1 UMR Transfrontalière BioEcoAgro N° 1158, Univ. Lille, INRAE, Univ. Liège, UPJV, ISA, Univ. Artois, Univ. Littoral Côte d’Opale, Institut CharlesViollette, Joint Laboratory CHIC41H, University of Lille-Florimond Desprez, Cité scientifique, 59655 Villeneuve d’ascq, France
Abstract
Background: Chicory (Cichorium intybus L.), a member of the Asteraceae family, is known for its numerous health benefits, including its prebiotic, digestive, antioxidant or anti-inflammatory effects. Used as a coffee substitute, chicory roots is also appreciated for its bitterness, which can prove to be a disadvantage for other uses in food. The bitterness of chicory is largely linked to the presence of sesquiterpene lactones (STLs) in the roots. Methods: In order to create less bitter industrial chicory varieties, CRISPR/Cas9 technology was used to inhibit the first two genes of the STL biosynthetic pathway: germacrene A synthase (CiGAS), short form, and germacrene A oxidase (CiGAO). To determine the impact of these reductions on the perception of bitterness, a sensory analysis of 13 field-grown chicories genotypes, contrasting for their STL composition, allowed the construction of obtain a bitterness scale by correlating STL content with perceived bitterness. The edited chicories were positioned on this scale according to their STL content. Results: Biallelic mutations in two of the copies of CiGAS-short form or in the CiGAO gene led to a reduction in STL content of edited chicories and a reduction in bitterness, or even an absence of perception, was obtained for some mutants. Conclusions: The use of the CRISPR/Cas9 tool as well as the choice of targets therefore makes it possible to modulate the bitterness of chicory.
Keywords
- Cichorium intybus L.
- bitterness
- sesquiterpene lactones
- CRISPR/Cas9
- hairy roots
- germacrene A synthase
- germacrene A oxidase
Cichorium intybus L., also known as chicory, is a species belonging to the Asteraceae family widely used in traditional medicine. Its biological activities are mainly attributed to the accumulation of inulin, vitamins, and specialized metabolites such as sesquiterpene lactones (STLs), flavonoids, coumarins, hydroxycinnamic acids as well as alkaloids present in different parts of C. intybus L. [1]. In Europe, different varieties of chicory are cultivated either for the consumption of their leaves or for the use of their roots. In the North of France, C. intybus var. foliosum is commonly used as a salad and the taproot of C. intybus var. sativum (or industrial chicory) is grown for industrial purposes. Roots of industrial chicory are of economic importance for roasted ingredients, beverage processing such as coffee substitute, and inulin production. The concept of functional ingredients is increasingly developing due to the attraction of consumers towards healthier food. Chicory inulin is already considered as a functional ingredient due to its health effects and recently a new trend describing chicory flour as such has been developed [2, 3, 4, 5, 6]. Chicory flour, produced from roots, is mainly employed as supplement in baking since it improves the taste, the preservation, and the texture of bakery products in addition to being low in calories and rich in fibers. However, its incorporation is limited to 5% because chicory bitterness becomes too perceptible in larger proportions [7]. Bitterness plays a major role in food consumption and can cause an aversive reaction in consumers. By offering less bitter varieties of chicory, it will be possible to produce less bitter ingredients from chicory and, as a consequence, to integrate more chicory flour in bakery to benefit from its nutritional and functional qualities. Industrial chicory bitterness is mainly due to the presence of STLs [8, 9, 10]. These compounds represent on average 0.42% of the dry weight of the roots, making them the major specialized metabolites found in the root [11]. They belong to the class of guaianolides and are: lactucin, lactucopicrin, 8-deoxylactucin, their 15-oxalated and 15-glycosylated forms, and their derivatives 11(S),13-dihydro [12, 13, 14]. The biosynthesis of the guaianolides starts with the conversion of farnesyl-pyrophosphate (FPP) into germacrene A by germacrene A synthase (GAS) [15]. Four copies of CiGAS gene were identified in the industrial chicory genome, including a single copy of CiGAS-long form and three expressed copies of CiGAS-short form [16]. Then cytochrome P450 enzymes germacrene A oxidase (GAO) and costunolide synthase (COS) act consecutively to produce costunolide [17, 18, 19, 20]. Recently, a new step of this biosynthetic pathway was revealed with the identification of the kauniolide synthase (KLS) gene cluster involved in the conversion of costunolide to kauniolide [21].
Genome editing approaches using CRISPR/Cas9 have developed rapidly in recent years in several plant species. They greatly accelerated gene function analysis or crop improvement to generate new varieties in a shorter time [22, 23, 24]. Its use allows to rapidly test strategy to reduce the bitterness of chicory in order to make it more acceptable for its use as a functional ingredient. In C. intybus, the CRISPR/Cas9 editing system was first successfully used by Bernard et al. [25] using two methods of transformation: Rhizobium rhizogenes-mediated transformation and protoplast transfection. Thereafter, the method was used to validate the function of genes involved in the biosynthesis of phenolamide in chicory pollen grains [26]. In parallel, CRISPR/Cas9 technology has been used in several studies to modify the level of STLs in C. intybus. First, De Bruyn et al. [27] edited CiGAS, CiGAO and CiCOS genes of C. intybus var. foliosum through PEG-mediated protoplast transfection but the effects on the chemical phenotype was not described. Then Cankar et al. [28] edited CiGAS genes of C. intybus var. sativum using the same editing transfection system and showed a strong reduction in the content of lactucin, lactucopicrin, 8-deoxylactucin and their oxalated forms. These results confirmed those of a previous study in which the CiGAS genes were silenced by an amiRNA approach [29]. Recently, Cankar et al. [21] showed that knockout (KO) of KLS genes led to a nearly complete elimination of the major chicory STLs and their oxalate forms in leaves and roots of genome-edited chicories. They also investigated the presence of intermediates in the STL pathway and showed that there was no accumulation of compounds in the leaves, in contrast to the taproot where costunolide, its conjugates and germacrene A acid were detected. Despite these works, no sensory analysis was correlated with these results to determine the effect of STLs reduction on bitterness perception.
In the present study, in order to induce a modulation in STL content, industrial chicories ChicBitter002, cultivated for their roots, were edited with CRISPR/Cas9 technology using R. rhizogenes-mediated transformation. The CiGAS-short form and CiGAO genes, involved in the STL pathway were targeted. In addition, to link the STL content to the bitterness level, a sensory analysis was carried out on roots of several field-grown chicories. A linear correlation was established between the bitterness score associated with each of these chicories and their STL content. A bitterness scale was then generated and used to evaluate the bitterness of the genome-edited chicories based on their STL content. The involvement of the different targeted genes in the synthesis of STLs was also discussed.
Field-grown chicories R01 to R13, furnished by Florimond-Desprez, are a breeding material chosen for their variability in the STL content based on the dosage of STLs conducted in 2018 and 2019 (unpublished data). They have been cultivated in 2020 (Coutiches, France) for their roots by Florimond-Desprez SA (Cappelle-en-Pévèle, France). As chicory is allogamous and not able to form stable lineages, each chicory R01 to R13 is a population sharing a common gene pool. For the sensory analysis, each field-grown chicory was furnished as a sample originating from a pool of all roots of three field plots. ChicBitter002, is an in vitro-propagated industrial chicory clone, which genome has been sequenced by the Gentiane platform (INRAE, Clermont-Fermond, France) for Florimond Desprez SA (Cappelle-en-Pévèle, France). The clone ChicBitter002 was used in a wild-type form (Ctrl) or transformed by R. rhizogenes (HR lines).
Several CiGAS gene candidates were identified in the industrial chicory genome ChicBitter002 with Blastp or tBlastn algorithms using the germacrene A synthase short form gene (Uniprot accession: Q8LSC2) and the germacrene A synthase long form gene (Uniprot accession: Q8LSC3) from Bouwmeester et al. [30] as queries and with 26,367 ‘high probability’ predictive gene models from the Chicbitter002 sequence (unpublished database, Florimond Desprez SA, Cappelle-en-Pévèle, France) and 31,631 predictive gene models from a witloof chicory genome (Cargese program) or their complete genome sequences as subjects. Nucleotide sequences of significant hits were aligned to the Chicbitter002 sequence using GMAP and their positions were compared to corresponding positions of predictive gene models of recently sequenced Asteraceae, such as Lactuca sativa, Cynara cardunculus var. scolymus, Artemisia annua and transcript sequences from unpublished RNA-seq data in industrial and witloof chicories [31, 32, 33, 34]. The sequence of the CiGAO gene (Uniprot accession: D5JBW8) was used to identify candidates using the same procedure as for CiGAS genes. The CiGAS gene candidates were used as queries in a tBlastn procedure by comparing them to the BAC clone sequences previously published [16]. Multiple protein sequence alignment from the whole set of CiGAS sequences was carried out using MAFFT (Supplementary Fig. 1) [35].
Guide RNAs (sgRNAs) for CiGAS-short form and CiGAO
genes were designed using the software CRISPOR (crispor.tefor.net) based on
their GC content and their Doench score [36]. Two sgRNAs were defined for each
gene except the gene CiGAS-S5 for which only one target could be defined
(Supplementary Table 1). Potential off-targets were checked with the
tool Cas-OFFINDER and verified on the chicory ChicBitter002 genome (unpublished
sequence, Florimond Desprez SA, France) to avoid the disruption of off-target
gene coding region [37]. Two binary expression vectors for CRISPR/Cas9 were
constructed as described previously [25, 38]. Briefly, each sgRNA was inserted by
digestion/ligation into an intermediate plasmid pKanCiU6-1p-sgRNAscaffold, which
includes C. intybus U6-1p promoter and a sgRNA scaffold. Four vectors
containing the cassette “CiU6-1p-Guide-sgRNAscaffold” were obtained. Adaptors
for Golden Gate restriction/ligation method were added to the end of each
cassette by PCR using PrimeStar HS polymerase (Takara Bio Europe,
St-Germain-en-Laye, France) using primers GG1-F & GG1-R (Supplementary
Table 1) for plasmid containing CiGAS_T1 and CiGAO_T1 guide and primers GG2-F
& GG2-R (Supplementary Table 1) for plasmid containing CiGAS_T2 and
CiGAO_T2 guide. PCR products were purified with NucleoSpin Gel and PCR Clean-up
kits (Macherey-Nagel, Düren, Germany) and were cloned in
pYLCRISPR/Cas9P
In vitro plants of C. intybus L. ChicBitter002 clone were used in this study. Plants were maintained on H10 solid medium as previously described [25]. Wild-type R. rhizogenes 15834 strains (provided by Marc Buée, INRAE, Nancy, France) were transformed with pYLCRISPR-GASsh or pYLCRISPR-GAO by electroporation. Wild-type (WT) strains or transformed strains of R. rhizogenes were used to generate hairy root (HR) lines from ChicBitter002 chicory. HRs were obtained and maintained in hormone-free media as previously described and spontaneous shoots regenerated from HRs were transfered in H10 medium [25]. A selection of HR mutant lines and control plants were grown in pots for a period of 3 months in a S2 greenhouse. Genomic DNA from HRs was extracted using NucleoSpin Plant II kit (Macherey-Nagel, Düren, Germany) using the manufacturer’s protocol. RolB specific primers were used to confirm R. rhizogenes infection, and Cas9 presence was checked by PCR using primer pair C9-F and C9-R (Supplementary Table 1).
