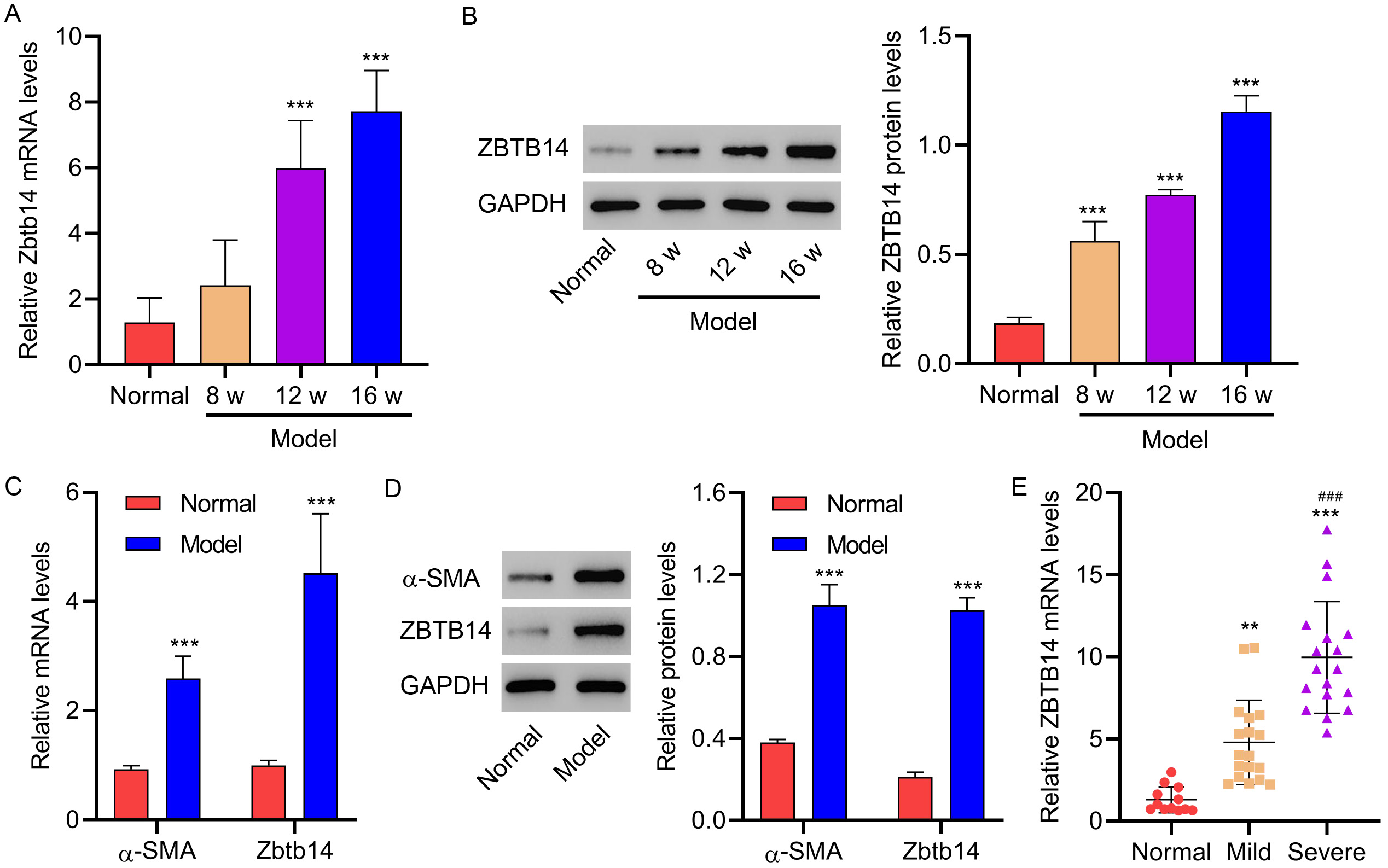 Fig. 2.
Fig. 2.Zbtb14 expression in gerbils and patients with fibrosis.
Gerbils were treated with the HFD for 8, 12, or 16 weeks (Model; n = 6 per
group). Untreated gerbils were used as the normal control group (Normal; n = 6).
