1. Introduction
Ovarian cancer is characterized by a high prevalence and high mortality rate and
ranks as the second leading factor causing gynecologic cancer-related deaths in
women worldwide [1]. Platinum-based chemotherapy is still the main treatment for
advanced ovarian cancer [2]. Although ovarian cancer mortality has declined in
recent years [3], ovarian cancer stemness, chemoresistance, and metastasis are
common reasons for treatment failure and poor patient prognosis [4]. Therefore,
identifying novel targets to overcome these tumorigenic properties is now an
urgent priority for this disease.
N-methyladenine (mA) RNA methylation is the most abundant RNA
modification type in non-coding RNAs and mRNAs and can regulate downstream
effectors to affect ovarian cancer progression [5]. The mA RNA methylation
process is mediated by many proteins including methyltransferases, demethylases,
and mA readers [6]. Previous studies have suggested that mA
regulator-mediated methylation modification patterns can affect cancer
progression [7, 8]. Copy number variation (CNV) gains are associated with an
increase in gene expression, whereas CNV deletions are correlated with a decrease
in gene expression. Moreover, advanced-stage cancers harbor more CNV events of
mA regulators [9], and patients with CNV gain of
N6-adenosine-methyltransferase 70 kDa subunit (METTL3) have a poor prognosis
[10]. METTL3 functions as a methyltransferase and plays an oncogenic role in
ovarian cancer [11, 12]. However, the dysregulation of downstream effectors of
these mA regulators remains largely undetermined.
Prostaglandin E2 receptor (subtype EP2), namely PTGER2, belongs to the G
protein-coupled receptor superfamily. The activation of PTGER2 results in the
increased activity of cAMP-dependent signaling [13]. Previous studies have
revealed that PTGER2 acts as an oncogene in colorectal cancer [14] and prostate
cancer [15] and serves as a tumor suppressor in neuroblastoma [16]. However, the
biological role and clinical significance of PTGER2 and the mechanism of PTGER2
dysregulation in ovarian cancer remain unclear.
Here, we elucidated the role of mA modification in ovarian cancer and
identified specific genes modulated by mA regulators using a public cancer
database. In particular, we focused on the mechanism by which METTL3-mediated
mA modification of PTGER2 mRNA results in its upregulation, and explored
the impact of these aberrant interactions on ovarian cancer cell stemness,
chemoresistance, proliferation, and metastasis. Our work further suggests the
potential application of PTGER2 as a reliable prognostic predictor and a novel
therapeutic target for ovarian cancer treatment by using ovarian cancer cell
lines and clinical samples.
2. Materials and Methods
2.1 Bioinformatics Analysis
Bioinformatics analysis was performed according to our previous study [17].
The genomic data, mRNA expression profiles, and clinical data of The Cancer
Genome Atlas (TCGA) ovarian serous cystadenocarcinoma (OV) database were
downloaded for conducting differential analyses, survival analyses, correlation
analyses, unsupervised clustering analyses, gene set variation analysis (GSVA),
and gene set enrichment analysis (GSEA). The gene set representing platinum drug
resistance was constructed using data obtained from the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database as previously reported [18]. Based on the best
cut-off value obtained from survival analysis, samples were assigned as low and
high expression groups.
2.2 Cell Cultures
Ovarian cancer cell lines OV90 and SKOV3 with short tandem repeat verification
were purchased from American Type Culture Collection (Manassas, VA, USA) and
cultured in Dulbecco’s Modified Eagle Medium (DMEM) harboring 10% fetal bovine
serum (FBS) at 37 °C with 5% CO. It was confirmed that the cell
lines did not have mycoplasma contamination.
2.3 Cell Transfections
Single hairpin RNA (shRNA) negative control and shRNA targeting PTGER2
(sh-PTGER2) were constructed and inserted into lentiviral particles by GeneChem
Corporation (Shanghai, China). Small interfering RNA (siRNA) against METTL3
(si-METTL3) was synthesized by RiboBio Corporation (Guangzhou, China). Polybrene
and Lipofectamine 3000 were used for lentivirus particle transfection and siRNA
transduction, respectively. After transfection or transduction for 48–72 h,
ovarian cancer cells were subjected to further investigation.
2.4 RT-PCR and Quantitative PCR
Total RNAs of ovarian cancer cells were used for reverse transcription with a
kit purchased from TaKaRa Corporation (Dalian, China). RT-PCR and quantitative
PCR (qPCR) were conducted with the generated cDNAs and specific primers. Bio-Rad
GelDoc XR+ (Hercules, CA, USA) was applied to capture images for RT-PCR, and the
2 method was used to determine the relative
expression of mRNA for qPCR.