Genotyping of HRs was realized to select mutant lines. Primer pairs GAO and S1
to S5 (Supplementary Table 1) labelled with different fluorescent dyes
(6-Fam or Hex) were designed to amplify the sequences of GAO gene or
each potentially expressed copy of CiGAS-short form gene. The PCR assay
was performed in a volume of 15 µL containing 1X PCR buffer with 2
mM MgCl
Roots of edited chicories (n = 3 for each HR lines), WT 3-month-old chicories (n
= 4) and field-grown chicories (R01 to R13) taproots were freeze-dried and
powdered. Hundred mg of powder were extracted with 1 mL of water (LC-MS grade)
under agitation (1400 rpm; Eppendorf ThermoMixer C, Hamburg, Germany) for 15 min
at 35 °C. The supernatants were collected by centrifugation (15 min, 4
°C, 21,000 g) and boiled for 10 min at 100 °C under agitation
(600 rpm). All aqueous extracts were finally filtered through a 0.45
µm PP Whatman UNIFILTER microplate (Cytiva, VWR, Rosny-sous-Bois,
France) and transferred to vials for LC-UV analysis. STLs content was determined
using an Ultimate 3000RS system equipped with an LPG-3400RS pump, a WPS-3000TRS
autosampler and a DAD-3000RS (Thermo Fisher Scientific, Waltham, MA, USA).
Chromatographic separation was achieved on an Uptisphere Strategy PHC4 column (3
µm, 150
STL compounds were identified using an Ultimate 3000RS system equipped with a
DAD-3000RS (Thermo Fisher Scientific, Waltham, MA, USA), interfaced with a
high-resolution quadrupole time-of-flight mass spectrometer and equipped with an
ESI source (Impact II, Bruker Daltonik GmbH, Bremen, Germany). One gram of
freeze-dried powdered chicory root from the pool of field-grown chicories
(R01-R13) was extracted with 10 mL of water (LC-MS grade) under agitation (1000
rpm) for 15 min at 35 °C. After centrifugation (15 min, 4 °C,
21,000 g) , the supernatant was immediately evaporated, and the residue was
resuspended in 2% acetonitrile (v/v) containing 0.1% formic acid (v/v) and
passed through a 0.2 µm hydrophilic PTFE syringe filter (Merck,
Darmstadt, Germany) and transferred to a vial for LC-MS analysis. Chromatographic
separation was achieved on an Uptisphere Strategy PHC4 column (2.2
µm, 150
The sensory evaluation was performed at the agrifood platform of Polytech Lille
(University of Lille, France) in laboratory rooms according to ISO 8589:2007. The
panel was constituted from volunteers recruited among the personnel of the
University of Lille and trained in accordance with ISO 8586:2012. Panelists were
first selected based on their ability to recognize bitterness, sweetness,
saltiness, and sourness. Four series of different concentrations of taste
solutions in water were prepared by dilution of stock solutions as followed:
bitter with caffeine (concentrations: 0.27, 0.35, 0.46, 0.59 g·L
An ANOVA and Student’s t-test were performed to determined panelist performance (discrimination ability, consensus in notation, repeatability) and to test for bitterness differences between the chicory root samples from the field-grown chicories (R01-R13). Comparison of STL content of edited and WT 3-month-old chicories and assessment of the differences in panelist perception of bitter taster were tested by non-parametric ANOVA (Kruskal-Wallis) followed by post-hoc uncorrected Dunn’s test or post-hoc paired Wilcoxon, respectively. The assumptions of normality, homogeneity of variance, and independence were checked by Shapiro-Wilk test, Bartlett test, and Durbin-Watson test, respectively. The bitterness correlation with STL content was established by Pearson’s correlation test. The significance threshold was set to 0.05 for all statistical tests. All analyses were done with R Statistics 4.0.3 (r-project.org) and GraphPad Prism 9 software (Windows Inc., San Diego, CA, USA).
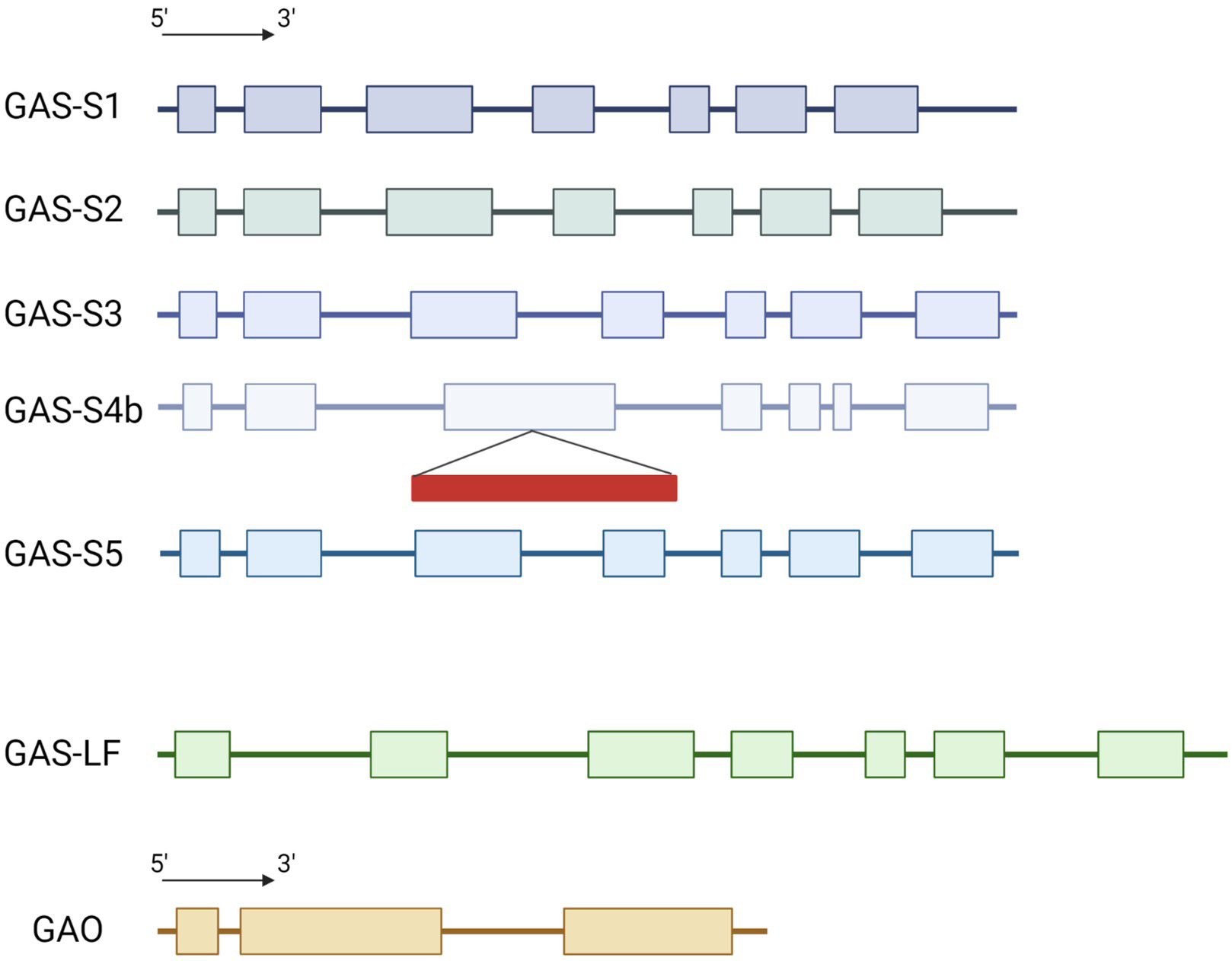
The genome of the industrial chicory ChicBitter002 was searched to identify CiGAS and CiGAO genes putatively involved in the STL pathway. A single copy of CiGAO gene was highlighted in the genome of ChicBitter002 (Supplementary Fig. 2). For CiGAS genes, quite similar data to Bogdanović et al. [16] were found with one gene for CiGAS-long form and five gene copies of CiGAS-short form. Sequence analysis confirmed the existence of GAS-S1, GAS-S2, GAS-S3 and CiGAS-S4b but CiGAS-S4a was not found. However a new CiGAS-short form candidate copy, named CiGAS-S5, was highlighted (Fig. 1). Sequence analysis confirmed that copies CiGAS-S1, CiGAS-S2, CiGAS-S3 and CiGAS-S5 could be expressed with a complete 7-exons structure and that copy CiGAS-S4b has a retrotransposon into the exon 3 that could hamper its expression. The five copies were identified at two different locations in the genome. On the one hand, CiGAS-S1, S2 and S3 were located on the same contig (contig_403_pilon) so they can be considered as three distinct genes but physically close on the chicory genome. Indeed, the CiGAS-S1 and the CiGAS-S3 are separated by ~85,000 bp, with CiGAS-S2 in between (Supplementary Fig. 3). It can also be noted that CiGAO gene is present on the same contig as the three CiGAS-short form genes, and ~85,000 bp away from the CiGAS-S3 copy (Supplementary Fig. 3). On the other hand, CiGAS-S4b and S5 also shared the same contig (contig_1740_pilon) and are ~76,000 bp apart. Based on these results, 4 copies of CiGAS-short can be considered as putatively expressed : copies S1, S2, S3 and S5 (Fig. 1). Since the copy S4b was presumably not expressed due to the presence of a retrotransposon, it was not targeted.