(A,B) Zbtb14 expression in liver tissues isolated from gerbils. (C,D)
| Characteristics | ZBTB14 mRNA expression | p value | ||
| Low (n = 18) | High (n = 18) | |||
| Gender | 0.502 | |||
| Male (n = 16) | 7 | 9 | ||
| Female (n = 20) | 11 | 9 | ||
| Age (years) | 0.172 | |||
| 9 | 5 | |||
| 9 | 13 | |||
| Type 2 diabetes n (%) | 10 (55.5%) | 15 (83.3%) | 0.070 | |
| Body mass index (kg/m |
30.9 |
31.8 |
0.420 | |
| Total cholesterol (mg/dL) | 191.6 |
186.4 |
0.438 | |
| HDL cholesterol (mg/dL) | 50.2 |
47.5 |
0.496 | |
| LDL cholesterol (mg/dL) | 103.3 |
111.8 |
0.536 | |
| Triglycerides (mg/dL) | 151.1 |
166.1 |
0.424 | |
| AST (U/L) | 37.9 |
56.0 |
0.012 | |
| ALT (U/L) | 49.5 |
72.8 |
0.014 | |
| HbA1c (%) | 6.19 |
6.63 |
0.042 | |
AST, aspartate aminotransferase; ALT, alanine aminotransferase; HDL, high density lipoprotein; LDL, low density lipoprotein; HbA1c, glycated hemoglobin. Differences between groups were determined by the Chi-square test or Mann-Whitney test.
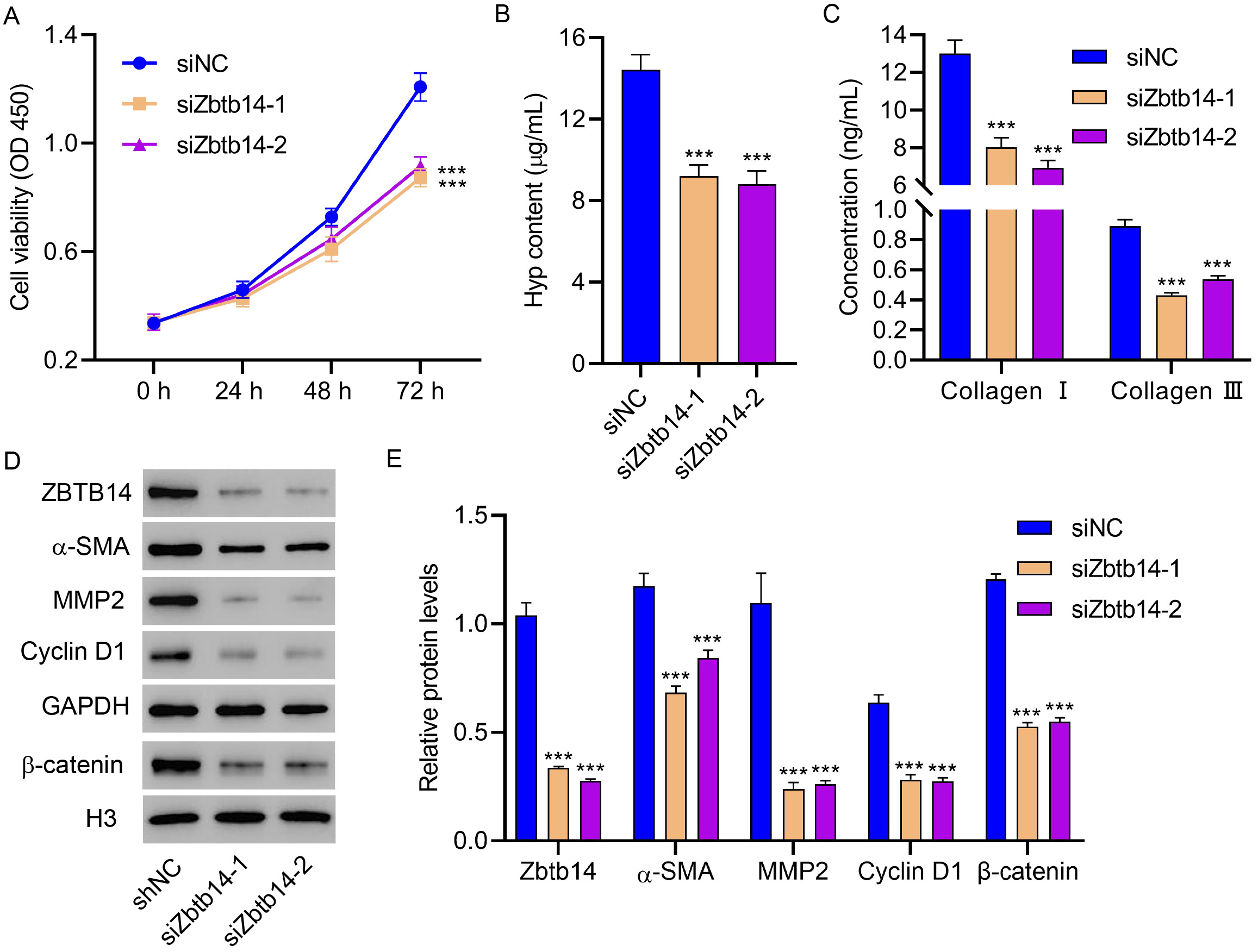
The role of aberrant expressed Zbtb14 in HSC activation was identified to
further study the function of Zbtb14 on NAFLD-associated fibrosis. HSCs
obtained from gerbils treated with the HFD for 16 weeks were transfected with the
siRNA against Zbtb14. Results revealed that the silenced Zbtb14
notably inhibited the cell viability of HSCs (p
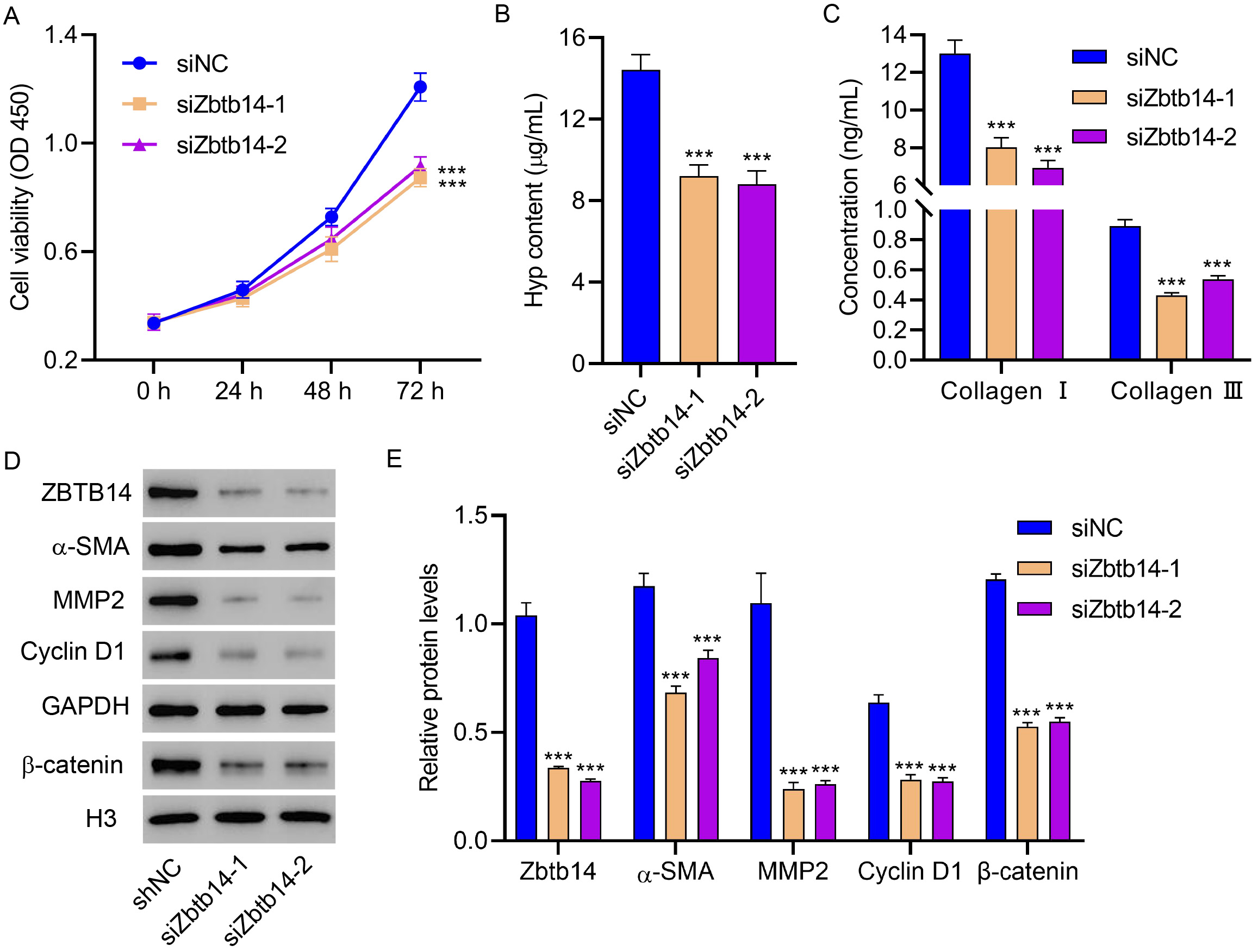 Fig. 3.