2.5 Western Blot Analysis
Total proteins of ovarian cancer cells in lysis buffer were quantified by the
BCA method as previously described in our investigation [19]. Then the separated
and transferred proteins were incubated with antibodies against the following
proteins: METTL3, PTGER2, Myc, cyclin D1 (CCND1), vimentin, and -actin.
The chemiluminescence method was adopted for protein detection, and the Bio-Rad
GelDoc XR+ was applied to record images.
2.6 Methylated RNA Immunoprecipitation
The Methylated RNA Immunoprecipitation (MeRIP) mA Kit
purchased from RiboBio Corporation (Guangzhou, China) was used to conduct MeRIP
as previously described in our investigation [8]. Briefly, total RNAs of ovarian
cancer cells were used to generate fragments that were pulled down with magnetic
beads harboring the mA antibody. Enriched RNA was purified and used for
RT-PCR and qPCR.
2.7 Luciferase Reporter Assay
PmirGLO luciferase reporter vectors (Promega Co., Madison, WI, USA) containing
wild-type or mutant 3’-untranslated region (UTR) of PTGER2 were transfected into
ovarian cancer cells. After transfection for 48 h, the luciferase activity in
cells was measured to evaluate the effects of METTL3 on PTGER2 transcriptional
levels on the BioTek Luminometer Synergy H1 (Winooski, VT, USA) using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
2.8 Tumorsphere Formation Assay
Ovarian cancer cells seeded on 6-well ultra-low-attachment plates were incubated
in serum-free DMEM/F12 (20 ng/mL epidermal growth factor, 20 ng/mL fibroblast
growth factor, and 2% B27). The images of tumorspheres were captured, and the
number of tumorspheres was counted after a 14-day culture. Three continuous
generations of tumorspheres number were calculated for analysis.
2.9 Immunofluorescence
Ovarian cancer cells seeded on coverslips were treated with paraformaldehyde
(4%) for fixation, incubated with Triton X-100 (0.2%) for permeabilization, and
subjected to incubation with antibodies against the following proteins: cluster
of differentiation 44 (CD44), CD133, and gamma-H2A histone family member X
(H2AX). DAPI was used to co-stain the cells, and the images of stained
cells were captured with a fluorescence confocal microscope.
2.10 Colony Formation Assay
To detect cell proliferation and chemoresistance, ovarian cancer cells were
subjected to treatment with carboplatin for 6 h at the indicated concentrations
and then seeded in wells for 10 days. The generated colonies were subjected to
fixation, staining, and recording for analysis.
2.11 Transwell Assays
The migration and invasion ability of ovarian cancer cells were detected by
transwell assays. For the migration assay, serum-free cells were seeded in the
upper chamber of a transwell without Matrigel coating. For the invasion assay,
serum-free cells were plated in the upper chamber of a transwell with Matrigel
coating. DMEM containing 10% FBS was added to the lower chambers of a transwell
were for both the migration and invasion assays. The migrated and invaded cells
were subjected to fixation, staining, and recording for analysis.
2.12 Patient Tissues
One hundred and fifty-eight ovarian cancer specimens described in our previous
study [19] were collected for analysis. All specimens had a pathological
diagnosis. Patient written consent and ethics approval from the Ethics Committee
of the hospital were obtained (K2020-036-01). All procedures strictly adhered to the policies
approved by the Institutional Ethics Committee and Declaration of Helsinki.
2.13 Immunohistochemistry
Immunohistochemistry (IHC) and staining evaluation were carried out as
previously described [19]. Paraffin-fixed sections from ovarian cancer specimens
were deparaffinized and rehydrated before antigen retrieval in citrate buffer.
Then the sections were subjected to the eradication of endogenous peroxidase
activity and blocking of non-specific antigens. After incubation with anti-PTGER2
antibody, the sections were treated with Diaminobenzidine substrate and subjected to the
evaluation of staining intensities. For statistical analysis, a score of 7–12
was assigned as high expression, and a score of 0–6 was considered low
expression.
2.14 Statistical Analyses
SPSS 22.0 (IBM SPSS Inc., Chicago, IL, USA) or RStudio 9.0 was applied for analysis
of the data expressed as the mean standard deviation from three
independent experiments. The Wilcoxon rank-sum test and Student’s two-tailed
t-test were used to test the statistical significance between two
groups. One-way analysis of variance was applied to reveal the statistical
significance among multiple groups. Spearman’s rank correlation test and
Chi-square test were applied for the correlation analyses. Survival analysis was
performed by plotting Kaplan-Meier survival curves with log-rank tests.