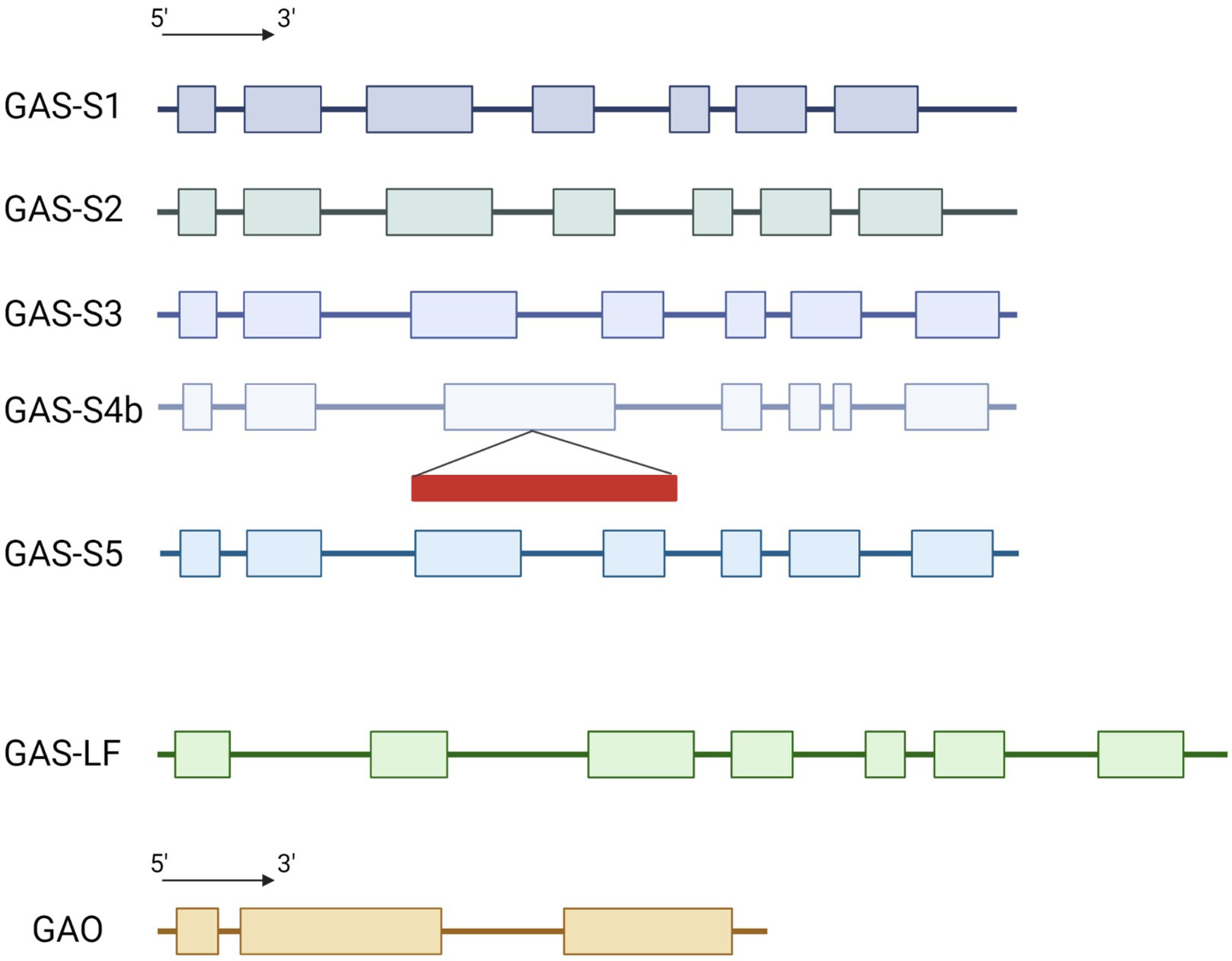 Fig. 1.
Fig. 1.Schematic representation of the structure for CiGAS and CiGAO genes. GAS-S stands for the CiGAS-short form and GAS-LF stands for the CiGAS-long form. The exons and introns lengths are proportional to the nucleotide sequences of the genes. Colored box: exon, line: intron, red rectangle: retrotransposon.
The objective was to perform a KO of the 4 putatively expressed copies of CiGAS-short form and of the CiGAO gene. In order to design sgRNAs for the four putatively expressed copies of CiGAS-short form, the homology of these genes was investigated by comparing their exonic sequences using ClustalOmega tool [40]. All the putatively expressed copies (CiGAS-S1 to S5) are relatively close since they share between 86 to 97% identity. CiGAS-S1 and CiGAS-S2 sharing the highest identity (Supplementary Fig. 4). The newly identified CiGAS-S5 copy shares 86–93% identity with the other copies, confirming its link to the multiple copies of CiGAS-short form (Supplementary Fig. 4). The high level of homology between the potentially expressed copies of CiGAS-short form did not allow specific targeting of each copy, but it was possible to design sgRNAs in the third exon to target all copies simultaneously. Since a single copy of CiGAO was identified, two sgRNAs were designed at the beginning of the first exon in order to disrupt the gene function.
The use of the CRISPOR tool to determine the potential targets gave a list of sgRNAs that were then analyzed using the Cas-OFFINDER tool to search for potential off-targets. The results were checked on the ChicBitter002 genome, to avoid selecting sgRNAs with off-targets in gene coding regions. Finally, two sgRNAs were identified in the first exon of CiGAO and two sgRNAs were identified in the third exon of CiGAS-S1, CiGAS-S2 and CiGAS-S3. For the CiGAS-S5 copy, a single sgRNA common to the other copies was defined (Supplementary Table 1). The GAS-long gene was not targeted because it has been demonstrated to have minimal impact on STLs production in chicory roots [28].
Chicory leaves of 14-day-old vitroplants were infected with R. rhizogenes strain 15834 transformed with the binary vectors pYLCRISPR-GASshort or pYLCRISPR-GAO. Two weeks after R. rhizogenes transformation, selection of HRs based on their phenotype was carried out. Eighty-five HR lines were collected with 54 lines potentially transformed with the vector targeting the CiGAS-short form genes and 31 lines potentially transformed with the vector targeting the CiGAO gene. After verification of the presence of the T-DNA of R. rhizogenes and the integration of the binary vector, the number of transformed lines was reduced to 43 for CiGAS-short and 30 for CiGAO (Table 1). As integration of the binary vector is not always synonymous of mutation, a preliminary sorting of the mutants was performed by genotyping. Indeed, the action of CRISPR/Cas9 can generate a variety of mutated alleles and because multiple copies of CiGAS-short were highlighted, it is possible that all copies are not mutated at the same time. Then, a Sanger sequencing of the mutant HR lines identified by genotyping was performed to obtain the sequences of the mutants. Out of the 43 lines transformed with pYLCRISPR-GASshort, analysis of the mutations revealed 6 lines with one copy of CiGAS gene mutated, 4 lines with two mutated copies and 3 lines with three mutated copies, resulting in a mutation rate of 30.2% (Table 1). HRs transformed with pYLCRISPR-GAO showed a mutation rate of 20% with 6 mutated lines out of 30 transformed lines (Table 1).
| Genes | CiGAS-short | CiGAO | Control | |
| Initial number of HR lines | 54 | 31 | 6 | |
| Number of lines with binary vector | Total | 43 | 30 | 6 |
| WT | 30 | 24 | 6 | |
| 1 copy mutated (KO) | 6 (3) | 6 (2) | 0 | |
| 2 copies mutated (KO) | 4 (1) | nc | nc | |
| 3 copies mutated (KO) | 3 (1) | nc | nc | |
| 4 copies mutated (KO) | 0 (0) | nc | nc | |
| Mutation frequency | 30.2% | 20.0% | 0.0% | |
| Gene KO frequency | 11.6% | 6.6% | 0.0% | |
HR, Hairy Root; WT, Wild-Type lines; KO, Knock-Out due to the onset of premature stop codon; nc, not concerned. Mutation frequency was calculated by dividing the sum of mutant lines by the total of lines with binary vector. Gene KO frequency was calculated by dividing the sum of (KO) by the total of lines with binary vector.
As the chicory genome is diploid, the CRISPR/Cas9 system can induce two types of mutations. On the one hand, a monoallelic mutation could occur. In such case, one allele is mutated and the other remains wild-type (heterozygous) leading to an absence of full KO of the targeted gene. On the other hand, editing could generate a biallelic mutation where both alleles are mutated, either with the same mutation on each allele (homozygous), or with a different mutation (also heterozygous) that can lead to a KO of the gene (Table 2 and Fig. 2). Most of the identified mutations were small insertions of one nucleotide (HR1, HR2, HR4, HR5, HR6, HR8, HR9, HR10, HR12, HR13, HR14, HR16, or HR18) or small deletions of less than 10 nucleotides (HR1, HR2, HR5, HR6, HR7, HR9, HR10, HR11, HR13, HR15, HR17, HR18, or HR19), but in some case, larger deletions were found between the two targets T1 and T2 as shown for HR4, HR7, HR8, HR12, HR12*, HR15 and HR17 (Fig. 2). Some mutations cause a change in the coding frame leading to the KO of a gene or a gene copy and the potentially premature termination of protein translation. This type of event was observed for 5 HR lines mutated on CiGAS-short genes: 3 lines mutated on a single copy (HR1, HR2 and HR4), 1 line mutated on two copies (HR8) and 1 line mutated on 3 copies (HR12*); and in 2 HR lines mutated on CiGAO (HR18 and HR19) resulting in a gene KO frequency of 11.6% and 6.6% respectively (Tables 1,2). The R. rhizogenes-mediated transformation is a stable transformation, meaning that the T-DNA is inserted into the plant genome. As a result, the Cas9 gene is integrated into the genome of HR lines and can continue to exert its action and cause mutations. This case can be observed for the lines HR12 and HR12*. Originally, HR12 had a mutation on both CiGAS-S1 and CiGAS-S2copies but after a few months, an additional mutation appeared on the CiGAS-S5 copy.
| CiGAS-short | |||||||||
| S1 | S2 | S3 | S5 | CiGAO | |||||
| Line | T1 | T2 | T1 | T2 | T1 | T2 | T1 | T1 | T2 |
| HR1 | –3/–3 | +1/+1 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR2 | WT/WT | –5/+1 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR3 | WT/WT | WT/–41 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR4 | WT/WT | WT/WT | +1/+1–45 | +1/–45 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR5 | WT/WT | WT/WT | WT/+1 | WT/–6 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR6 | WT/WT | WT/WT | WT/+1 | WT/–6 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR7 | WT/–31 | WT/–31 | WT/WT | WT/–10 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR8 | WT/–29 | +1/–29 | WT/WT | +1/–1 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR9 | WT/WT | WT/–5 | WT/+1 | WT/–6 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR10 | WT/WT | WT/–5 | WT/+1 | WT/–6 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR11 | WT/WT | WT/–10 | WT/WT | WT/–3 | WT/WT | WT/WT | WT/–7 | WT/WT | WT/WT |
| HR12 | WT/–29 | +1/–29 | WT/WT | +1/+1 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT |
| HR12* | WT/–29 | +1/–29 | WT/WT | +1/+1 | WT/WT | WT/WT | +1/–1 | WT/WT | WT/WT |
| HR13 | WT/WT | WT/+1 | WT/WT | +1/–41 | +1/–3 | WT/WT | WT/WT | WT/WT | WT/WT |
| HR14 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/+1 |
| HR15 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | –12/–22 | WT/–7 |
| HR16 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/+1 | WT/WT |
| HR17 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/–22 | WT/–6 |
| HR18 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | +1/–8 | WT/WT |
| HR19 | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | WT/WT | –7/–7 |
Mutation leading to a change in the coding frame are in red. Larger deletion between the two targets (inter-guide deletion) are underlined. HR12* correspond to the line HR12 but with an additional mutation as a result of the CRISPR/Cas9 action. T1, Target 1; T2, Target 2; WT, Wild-Type; +, insertion; -, deletion.