Fig. 3.Zbtb14 knockdown inhibits primary HSC activation.
Primary HSCs isolated from gerbils treated with the HFD for 16 weeks were
transfected with Zbtb14 siRNA, and the (A) cell viability, (B) Hyp, (C)
type I collagen, and type III collagen levels, and (D,E) Zbtb14,
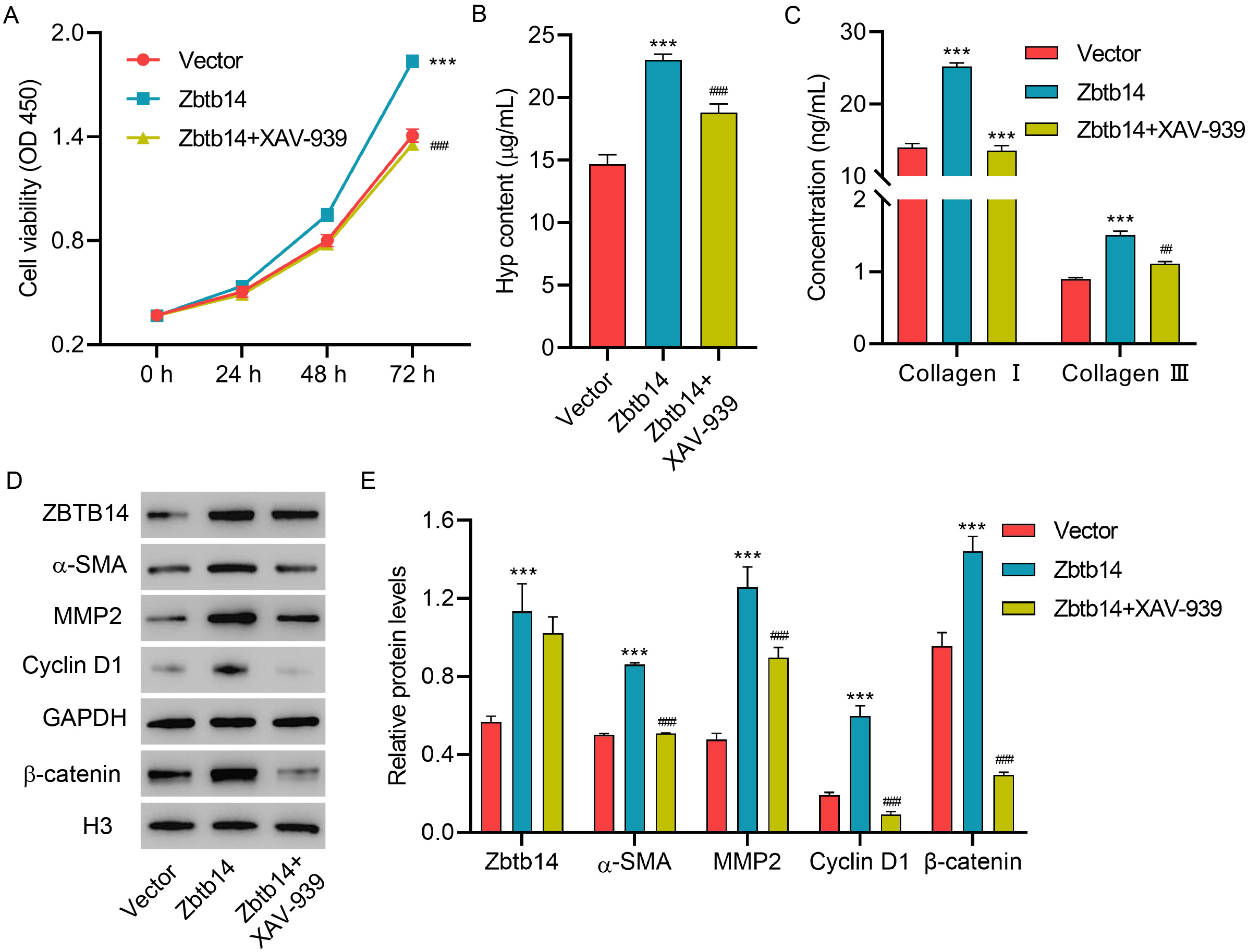
To better determine the function of Zbtb14 on HSC activation and the underlying
mechanism, the HSCs were transfected with Zbtb14 overexpression vector
with or without XAV939 (inhibitor of
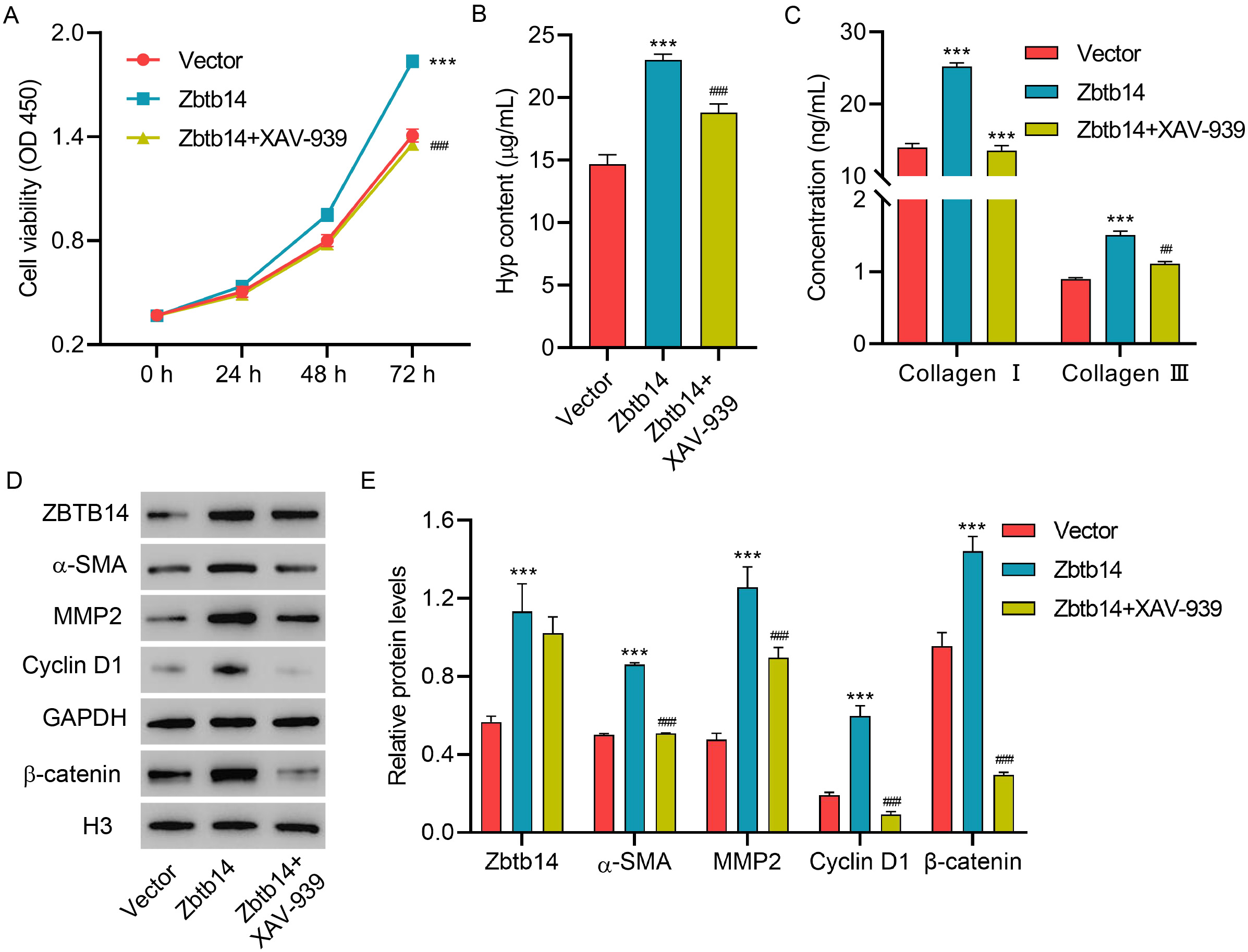 Fig. 4.