Univariate and multivariate Cox regression models were applied to identify the
correlation between PTGER2 expression and the overall survival of patients with
ovarian cancer. Statistical significance was achieved with p 0.05.
3. Results
3.1 Bioinformatics Analyses Indicate the Important Role of mA
RNA Methylation in Ovarian Cancer Progression
To explore the role of mA regulators in ovarian cancer
progression, we adopted 9 mA writers (including Cbl Proto-Oncogene Like 1
[CBLL1], METTL3/14/16, RNA-binding motif protein 15/15B [RBM15/15B], Vir-like
mA methyltransferase associated, WT1-associated protein [WTAP], and zinc
finger CCCH-type containing 13 [ZC3H13]), 2 mA erasers (AlkB homolog 5, RNA
demethylase [ALKBH5] and fat mass and obesity-associated protein), and 15
mA readers (including eukaryotic translation initiation factor 3 subunit A,
ELAV like RNA-binding protein 1 [ELAVL1], fragile X messenger ribonucleoprotein 1
[FMR1], heterogeneous nuclear ribonucleoprotein A2/B1 [hnRNPA2B1], hnRNPC,
insulin-like growth factor 2 mRNA-binding protein 1/2/3 [IGF2BP1/2/3], leucine
rich pentatricopeptide repeat containing, YTH domain-containing protein 1/2
[YTHDC1/], YTHDF1/2/3 and RNA-binding motif protein X-linked), which are
dysregulated in human cancers. Since the deletion or amplification of gene copy
number affects the corresponding gene expression and is regarded as the trigger
of cancer development [8], we applied bioinformatics analyses based on TCGA OV
database to identify CNVs in these 26 mA regulators and found that 23
mA regulators contained CNVs in ovarian cancer (Supplementary Fig.
1). Then the 23 CNV-carrying genes in TCGA database were used to continue our
analysis of the clinical significance of these mA regulators in ovarian
cancer. Gene interaction network analysis suggested that the expression of
ALKBH5, CBLL1, ELAVL1, FMR1, HNRNPA2B1, HNRNPC, IGF2BP2, METTL14, METTL16,
METTL3, YTHDC2, YTHDF2, and WTAP was correlated with the overall survival of
patients with ovarian cancer and indicated a strong correlation among the
expression of 23 mA regulators (Supplementary Fig. 2A),
suggesting these dysregulated mA regulators as contributors to ovarian
cancer development and progression.
To further examine how the 23 mA regulators exert their clinical
significance in ovarian cancer, we performed unsupervised clustering and
identified two distinct patterns of mA modification in TCGA OV patients
according to the expression of 23 mA regulators (Supplementary Fig.
2B). Survival analyses showed the superior prognosis of patients with ovarian
cancer in cluster A patterns than in cluster B patterns (Supplementary
Fig. 2C). Differently enriched Gene Ontology (GO) and KEGG terms between these
two clusters were elucidated by conducting GSVA. The analysis verified that
cluster B was involved in RNA processing (Supplementary Fig.
2D) and cancer-associated signaling pathways (Supplementary Fig. 2E), such as the Notch and Wnt pathways, as well as DNA
replication, cell cycle, base excision repair, nucleotide excision repair,
homologous recombination, non-homologous end joining, and mismatch repair
compared to cluster A patterns, further indicating the critical role of mA
modification in ovarian cancer progression.
3.2 PTGER2 Expression is Associated with Aberrant mA RNA
Methylation and Functions as a Tumorigenic Gene in Ovarian Cancer
To identify specific genes that could be affected by mA RNA methylation in
ovarian cancer, we carried out differential expression analyses to compare the
identified cluster A with cluster B and elucidated 492 differentially expressed
genes (fold change 2.0, p 0.05). Then we integrated and
analyzed the differential expression data with another database to identify
specific genes participating in ovarian cancer progression. First, we calculated
the index of platinum drug resistance by GSVA in TCGA OV patients and performed
correlation analyses to elucidate genes associated with chemoresistance (Geneset
A). Furthermore, we conducted survival analyses to reveal genes correlated with
the prognosis of TCGA OV patients (r 2.0,
p 0.05) (Geneset B). In addition, we extracted genes potentially
modified by mA RNA methylation as suggested by the mA-Atlas database
with experimental evidence (Geneset C). Interestingly, the intersection of these
three genesets only identified PTGER2 as an overlapping gene (Fig. 1A).