 Fig. 2.
Fig. 2.Examples of sequences obtained by CRISPR/Cas9 editing. (A) Sequence analysis of the two alleles of hairy root lines transformed with pYLCRISPR-GASshort construct. (B) Sequence analysis of the two alleles of hairy root lines transformed with pYLCRISPR-GAO construct. The target sequences are in blue, and the PAM sequences are underlined. Mutations are highlighted in red and the changes in the nucleotide sequences are shown on the right of each allele. HR.x.x, Hairy Root x allele x; WT, Wild-Type; T1, Target 1; T2, Target 2; +, insertion; -, deletion.
For the rest of the paper, we will focus on only few mutants (HR2, HR3, HR9, HR12, HR12*, HR16 and HR18) to see which mutation event can affect the STL production.
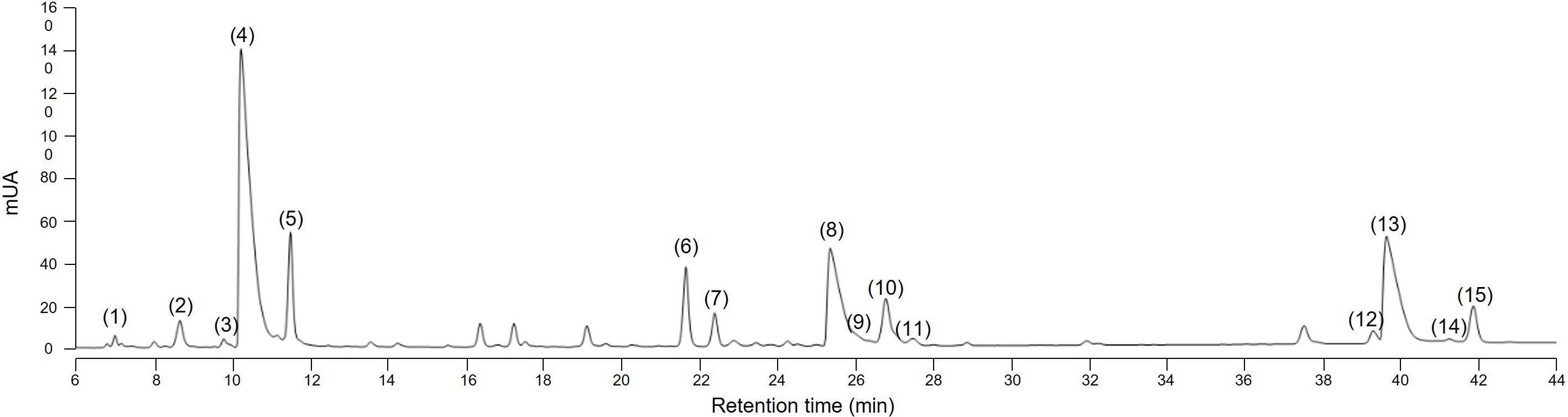
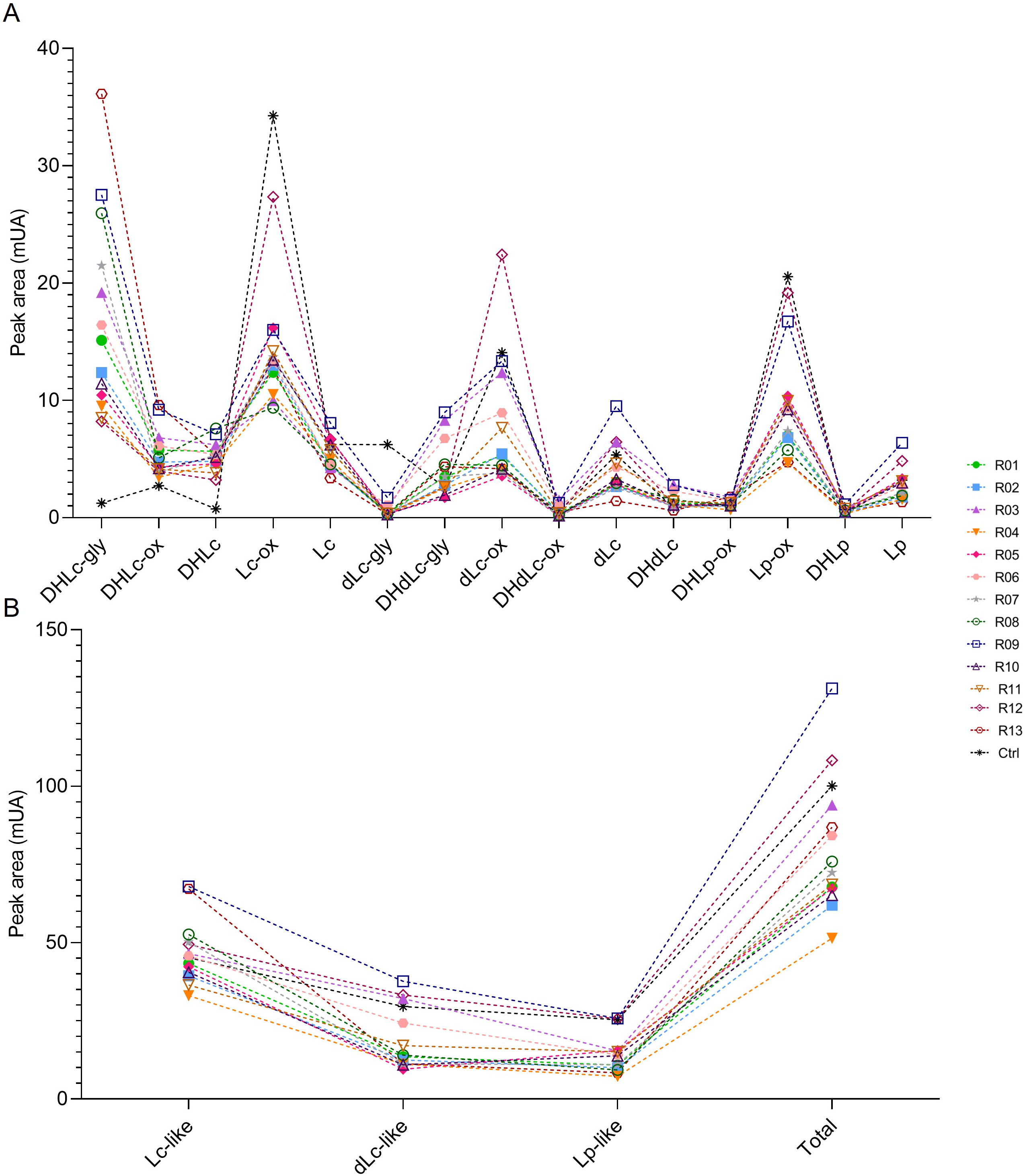
Several methods to extract sesquiterpene lactones from chicory roots have been described using various solvents, but a method more representative of physiological conditions of human consumption was desired, since the objective of our project is to use industrial chicory root as functional eaten ingredient. Using water, an aqueous extract rich in sesquiterpene lactones was obtained with 15 compounds detected at a wavelength of 254 nm in the WT chicory ChicBitter002: lactucin, 8-deoxylactucin, lactucopicrin and their 11(S),13-dihydro derivatives, in addition of their oxalated forms and some glycosylated forms (Fig. 3). The same extraction method was used for the field-grown chicories used in sensory analysis and their STL composition was compared to that of the WT chicory ChicBitter002 “Ctrl” grown under controlled conditions (in vitro then in greenhouse). Fifteen identified compounds are present in all chicories, but a quantitative disparity was observed between the field-grown chicories (R01-R13) and the chicory “Ctrl” (Fig. 4A). As a general trend, the lactucin-15-oxalate and 8-deoxylactucin-15-glycoside content appeared to be higher in “Ctrl” plants compared with the field-grown chicories, whereas the different forms of 11(S),13-dihydrolactucin were in lower quantities. These differences could be due to the growing conditions or genotypic. However, if the distribution of STL content was examined according to the lactucin-like, 8-deoxylactucin-like, and lactucopicrin-like groups, the difference between chicories would fade, as would the total STL content (Fig. 4B). In the following analyses, the STL content of the chicories were therefore compared by considering their total STLs obtained by the sum of the 15 compounds detected in Fig. 3. This is based on the hypothesis that taking into account all STLs is a good indicator of bitterness. Indeed, the different studies on chicory have not clearly established a compound more involved than another in bitterness and when consuming chicory-based products, the consumer doest not taste the STLs separately in his mouth but perceives them as a whole and detects a bitter taste.
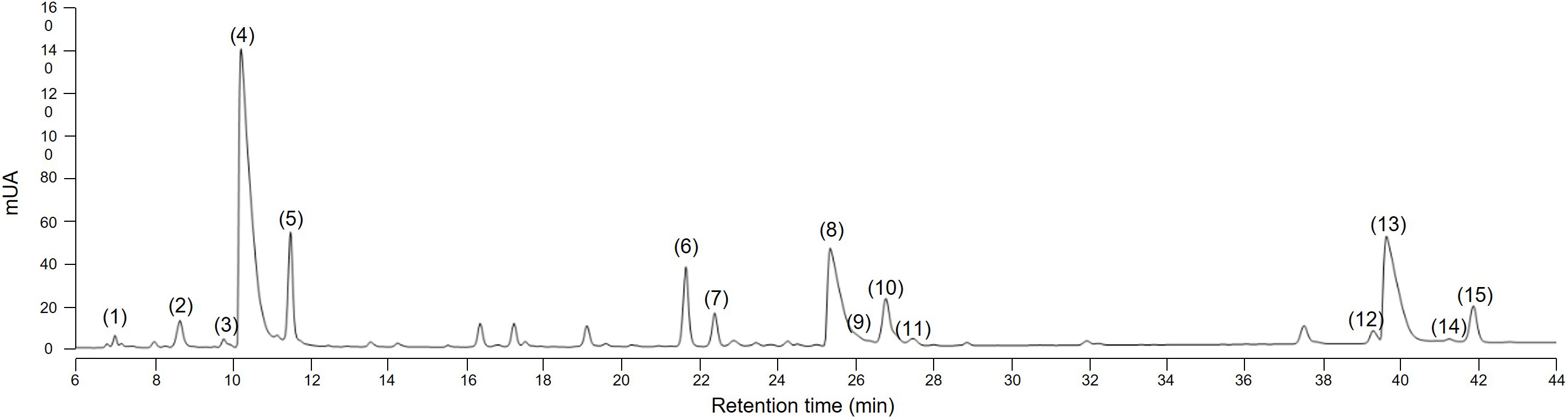 Fig. 3.