Fig. 4.Zbtb14 overexpression promotes primary HSC activation
via the
NAFLD is a popular chronic liver disorder with high morbidity worldwide [1, 2, 3]. Currently, no drug is approved for NAFLD treatment [6]. Liver fibrosis is considered an important predictor of the efficacy of drugs for NAFLD therapy [10, 11]. Hence, the possible mechanisms of liver fibrosis should be elucidated and targets to prevent liver fibrosis and NAFLD should be determined.
The gerbil model of NAFLD with fibrosis was first established to study the mechanism of NAFLD-associated liver fibrosis. In this model, denaturation and necrosis were observed in liver tissues. Additionally, liver function was evaluated by determining serum ALT and AST levels. ALT is an abundant enzyme in hepatocytes [21]. The serum ALT level increased after hepatocytes were injured [21]. Therefore, ALT usually indicates hepatic inflammation and injury in NAFLD [21]. AST is another biochemical marker of liver injury, which is released into the bloodstream after liver injury [22]. This study revealed increased ALT and AST levels in the NAFLD-associated fibrosis model, indicating that liver injury was induced in the gerbil.
Furthermore, obvious fibrosis was presented in the liver tissues of the gerbil model. The Hyp level is determined to confirm the finding. Hyp determination was a common strategy for assessing tissue fibrosis and collagen deposition [23, 24]. Results revealed elevated Hyp levels in the gerbil NAFLD model, consistent with the previous study [25]. These findings demonstrated a successful NAFLD-asLociated fibrosis model construction.
DEGs in the NAFLD-associated fibrosis model were screened using high-throughput mRNA sequencing to explore the pathogenesis of the NAFLD-associated fibrosis model. GO enrichment results proved that DEGs were mainly gathered in “negative regulation of hemopoiesis”, “response to interleukin-1”, and “granulocyte migration”. Shvarts et al. [26] revealed suppressed hemopoiesis in CCl(4)-induced hepatic fibrosis. Additionally, the production of the members of the interleukin-1 family exerted a critical function in NAFLD [27]. Furthermore, neutrophils, which are the most abundant type of granulocyte, were activated and migrated from the blood to the tissues in NAFLD [28]. The above evidence suggested that the biological processes mainly enriched by DEGs, including “negative regulation of hemopoiesis”, “response to interleukin-1”, and “granulocyte migration”, were significant for NAFLD-associated fibrosis.
ZBTB14, also known as ZNF478 and ZFP161, is a zinc finger protein classified as
the ZBTB family [29]. This study revealed Zbtb14 as one of the top 10 upregulated
genes in the NAFLD-associated fibrosis model. Interestingly, Zbtb14 was
demonstrated to modulate hemopoiesis [30]. Therefore, Zbtb14 was selected to
further study its role in NAFLD-associated fibrosis. Western blot and RT-qPCR
results, consistent with high-throughput mRNA sequencing, verified that Zbtb14
was enhanced in the NAFLD-associated fibrosis model and patients with liver
fibrosis. Additionally, results revealed that Zbtb14 was elevated in activated
HSCs. The function of Zbtb14 in HSCs activation was investigated by determining
cell viability and activation markers because HSCs are vital in liver fibrosis
development [12, 13]. Results revealed that Zbtb14 knockdown suppressed
primary HSCs viability and the
The
The additional diet cholesterol may cause some metabolism differences, which is the remaining limitation of the gerbil model. Further studies are required to address whether alterations in diet composition can lead to a refined model, which can completely reproduce the human disease mechanism. Additionally, a large portion of expressed genes generally show sex-specific differences in the liver, and diet effects could conceivably be confounded by sex effects. Only male gerbils were used in the present study. Therefore, further examination of NAFLD-associated fibrosis in female gerbils would be meaningful and more reliable.
In conclusion, this study identified the DEGs in NAFLD-associated fibrosis and
reported Zbtb14 modulated NAFLD-associated fibrosis in male gerbils via the
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
GC and XC designed the research study. XW, YZ, and HH performed the research. GC, XW, and XC provided help and advice on conception, acquisition of data, and supervision. YZ, HH, and XC analyzed the data. GC wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
The study was performed in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the Sir Run Run Shaw Hospital, Affiliated with the School of Medicine, Zhejiang University (approved number 2023-0062). The animal experiments were approved by the local Ethics Committee of the Zhejiang Academy of Medical Sciences (approved number 2019-056) and were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All participants provided written informed consent to participate in the study after the procedures had been completely explained.
Not applicable.
This research was funded by the National Natural Science Foundation of China (31970511) and Zhejiang province commonweal projects (LGD20C040003).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.