 Fig. 1.
Fig. 1.
PTGER2 expression serves as an mA RNA methylation-mediated
oncogene in ovarian cancer as per The Cancer Genome Atlas (TCGA) database. (A) Venn diagram showing genes
correlated with chemoresistance, mA modification, and patient prognosis in
ovarian cancer. (B–H) The associations among Myc, CD44, CD133, L1CAM, p53,
CCND1, vimentin, and PTGER2 expression. (I,J) Gene set enrichment analysis (GSEA) of PTGER2 showing enrichment
of dysregulated pathways and processes in Hallmark (Top 30) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Top 30).
(K) Comparison of PTGER2 expression in ovarian cancer patients with or without
distant metastasis. (L) The Kaplan-Meier curve delineating the overall survival
of patients with ovarian cancer based on PTGER2 expression.
Subsequent correlation analyses revealed a positive correlation between PTGER2
expression and the expression of Myc, CD44, CD133, L1 cell adhesion molecule
(CAM), CCND1, and vimentin, which serve as stemness, proliferation, and
epithelial-mesenchymal transition (EMT)-associated markers, and showed a negative
association between PTGER2 expression and expression of the DNA damage
repair-associated marker p53 (Fig. 1B–H). Moreover, GSEA confirmed that PTGER2
positively participated in oncogenic processes and pathways such as DNA repair,
E2F targets, EMT, Myc targets, Wnt/-catenin signaling, CAMs, focal
adhesion, and the extracellular matrix receptor interaction (Fig. 1I,J).
Importantly, PTGER2 expression was upregulated in ovarian cancer patients with
distant metastasis compared to those without distant metastasis (Fig. 1K), and
high PTGER2 expression predicted poor patient prognosis in TCGA (Fig. 1L). These
findings collectively suggest PTGER2 as a potential oncogene in ovarian cancer.
3.3 METTL3-Mediated mA Modification Elevates PTGER2
Expression
Based on the findings of the above-mentioned bioinformatics analyses, we further
investigated whether PTGER2 expression is modulated by mA modification.
PTGER2 was potentially modified by METTL3, METTL14, and IGF2BP1 in the m6A2Target
database (http://m6a2target.canceromics.org/). Since our above survival analysis
suggests METTL3 as a risk factor for the prognosis of ovarian cancer patients and
PTGER2 serves as a potential oncogene, we explored the impact of METTL3 on PTGER2
and found that silencing METTL3 downregulated the mRNA and protein levels of
PTGER (Fig. 2A–C). Moreover, mA individual-nucleotide-resolution
cross-linking and immunoprecipitation and mA MeRIP-sequencing data
collected in the RMVar database (http://rmvar.renlab.org) revealed three
potential mA modification sites in the coding sequence (CDS) and 3 UTR
of PTGER mRNA (Fig. 2D). Considering the location relationship of these three
sites, we synthesized two pairs of primers covering site 1, and sites 2 and 3,
respectively. Furthermore, mA MeRIP analyses validated that PTGER mRNA was
immunoprecipitated by anti-mA antibody and could be detected by qPCR with
the primers covering sites 2 and 3, but not site 1 (Supplementary Fig.
3). Moreover, the decrease in mA modification of PTGER mRNA was detected
in METTL3-silenced ovarian cancer cells (Fig. 2E,F). To further elucidate the
site responsible for mA modification of PTGER mRNA, we constructed plasmids
harboring mutant sites 2 or site 3 (Fig. 2G). Luciferase reporter assays further
indicated that METTL3 knockdown can inhibit the transcription of wild-type PTGER,
whereas mutation at site 2 but not at site 3 abolished this suppression (Fig. 2H). These findings demonstrate that METTL3-mediated mA RNA methylation
contributes to controlling PTGER2 expression in ovarian cancer.