Fig. 3.HPLC chromatogram at 254 nm of sesquiterpene lactones in root of the wild-type chicory ChicBitter002. (1) 11(S),13-dihydrolactucin-15-glycoside (DHLc-gly); (2) 11(S),13-dihydrolactucin-15-oxalate (DHLc-ox); (3) 11(S),13-dihydrolactucin (DHLc); (4) Lactucin-15-oxalate (Lc-ox); (5) Lactucin (Lc); (6) 8-deoxylactucin-15-glycoside (dLc-gly); (7) 11(S),13-dihydro-8-deoxylactucin-glycoside (DHdLc-gly); (8) 8-deoxylactucin-15-oxalate (dLc-ox); (9) 11(S),13-dihydro-8-deoxylactucin-15-oxalate (DHdLc-ox); (10) 8-deoxylactucin (dLc); (11) 11(S),13-dihydro-8-deoxylactucin (DHdLc); (12) 11(S),13-dihydrolactucopicrin-15-oxalate (DHLp-ox); (13) Lactucopicrin-15-oxalate (Lp-ox); (14) 11(S),13-dihydrolactucopicrin (DHLp); (15) Lactucopicrin (Lp). Identity of the molecules was confirmed by mass spectrometry.
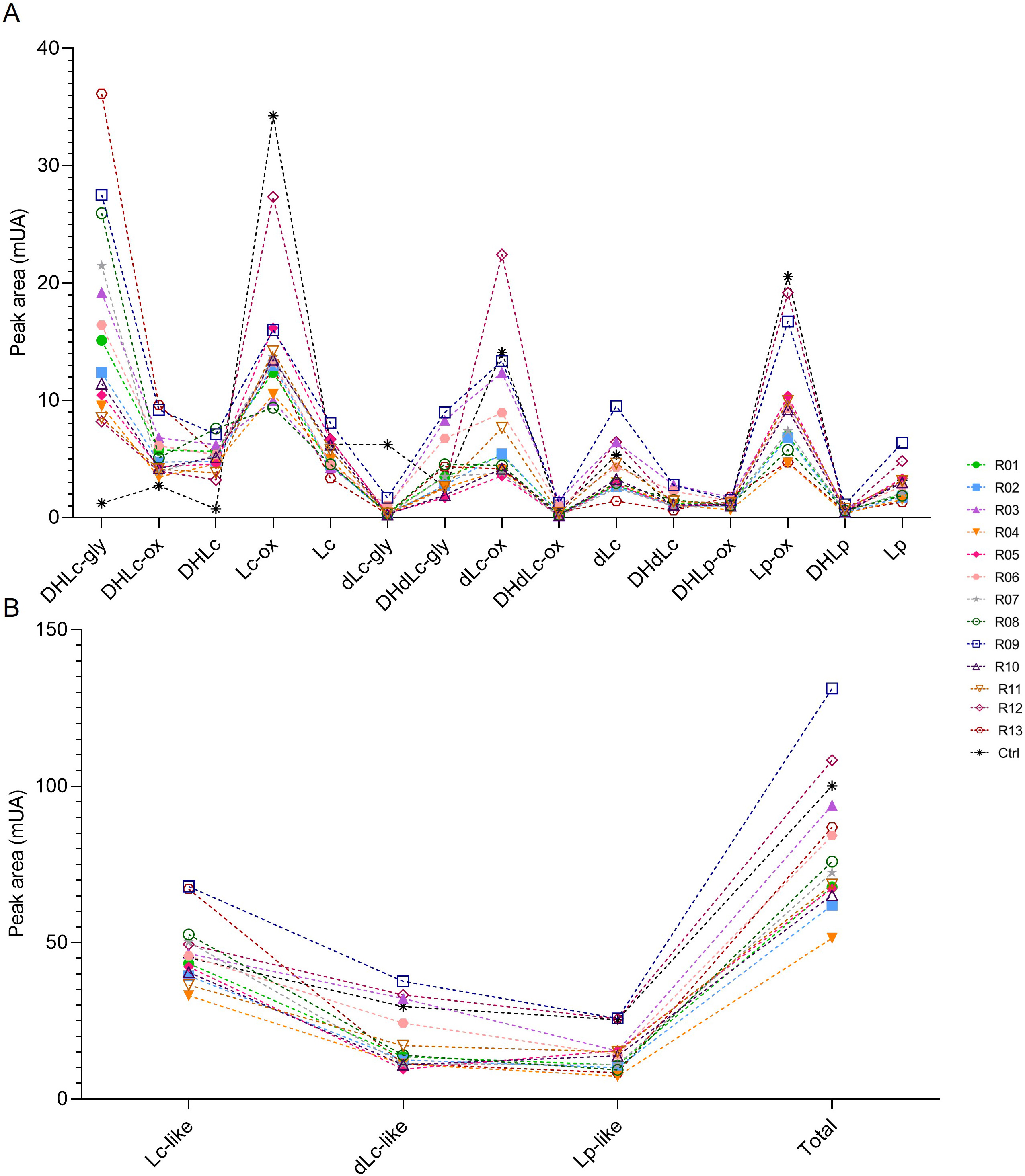 Fig. 4.
Fig. 4.STL content in roots of chicories according their peak area (mUA) obtained by HPLC analysis. (A) represents the distribution of each STL into the roots of chicories. (B) represents the STL levels of each chicories according to their structural group (Lc-like, dLc-like or Lp-like) and the total STL content of them. R01 to R13 are field-grown chicories used for sensorial analysis. Ctrl is wild-type ChicBitter002 grown under controlled condition. DHLc-gly, 11(S),13-dihydrolactucin-15-glycoside; DHLc-ox, 11(S),13-dihydrolactucin-15-oxalate; DHLc, 11(S),13-dihydrolactucin; Lc-ox, Lactucin-15-oxalate; Lc, Lactucin; dLc-gly, 8-deoxylactucin-15-glycoside; DHdLc-gly, 11(S),13-dihydro-8-deoxylactucin-glycoside; dLc-ox, 8-deoxylactucin-15-oxalate; DHdLc-ox, 11(S),13-dihydro-8-deoxylactucin-15-oxalate; dLc, 8-deoxylactucin; DHdLc, 11(S),13-dihydro-8-deoxylactucin; DHLp-ox, 11(S),13-dihydrolactucopicrin-15-oxalate; Lp-ox, Lactucopicrin-15-oxalate; DHLp, 11(S),13-dihydrolactucopicrin; Lp, Lactucopicrin. Lc-like, sum of all DHLc and Lc forms; dLc-like, sum of all DHdLc and dLc forms; Lp-like, sum of all DHLp and Lp forms; Total, sum of the 15 identified STLs.
A particularity of chicory hairy root lines is that they are able to regenerate spontaneous shoots. Thus, from the mutated hairy root lines we were able to regenerate whole plants that were grown in a greenhouse for 3 months. Applying the previously described extraction method, 3-month-old edited chicory roots (n = 3 for each HR lines selected) were analyzed to evaluate the impact of the CRISPR/Cas9 mutation on STL accumulation.
All the analyzed plants were derived from ChicBitter002 clone, with “Ctrl”
corresponding to WT plants and “Ctrl_HR” corresponding to R.
rhizogenes-infected chicory plants with no pYLCRISPR/Cas9P
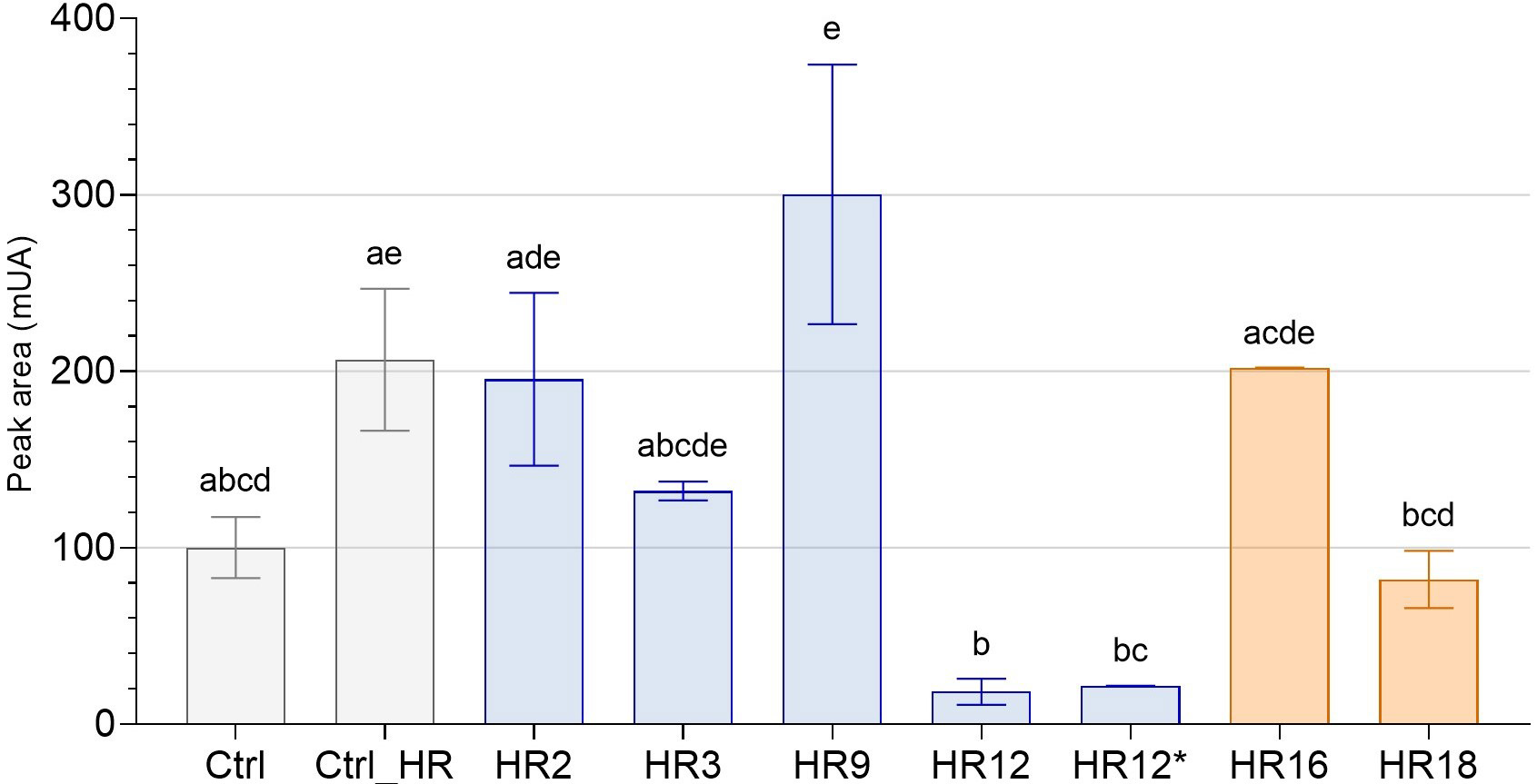
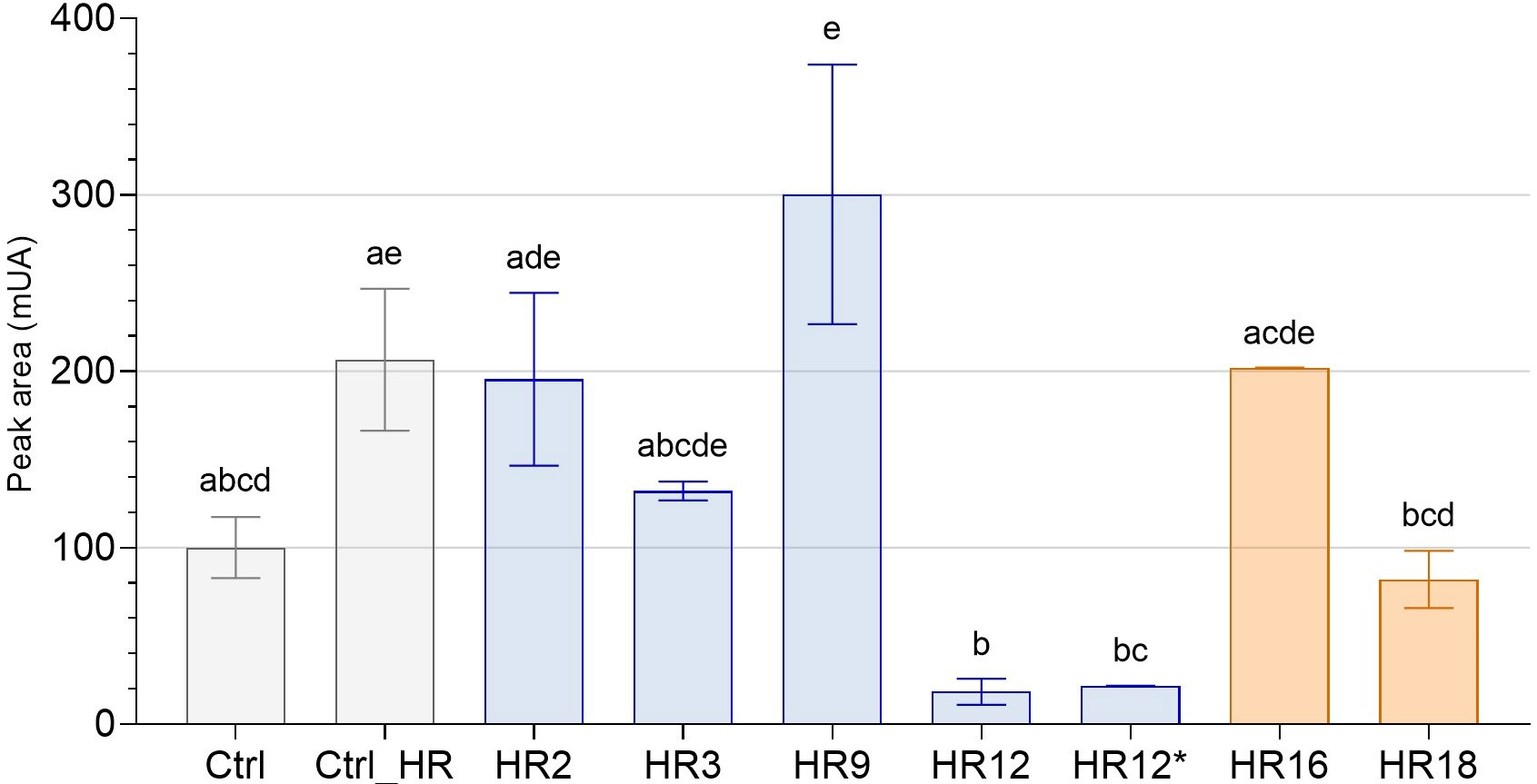 Fig. 5.
Fig. 5.Total sesquiterpene lactone content of roots of edited chicory
regenerated from HR lines by HPLC. The peak areas (mUA) of STLs in the roots of
7 genome edited chicory regenerated from HR lines were analyzed by HPLC and
compared to WT chicory lines (Ctrl and Ctrl_HR). Gray bars correspond to control
chicory lines, blue bars to CiGAS-short edited lines and orange bars to
CiGAO mutants. The histogram shows the modulation of total STL content
(sum of all identified STLs) as a function of mutation on chicory lines. HR12 and
HR12* are originally the same hairy root line except that HR12 is mutated for
only CiGAS-S1 and CiGAS-S2 and HR12* is mutated for
CiGAS-S1, CiGAS-S2 and CiGAS-S5. The letters indicate significantly different groups according to non-parametric one-way ANOVA and Dunn’s post-hoc test (p
For the CiGAS-short form mutants, 5 lines have been selected: line HR3, a monoallelic mutant which carries a mutation in a single allele of CiGAS-S1; line HR2, a heterozygous biallelic mutant for CiGAS-S1 where editing events induce a change in the coding frame; line HR9, a monoallelic mutant for both CiGAS-S1 and CiGAS-S2, meaning that only one allele is mutated in each of the two genes; line HR12, a heterozygous biallelic mutant for CiGAS-S1 and a homozygous biallelic mutant for CiGAS-S2, meaning there is a change in the coding frame in both of these gene copies (Supplementary Figs. 5,6), and line HR12* which is the same as line HR12 except that there is an additional homozygous biallelic mutation for CiGAS-S5 (Supplementary Figs. 5–7). For the CiGAO mutants, the 2 selected lines were: line HR16, a monoallelic mutant and line HR18, a heterozygous biallelic mutant with a change of the coding frame of its gene (Supplementary Fig. 8). The nucleotide sequences of these lines are described in Fig. 2.
By comparing the total STL content of all these lines with the “Crtl_HR”, only three lines showed significative reduction in STL content: HR12, HR12* and HR18. All three share biallelic mutations. All other CiGAS-short and CiGAO edited lines showed no significant difference in total STL content (Fig. 5). Most are monoallelic mutants, and one is a biallelic mutant only for one copy of the GAS gene (CiGAS-S1). Analysis of the STL content of a chicory edited for both alleles of CiGAS-S2 with a change in the coding frame was also performed and showed no significant difference, but we did not have enough biological replicates to include this result in our data. Regarding the CiGAS-short form mutants, both the HR12 and HR12* edited line showed similar profiles where the total STL content was strongly reduced (Fig. 5). This trend was confirmed in total free forms and total oxalate forms of STLs (Supplementary Fig. 9).
All these data suggest that the two copies (S1 and S2) of CiGAS-short are important for STL production in our plants and must be simultaneously KO to significatively reduce the STL content. The CiGAO gene also plays an important role in STL production, but the impact was not as great as for the two copies of CiGAS-short.