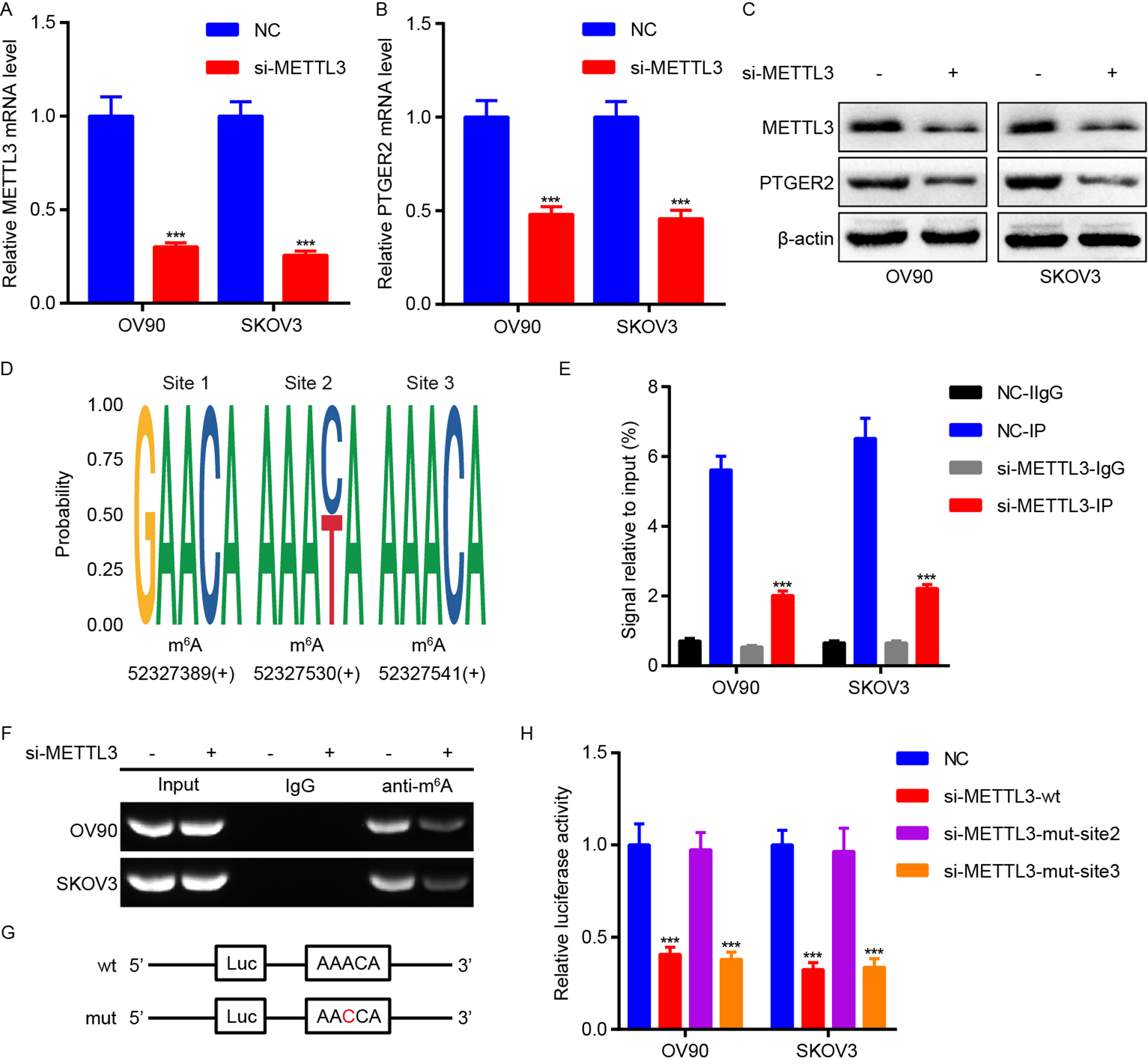 Fig. 2.
Fig. 2.
METTL3 is responsible for mA modification of PTGER2 mRNA.
(A) quantitative PCR (qPCR) analysis determining the efficiency of METTL3 knockdown in OV90 and
SKOV3 cells. (B) qPCR analysis showing the effect of METTL3 knockdown on PTGER2
mRNA expression in OV90 and SKOV3 cells. (C) Western blot analysis suggesting the
impact of METTL3 knockdown on PTGER2 protein expression in OV90 and SKOV3 cells.
(D) Bioinformatics analysis identifying mA modification sites within the
CDS and 3’UTR regions of PTGER2 mRNA. (E,F) mA MeRIP assays measuring the
influence of METTL3 knockdown on mA modification of PTGER2 mRNA in OV90 and
SKOV3 cells. The enriched RNAs were further subjected to qPCR (E) and RT-PCR (F).
(G) The sequences of wild-type and mutant mA modification sites. (H)
Luciferase reporter assays revealing the impact of METTL3 depletion on the
post-transcriptional inhibition of PTGER2 in OV90 and SKOV3 cells. p 0.001. NC, negative control.