Sensory analysis was conducted on the root powder of thirteen field-grown
chicory plants (R01 to R13). Each chicory was assigned a bitterness score from 0
to 10, and panelists determined that R09 and R06 were the most bitter chicories
(score of 7.83 and 6.13, respectively) and R04 and R13 were perceived as the
least bitter chicories (score of 2.21 and 2.49, respectively) (Fig. 6,
Supplementary Table 2). First, a correlation between bitterness and each
STL of the field-grown chicories was sought to assess whether a single or
multiple compounds are involved in bitterness. Of the fifteen STLs identified in
this paper, six were not correlated with bitterness: DHLc-gly, DHLc-ox, DHLc,
Lc-ox, dLc-ox and, DHLp-ox (Supplementary Table 3). The remaining nine
were more or less correlated with an Pearson’s r ranging from 0.5587 to 0.9223
(Supplementary Table 3). Since bitterness cannot be attributed to a
single compound, it is assumed that all STLs can be considered indicative of
chicory bitterness. Next, the total STL content of each tasted chicory was
assessed and a linear regression analysis was carried out to estimate whether the
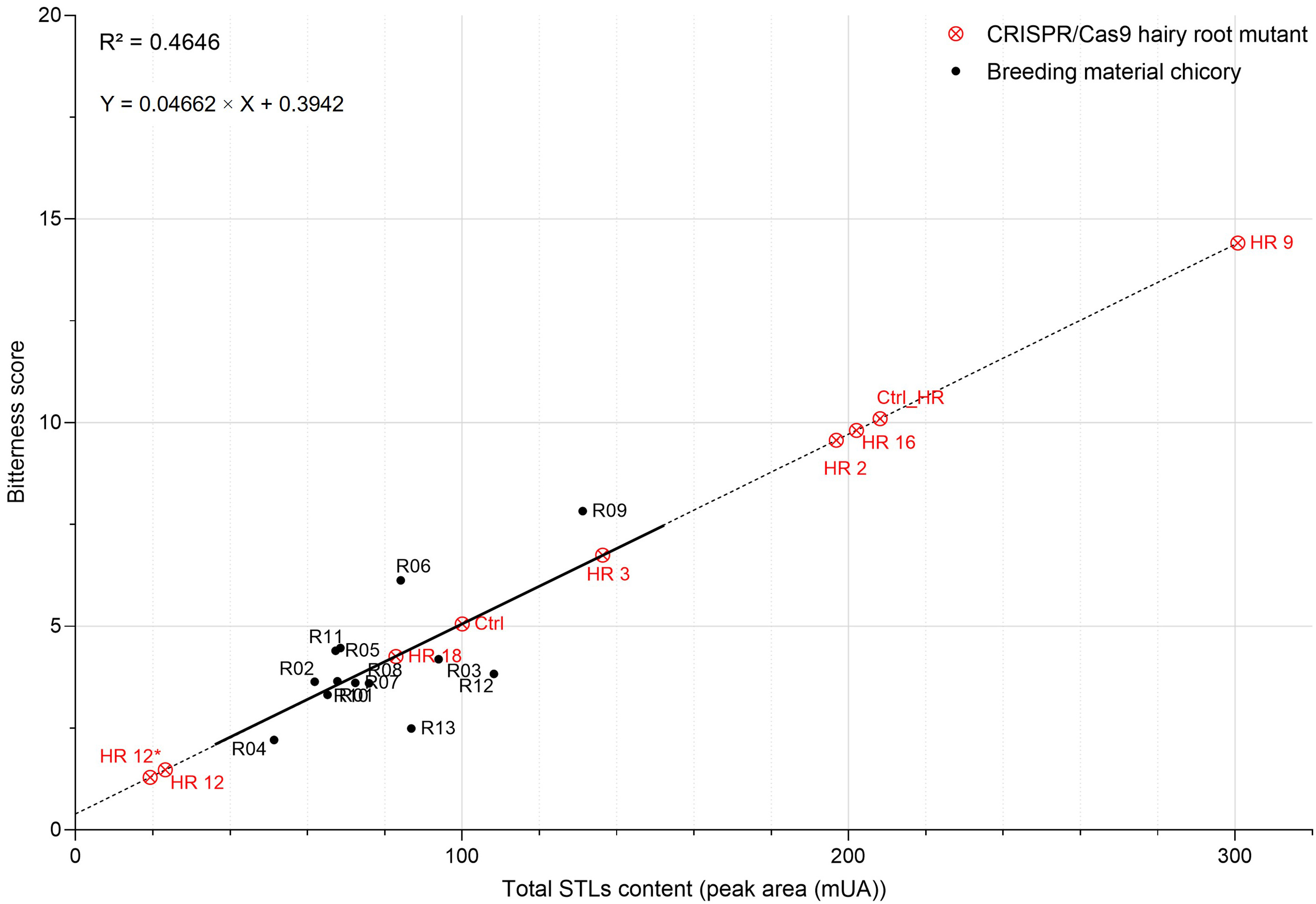
STL content can be correlated with the bitterness score (Fig. 6). A Pearson
correlation allowed to establish a positive correlation (R
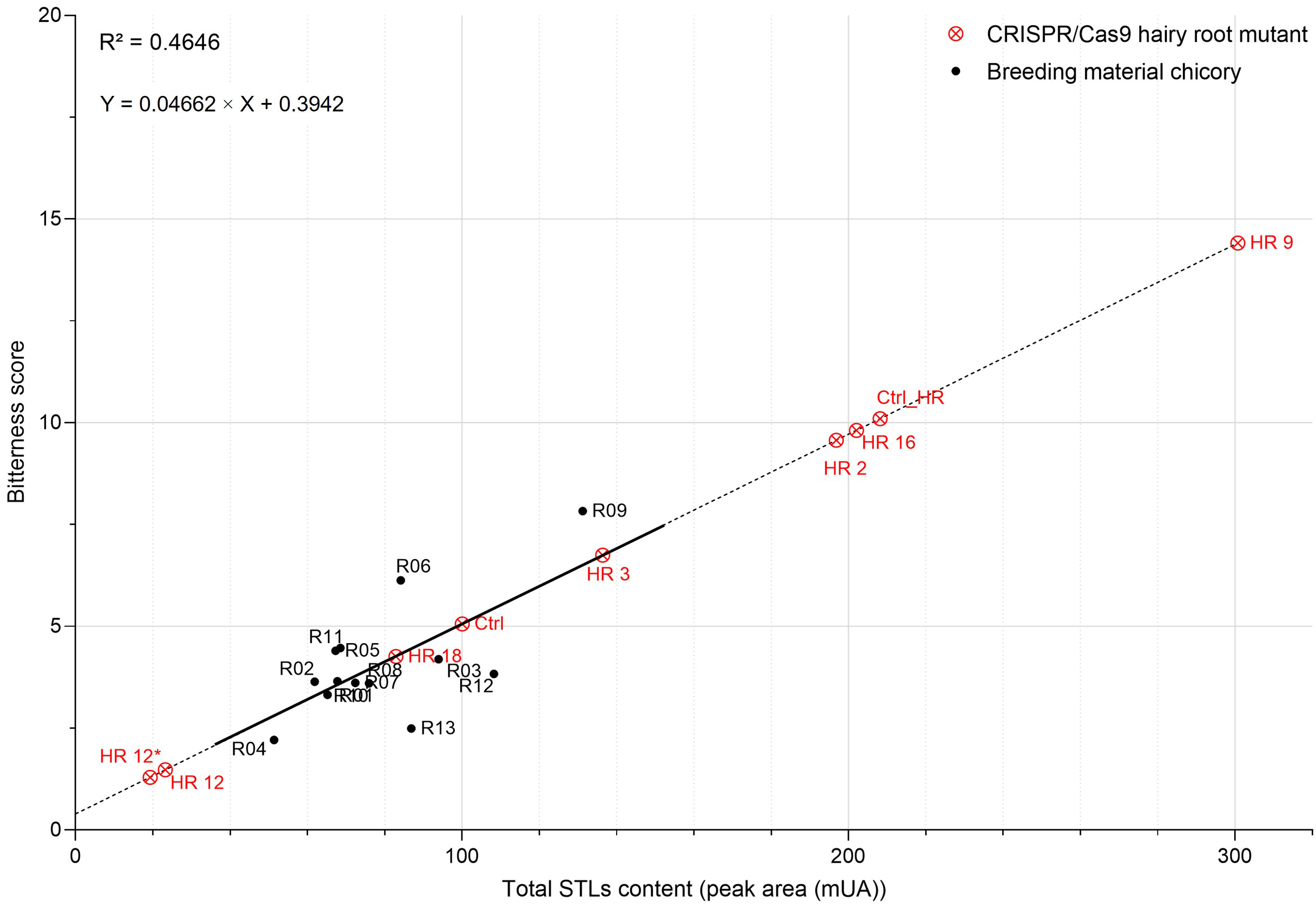 Fig. 6.
Fig. 6.Relationship between the total STL content and the bitterness
score of chicories. Positive correlation (r = 0.68) between these two parameters
was established using thirteen field-grown chicories (in black). CRISPR/Cas9
hairy root mutants (in red) were projected onto the linear regression curve using
the equation Y = 0.04662
It can be concluded that a complete KO of CiGAS-S1 and CiGAS-S2
genes could be sufficient to cause a drastic decrease in STL content that would
be reflected in the bitterness score. HR12 and HR12* edited chicory lines almost
lost their bitterness with a diminution of 86.35
In this work, CRISPR/Cas9 system was successfully used to inactivate CiGAS-S and CiGAO. As a result, a reduction in STL content and hence presumed bitterness of chicory were obtained.
In the past years, the CRISPR/Cas9 system has been already applied in C. intybus to abolish functions of numerous genes. The transformation methods used were either R. rhizogenes infection or protoplast transfection, both techniques having advantages and disadvantages [21, 25, 26, 27, 28, 29]. In this work, R. rhizogenes-mediated CRISPR/Cas9 editing was used for its efficiency (mutation frequency of 26%, calculated by the sum of mutant hairy root lines over the sum of total hairy root lines with binary vector), for its ability to generate a large amount of biological material and for the particularity of ChicBitter002 chicory HRs to spontaneously regenerate shoots as described in literature [41, 42]. HR-derived plants have a slightly modified chicory phenotype and can flower early without vernalization as it is required for WT plants [29]. The presence of T-DNA of R. rhizogenes also seems to activate the production of specialized metabolites as previously observed [43, 44, 45]. Indeed, an increase in STL content was observed in “Ctrl_HR” plants when compared to “Ctrl” plants (Fig. 5) and most of the HR mutant lines contained the same amounts of total STLs. However a significant decrease in total STL content was observed when biallelic mutants were obtained (HR12, HR12* and HR18). It is the case of the HR12 line, a biallelic mutant for both CiGAS-S1 and CiGAS-S2, who has a significant decrease in total STL content. These results are consistent with those published by Cankar et al. [28] where a decrease in STL content could be observed for bi-allelic mutants for at least two copies of CiGAS-S. However, this decrease in STL content is only significant when all 3 copies of GAS-S, or all copies of CiGAS-S and CiGAS-L are mutated, and not significant when only two copies of CiGAS-S are mutated (RN8 and RN10 plants), whereas in our study, the reduction is significant from the moment when two copies of CiGAS-S are mutated. Although only few mutants were analyzed in this article, in order to present a panel of mutation events, our results are slightly different from those of Cankar et al. [28] and would deserve analyses on a larger number of mutants. In addition, the HR12* line also showed a drastic decrease in total STL content. This line also has a biallelic mutation on both CiGAS-S1 and CiGAS-S2 but an additional mutation was observed on CiGAS-S5. However, no difference was observed between the total STL content of HR12 and HR12*. The explanation for this result could be that (1) the copy CiGAS-S5 was expressed at low in our growing conditions or in the root tissue, not compensating for the inhibition of CiGAS-S1 and CiGAS-S2, or was not expressed; or (2) the level of STLs in these HR lines was already very low and the effect of the mutation on CiGAS-S5 cannot be seen. To discriminate between these hypotheses, it could be considered to analyze the STL content of plants mutated only for CiGAS-S5 or, on the contrary, mutated for all CiGAS-short form except CiGAS-S5, or to realize qPCR in order to determine the expression levels of this copy but this could be difficult because the different copies of CiGAS-short share high sequence identity. A decrease was also observed for biallelic mutant for CiGAO (line HR18), but the impact was not as important as the decrease caused by the biallelic mutation of CiGAS-S1 and CiGAS-S2. Indeed, contrary to our expectations the production of STLs for HR18 line was not completely inhibited but only reduced by 60.1% compared to “Ctrl_HR”. This is the first time that the effect on STL production was described when CiGAO is mutated. The only partial reduction in STL content may suggest that other gene confers the ability to initiate STL synthesis, such as unidentified GAO-like genes or CYP71AV member genes that may perform the same function as GAO. In the literature, several expressed copies of CiGAO have been identified in C. endivia and only one functional gene has been described for CiGAO in C. intybus [17, 46, 47]. Given the existence of gene clusters for the CiGAS-S and CiKLS genes involved in the STL biosynthetic pathway, the hypothesis that multiple active copies of CiGAO would exist seems more than likely. However, it should be considered that CiGAO is a gene belonging to the large family of cytochromes P450, especially the CYP71AV subfamily [17, 48]. It has been shown that members of this subfamily can catalyze the conversion of several sesquiterpenes such as the valencene oxidase (CYP71AV8) which is able to convert germacrene A to its acid in vitro [18] or the CYP71AV9, a GAO gene identified in Cynara cardunculus, which is capable of partially converting (+)-germacrene D, cascarilladiene and amorpha-4,11-diene to their oxidized products [49]. Since many genes belong to this family, it is possible that some of them take over when CiGAO is inhibited. Taking these arguments into account, we can also hypothesize that if the CiGAS-S1 and CiGAS-S2 gene mutations have a greater impact on STLs content than the CiGAO gene mutation, this may be due to the fact that the CiGAS genes are terpene synthases that appear to have greater substrate specificity than cytochrome P450. Therefore, it can be assumed that even if other terpene synthases are present in the genome, they do not appear to compensate for the loss of several copies of CiGAS-short, resulting in greater inhibition of STLs synthesis.