3.4 PTGER2 Knockdown Inhibits Ovarian Cancer Stemness,
Chemoresistance, Proliferation, and Metastasis
According to the results obtained from the above bioinformatics analyses, we
determined the impact of PTGER2 on ovarian cancer cell stemness, chemoresistance,
proliferation, and metastasis. First, shRNA targeting PTGER2 (sh-PTGER2) was used
for PTGER2 silencing in ovarian cells. Then tumorsphere formation and
immunofluorescence assays showed that PTGER2 knockdown suppressed the stemness of
ovarian cancer cells and impaired the expression of stem cell markers CD44 and
CD133 in ovarian cancer cells (Fig. 3A–C). Colony formation assays indicated
that PTGER2 depletion inhibited the carboplatin resistance and proliferation of
ovarian cancer cells (Fig. 3D,E), and transwell assays measured the inhibition of
migration and invasion by PTGER2 knockdown in ovarian cancer cells (Fig. 3F,G).
Furthermore, immunofluorescence assays detected the upregulation of
H2AX by PTGER2 depletion in ovarian cancer cells treated with
carboplatin, indicating enhanced DNA damage (Fig. 3H). Moreover, the expression
of cell stemness, proliferation, and EMT-associated proteins including Myc,
CCND1, and vimentin were impaired in PTGER2-silenced ovarian cancer cells (Fig. 3I). These findings demonstrate that PTGER2 functions as an oncogene by
stimulating cell stemness, chemoresistance, proliferation, and metastasis.
 Fig. 3.
Fig. 3.
PTGER2 increases ovarian cancer stemness,
chemoresistance, proliferation, and metastasis. (A–G) Tumorsphere formation
(A,B), immunofluorescence (scale bar: 12.5 µm) (C), Clonogenic (D,E),
transwell (Scale bar: 50 µm) (F,G) assays were adopted for detecting the
stemness, chemoresistance, proliferation, and metastasis of PTGER2-depleted OV90
and SKOV3 cells and the controls. (H) Immunofluorescence analysis (scale bar: 8
µm) was applied to measure H2AX expression in PTGER2-depleted
OV90 cultured with carboplatin, PTGER2-depleted SKOV3 cells cultured with
carboplatin, and the controls. (I) Western blot analysis showing the expression
of proteins correlated with stemness and chemoresistance (Myc), proliferation
(CCND1), and metastasis (vimentin) in PTGER2-silenced OV90 and SKOV3 cells and
the controls. p 0.01, p 0.001 (compared
with the control group). p 0.01, p
0.001 (compared with the carboplatin group). NC, negative control.
3.5 PTGER2 Expression is Associated with the CLinical and
Pathological Characteristics of Patients with Ovarian Cancer
To gain further knowledge of the vital role of PTGER2 in the progression of
ovarian cancer, we performed IHC in 158 ovarian cancer tissues. IHC showed the
different levels of PTGER2 protein expression in the detected ovarian cancer
samples (Fig. 4A), and PTGER2 protein expression was upregulated in ovarian
cancer samples with metastasis compared to those without metastasis (p =
0.021) (Fig. 4B, Supplementary Table 1). However, PTGER2 protein
expression had no significant correlation with other clinical and pathological
characteristics. Survival analysis was performed and revealed
that high PTGER2 protein expression conferred poor overall
survival for patients with ovarian cancer (p = 0.009, log-rank test)
(Fig. 4C). Univariate analyses were subsequently conducted and elucidated the
positive associations between high PTGER2 protein expression, old age, advanced T
classification, advanced N classification, distant metastasis,
and poor overall survival of patients with ovarian cancer (Fig. 4D). Multivariate
analyses were performed and revealed that PTGER2 protein expression (HR = 1.095,
95% confidence interval [CI] 1.061–1.180, p = 0.027), T
classification, and lymph node metastasis served as independent and unfavorable
prognostic factors for patients with ovarian cancer (Fig. 4E).