In this work, additional information on the players in the STL biosynthesic pathway have also been provided. Bogdanović et al. [16] had previously described the genomic organization of the CiGAS genes with the identification of four copies of CiGAS-short form and a single gene for CiGAS-long form. The analysis of the industrial chicory ChicBitter002 genome carried out in this work enabled the identification of an additional copy named CiGAS-S5. However, the action of this newly identified gene copy on root STL content could not be confirmed in C. intybus. Over the last ten years, it has been shown that genes involved in the same biosynthetic pathway are sometimes colocalized in one region of the genome in several plants [50]. This gene clustering may cover a complete or near-complete biosynthetic pathway, or it may only be partial, involving a cluster of 2 or 3 genes encoding enzymes from 2 or 3 consecutive steps in a biosynthetic pathway [51]. For the biosynthetic pathway of STLs, it has been previously published that CiGAS-S1, CiGAS-S2 and CiGAS-S3 were mapped on the same linkage group (LG3), confirming an initial localization of one CiGAS-short form gene in the genetic reference map for chicory [16, 52]. In our case, we found the same data with the first three copies of CiGAS-short form colocalized with CiGAO on the same contig (Supplementary Fig. 3). Therefore, we physically established the colocalization of 3 active copies of CiGAS-short form genes and one active copy of the CiGAO gene, two genes involved in the consecutive enzymatic transformation of FPP into germacrene A for the first step and to germacrene A to germacrene A acid for the second step. Moreover, preliminary Blastp alignements of our CiGAS-S5 sequence to the nr database (Asterid section) under the NCBI site have indicated one candidate sequence (Protein_id=KAI3765470.1) from the C. intybus genome of Fan et al. [53], which is different from the sequence revealed with 3 CiCSAS-S1, S2 and S3 as entries (Protein_id=KAI3781194.1). According to the NCBI database informations, both sequences are on separate linkage group localizations, LG3 and LG2 for KAI3765470.1 and KAI3781194.1, respectively. Further, KAI3765470.1 (supposed CiGAS-S5 locus) is located to 60.1kb of a region with two predicted sequences (KAI3765465.1 and KAI3765466.1) with strong sequence homologies with our CiGAS-S4b, including the interruption in the exon 3 (data not shown). Overall, there is some evidence that CiGAS-S5/CiGAS-S4b, which were on a separate contig from the 3 CiGAS-S1 to CiGAS-S3 copies according to our data, are actually on different chicory chromosomes.
With the goal of using chicory as a functional ingredient, bitterness modulation represents a great importance for manufacturers and STLs are known to contribute predominantly to this bitterness in C. intybus [8, 9, 10]. In our work, the determination of the STL content was performed using an extraction mimicking food consumption condition and we considered that the total STL content was a good marker to assess bitterness, as confirmed by the correlation established in our sensory analysis. In fact, studies combining sensory analysis and identification of the most bitter compound are not always in agreement: on the one hand, Price et al. [8] consider lactucin glycoside to be the most bitter STL, while on the other hand, Van Beek et al. [9] states that 11(S),13-dihydrolactucopicrin is the most bitter due to its very low perception threshold. Furthermore, the various studies conducted on the characterization of STLs do not establish that a particular abundance of any form of STLs can influence the perception of bitterness. For example, Ferioli et al. [54] reported that glycosylated STLs were the most abundant forms in chicory, accounting for an average of 60% of the total STL content, and Graziani et al. [14] claimed that oxalated forms were the least abundant STLs in chicory, with the concentration of 11(S),13-dihydrolactucopicrin-15-oxalate accounting for about 0.2 to 2% of the total content of STLs. These data were contradicted by Kips [55] who reported that oxalate forms were the most abundant STLs in chicory, which was confirmed by Bogdanović et al. [29] and Twarogowska et al. [56] who found a two-to-four-fold higher oxalate content than the other STLs, apart for 11(S),13-dihydrolactucin and 11(S),13-dihydro-8-deoxylactucin. In the end, there is no consortium on the most bitter compound, but it is accepted that all STLs contribute strongly to bitterness. These inconsistencies may be due to various reasons such as cultivar, growing conditions and techniques, storage conditions and duration, or the method of drying or extraction of the chicory root. In addition, in most papers, chicory root extracts are hydrolyzed or undergo enzymatic treatment to release the bound forms of STL and allow the evaluation of a total content that better correlates with sensory bitterness as the STL content pool is closer to what the consumer perceives when chicory products are consumed [16, 46, 57, 58].
In addition of their involvement in bitterness, STLs play a role in plant defense such as antibacterial or antifungal activities. The lettucenin A, a STL of lettuce, has been shown to have antimicrobial activity against Bremia lactucae, Botrytis cinerae and Pseudomonas syringae phytopathogens [59] and the study of Wedge et al. [60] showed the antifungal activity of 6 STLs against Colletotrichum acutatum, Phytophtora fragariae, Phomopsis sp. and B. cinerae phytopathogens. To date, no study has tested the antimicrobial activity of chicory root extracts against phytopathogens but only against human pathogenic strains. However the presence of hydroxycinnamic acids, in particular chlorogenic acid and isochlorogenic acid, in large quantities in chicory roots do not allow to conclude the involvement of STLs [61, 62, 63]. STLs are also known for their antiparasitic activities [64], and for this purpose, chicory is used as a forage plant. Since these chicories require high levels of STLs, CRISPR/Cas9 could also be used not to reduce bitterness but rather to increase the levels of STLs, perhaps by acting directly on transcription factors.
Interest in using C. intybus as a functional ingredient has increased in the last few years and chicory flour has recently been shown to have multiple health benefits [5, 6]. Many articles have focused on the use of chicory root as a functional ingredient in food products such as yogurts and biscuits but also as chicory flour to be added to crackers, cakes and bread [7, 65, 66, 67, 68, 69, 70]. However, the amount of chicory used in these products never exceeds 5% because its bitterness causes a rejection by consumers. Several debittering methods have been investigated in chicory over the past years. For example, a change in the growing conditions can be responsible for a less bitter chicory taproot, chicory roots can be bleached or soaked into water for several hours, or low bitter taproot chicories can be bred by classical breeding approach [56, 71, 72]. However genetic engineering can also be used in chicory to reduce STL content in Cichorium intybus L. CRISPR/Cas9 is a useful tool for elucidating gene function that has been used to help breeding and to improve crops such as maize, rice, or wheat [23, 73, 74, 75, 76]. In this work, we successfully used the CRISPR/Cas9 system to inactivate CiGAS-short form and CiGAO genes and showed that biallelic mutations are required to reduce STL content. Given the limitations of current European legislation on the use of CRISPR/Cas9-modified plants, it was not possible to perform a sensory analysis and establish a direct link between bitterness and the amount of STLs in HR chicory lines. Therefore, an indirect link was established by performing sensory analysis of thirteen field-grown chicories, which allowed us to define a bitterness scale based on STLs content and to establish a theoretical bitterness score for the HR mutant lines. We identified three HR mutant lines with significantly reduced STLs content: the HR12 and HR12* lines, that have a theoretical bitterness score between 1 and 2 meaning that bitterness is almost no more perceived, and the HR18 line which has a bitterness score decreased by 57.8% compared to the “Ctrl_HR” which is much more bitter than industrial chicories grown in fields because of the integration of T-DNA in ChicBitter002. Using the CRISPR/Cas9 tool, we were able to modulate the bitterness of C. intybus and identify the genes responsible for this. To our knowledge, this is the first time that the effect of CRISPR/Cas9 edited chicory on bitterness can be assessed, even indirectly. The new CRISPR/Cas9-based biotechnology has proven once again to be more efficient and accurate, less energetic and less expensive in time than the other techniques of debittering. As plants obtained by using CRISPR/Cas9 technology are considered to be genetic modified organisms by the current European legislation they cannot be used directly but will provide useful data to breeders to develop and propose less bitter varieties of chicory by molecular marker-assisted breeding. The mutants are also an excellent research material to study the biological activities of STLs.
The witloof sequence used in the identification of the sequences in the current article, also called Cargese sequence, was obtained in the framework of the Cargese program (2012–2014). It aimed to obtain an annotated chicory sequence through a partnership of 6 private companies (Rijk Zwaan, Enza Zaden, Bejo, Vilmorin, Hoquet Endives, Florimond Desprez) and the CEA (Genoscope, Commissariat à l’Energie Atomique, Evry, France). The RNA-seq data were obtained as part of the Qualichic program (2012–2016), a result of a collaboration between the University of Lille and the company Florimond Desprez.
COS, costunolide synthase; FPP, farnesyl-pyrophosphate; GAO, germacrene A oxidase; GAS, germacrene A synthase; HR, hairy root; KLS, kauniolide synthase; KO, knockout; LG, linkage group; sgRNA, single-guide RNA; STL, sesquiterpene lactone; WT, wild-type.
The genome of chicory ChicBitter002 used during the current study are not publicly available due to their belonging to private consortium. The protein sequences of GAS-S and GAO analyzed during the current study are available under the accession number given in this paper.
TC, PH and CR designed the research study. JD, MT, AE and HADS performed the research. JD and TC analyzed the genomic data. MT and PH analyzed the sensorial data. JD and PH performed the statistical analysis. JD and CR conceptualized and drafted the manuscript. DG and JLH contribute to the conception of the work, and to the analysis, the interpretation of data for the work. JD, TC, PH, DG, and CR reviewed and edited the manuscript. CR supervised the project. JLH and CR provided fundings and administrated the project. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
The study was conducted in accordance with the Declaration of Helsinki and informed consent was obtained from all subjects involved in the study.
We would like to thank Florimond Desprez SA for providing chicory products and access to the chicory ChicBitter002 genome database and the agrifood platform of Polytech Lille for the access to the laboratory rooms used for sensory analysis.
This work has been supported by Région des Hauts-de-France via FEDER for CHIC41H 2019-2021 programs and via the Alibiotech CPER program. JD was supported by a doctoral fellowship from a convention ANRT/Florimond Desprez SA.
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/j.fbl2809201.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.