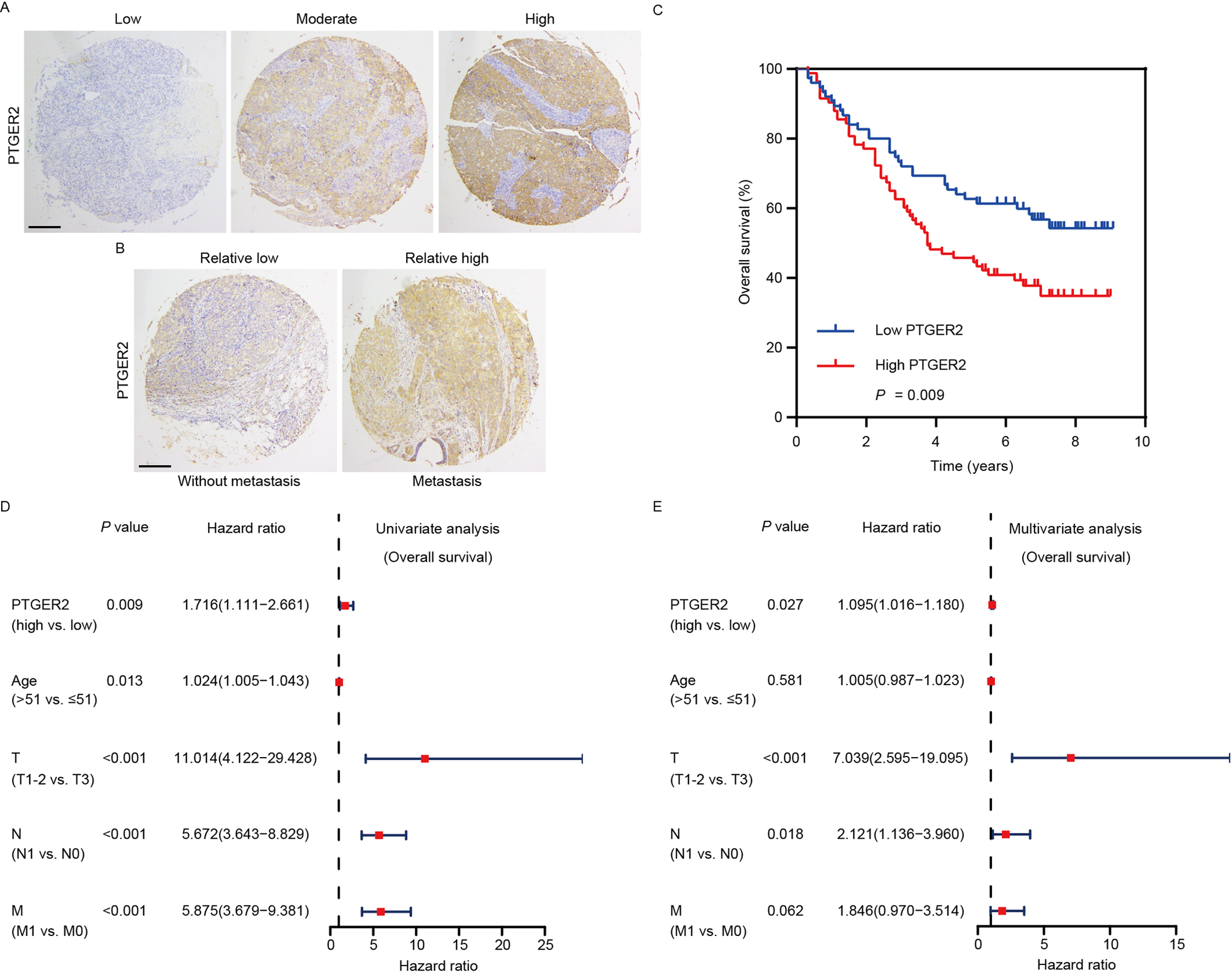 Fig. 4.
Fig. 4.
PTGER2 expression predicts the prognosis of ovarian
cancer patients. (A) Images representing the differential PTGER2 expression in
ovarian cancer tissues (scale bar: 125 µm). (B) Images representing the
differential PTGER2 expression between ovarian cancer patients with or without
distant metastasis (scale bar: 125 µm). (C) Kaplan-Meier curve displaying
the overall survival of patients with ovarian cancer based on PTGER2 protein
levels. (D) Univariate analysis was used to correlate PTGER2 protein levels,
clinicopathological parameters, and overall survival of patients with ovarian
cancer. (E) Multivariate analysis was conducted to correlate PTGER2 protein
levels, clinicopathological parameters, and overall survival of patients with
ovarian cancer.
4. Discussion
Developing new biomarkers for cancer diagnosis, treatment, and prognosis
evaluation are the key issues in cancer research [20, 21]. Stemness and
chemoresistance are responsible for proliferation and metastasis and are regarded
as contributors to poor prognosis of patients with ovarian cancer [19]. mA
modification plays a key role in cancer development and progression by regulating
many biological and pathological processes including cell stemness, drug
resistance, proliferation, and metastasis [22]. In this investigation, notable
links between mA modification and ovarian cancer progression were
identified using bioinformatics analyses. In addition, our study suggested that
METTL3-mediated mA RNA methylation modulated the expression of PTGER2,
which subsequently promoted ovarian cancer stemness, chemoresistance,
proliferation, and metastasis by affecting the expression of associated key
regulators. Importantly, high PTGER2 expression predicted the poor overall
survival of patients with ovarian cancer.
Distinct mA modification patterns can affect the prognosis of cancer
patients [7]. In this work, we conducted bioinformatics analyses and established
two mA modification patterns according to 23 CNV-harboring mA
regulators. The expression of 23 mA regulators was affected by DNA CNV,
suggesting the significance of their gene expression in cancer progression [23].
The aberrant expression of these mA regulators together with their
prognostic role in ovarian cancer further revealed the importance of mA
modification in cancer progression. Based on the vital value of mA
modification in ovarian cancer, we further identified PTGER2 as an oncogene
potentially modified by mA modification in a subsequent investigation.
Previous studies have shown that METTL3 promotes ovarian cancer proliferation,
migration, and invasion [24, 25]. In addition, METTL3 is an inducer of cancer
stem cell self-renewal and chemoresistance in human cancers [26, 27]. In this
work, the regulatory impact of mA RNA methylation on the expression of
PTGER2, which served as a promoter of ovarian cancer stemness, chemoresistance,
proliferation, and metastasis, was experimentally verified by the observation
that METTL3 enhanced PTGER2 expression through mA modification, further
suggesting the oncogenic role of METTL3 in ovarian cancer. Comprehensive analysis
has suggested that METTL3 mainly regulates mA RNA methylation in the CDS
region, stop codon, and 3’ UTR of its targets [28]. In this investigation, we
also found that METTL3 modulated mA RNA methylation of PTGER2 in the 3’ UTR
region, thus enhancing PTGER2 expression. However, some experts and scholars have
proposed that mA-sensitive PCR and MeRIP mapping of specific mRNA do not
provide definitive evidence of mA. Therefore, additional studies are needed
to confirm the mA RNA methylation of PTGER2 mRNA.
PTGER2 plays dual roles as an oncogene or a tumor suppressor in human cancers
[13, 15, 16]. PTGER2 facilitates cancer stemness, chemoresistance, proliferation,
and metastasis [14, 29]. In the present work, we also revealed the role of PTGER2
in promoting ovarian cancer stemness, chemoresistance, proliferation, and
metastasis. Cancer stem cell self-renewal properties contribute to regulating the
EMT process and DNA damage repair [30]. The facilitation of cancer
chemoresistance can be regulated by several mechanisms, including modulation of
cancer stemness, DNA damage repair, and the EMT [31]. In our investigation, we
further discovered that PTGER2 increased cancer stem cell self-renewal
properties, the EMT, and DNA damage repair to enhance cell stemness, resistance
to carboplatin, proliferation, and metastasis, thus potentiating ovarian cancer
progression. Moreover, bioinformatics analysis of TCGA ovarian cancer samples and
immunohistochemical assays in our collected clinical samples confirmed that
PTGER2 functioned as an oncogene and was correlated with ovarian cancer distant
metastasis. Importantly, PTGER2 expression was identified as an independent and
unfavorable factor in patient tissues, further supporting the conclusions drawn
from ovarian cancer cells.
5. Conclusions
Overall, our study provides a new regulatory axis consisting of METTL3 and
PTGER2 in the modulation of ovarian cancer progression. METTL3-mediated mA
RNA methylation of PTGER2 is responsible for the oncogenic role of PTGER2 in
ovarian cancer. This work delineates the role of mA RNA methylation in a
specific network of signal transduction and highlights the foundation for the
clinical translation of PTGER2 both as a prognostic predictor and as a novel
target for the management of ovarian cancer.
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from
the corresponding author on reasonable request.
Author Contributions
YL and BX conceived and designed the present study. YL wrote the manuscript. BX checked
and revised the manuscript. Both authors contributed to the article and approved
the submitted version. Both authors read and approved the final manuscript. Both
authors have participated sufficiently in the work and agreed to be accountable
for all aspects of the work.
Ethics Approval and Consent to Participate
Patient written consent and ethics approval from the Ethics Committee
of the hospital were obtained (K2020-036-01).
Acknowledgment
Not applicable.
Funding
This work was founded by Startup Fund for scientific research, Fujian Medical
University (Grant number: 2021QH1141).
Conflict of Interest
The authors declare no conflict of interest.
 Fig. 1.
Fig. 1. Fig. 2.
Fig. 2. Fig. 3.
Fig. 3. Fig. 4.
Fig. 4.