1 Department of Biology, Faculty of Medicine, Aix-Marseille University, INSERM UMR_S 1072, Marseille, France
2 School of Medicine, University Hospital of Wales Heath Park, 14 4XN Cardiff CF, UK
3 Neuroscience Center—HiLIFE, Helsinki Institute of Life Science, University of Helsinki, 00014 Helsinki, Finland
Abstract
Background: Plasmolipin (PLLP) is a membrane protein located in lipid rafts that participates in the formation of myelin. It is also implicated in many pathologies, such as neurological disorders, type 2 diabetes, and cancer metastasis. To better understand how PLLP interacts with raft components (gangliosides and cholesterol), we undertook a global study combining in silico simulations and physicochemical measurements of molecular interactions in various PLLP-ganglioside systems. Methods: In silico studies consisted of molecular dynamics simulations in reconstructed membrane environments. PLLP-ganglioside interaction measurements were performed by microtensiometry at the water-air interface on ganglioside monolayers. Results: We have elucidated the mode of interaction of PLLP with ganglioside GM1 and characterized this interaction at the molecular level. We showed that GM1 induces the structuring of the extracellular loops of PLLP and that this interaction propagates a conformational signal through the plasma membrane, involving a cholesterol molecule located between transmembrane domains. This conformational wave is finally transmitted to the intracellular domain of the protein, consistent with the role of PLLP in signal transduction. Conclusions: This study is a typical example of the epigenetic dimension of protein structure, a concept developed by our team to describe the chaperone effect of gangliosides on disordered protein motifs which associate with lipid rafts. From a physiological point of view, these data shed light on the role of gangliosides in myelin formation. From a pathological point of view, this study will help to design innovative therapeutic strategies focused on ganglioside-PLLP interactions in various PLLP-associated diseases.
Keywords
- lipid rafts
- ganglioside
- cholesterol
- neurological disorder
- disease
- signal transduction
- plasmolipin
Proteolipids are a subclass of membrane proteins soluble in a
chloroform–methanol mixture [1]. The proteolipid plasmolipin (PLLP), also known
as plasma membrane proteolipid (PMLP) or transmembrane 4 superfamily, member 11
(TSF11) [2], is a main component of the myelin sheath [3, 4]. Moreover, the human
PLLP (encoded by the PLLP gene) [5], an 18-kD protein, is presumably
involved in various pathological conditions such as Alzheimer’s disease [6], type
2 diabetes [7], and cancer metastasis [8]. PLLP is involved in intracellular
transport [9], lipid raft formation [10], and Notch signaling [2]. Finally, PLLP
was also identified as a virus entry receptor [11]. An intriguing aspect of
proteolipids such as PLLP is that most of the protein structure is embedded in
the membrane [2], leaving very few surfaces of contact for the polar headgroups
of glycosphingolipids such as gangliosides, which are key lipid raft components
[12, 13]. The predicted topology of human PLLP is a tetraspan protein with both
N-terminal and C-terminal domains located intracellularly [2]. Correspondingly,
the extracellular areas of PLLP are limited to two loops, encompassing amino acid
residues 125–144 (AVDLTSLRGTRPYNQRAAAS), referred to as loop 1 (the largest one)
and 57–64 (DTPYHLYP), referred to as loop 2 (the smallest one). Alpha-Fold has
not satisfactorily resolved the structure of these loops [14, 15], consistent with
secondary structure predictions that consider these regions as disordered domains
[16]. Indeed, apart from the transmembrane (TM) domains, the AlphaFold predictions for human PLLP
that are available through the UniProt database (entry Q9Y342,
https://www.uniprot.org/uniprotkb/Q9Y342/entry) have either low (70
Gangliosides are important components of the myelin sheath [17]. Knockout mice lacking major brain gangliosides display decreased central myelination and axonal degeneration in both the central and peripheral nervous systems [18]. However, although these data suggest that gangliosides are functionally important for myelin biogenesis [19], the molecular basis of this concept remains poorly understood. Solving this problem requires understanding the molecular interactions between PLLP and gangliosides. However, disordered loops in the proteolipid make this analysis particularly difficult. Since PLLP is found within lipid rafts [10, 20], these membrane domains may be a privileged site of ganglioside-PLLP interactions. The present study aimed to assess whether the disordered loops of PLLP could be structured upon binding to the sugar head group of gangliosides, according to the lipid chaperone theory [21, 22]. To this end, we studied the interaction of a synthetic peptide derived from the largest extracellular loop of PLLP with ganglioside monolayers using the Langmuir microtensiometry technology. Then we performed a series of molecular dynamics simulations of human PLLP in a lipid raft environment. Our results give new insights into how proteolipids interact with lipid rafts and how gangliosides control the folding of this intriguing class of membrane proteins through a limited set of molecular interactions at the plasma membrane-water interface.
Ganglioside binding domain (GDB) predictions were performed according to the
algorithm developed for amyloid proteins [23]. This allowed us to identify the
RGTRPYNQR motif in the second extracellular loop of human PLLP. A synthetic
peptide encompassing this motif (SLRGTRPYNQRA) was synthesized by Schafer-N
(Copenhagen, Denmark) at a purity
The synthetic peptide SLRGTRPYNQRA was dissolved in pure water at a
concentration of 1 mM and used at a final concentration of 10 µM in
Langmuir microtensiometry studies. Surface pressure measurements were recorded
with an automated Langmuir trough (Microtrough X, Kibron, Helsinki, Finland)
[24, 25, 26]. The peptide was added in the aqueous phase underneath a monolayer of the
indicated ganglioside (GM1, GD1a, or GT1b, all purchased from Matreya) at the
initial surface pressure
The three-dimensional structure of the human PLLP was modelized via an
ab initio method using the protein structure prediction service Robetta
(https://robetta.bakerlab.org/) [29]. The first thirty-three residues
corresponding to an intrinsically disordered region exposed to the intracellular
environment were removed from the structure. We simulated PLLP in two different
settings, (i) PLLP inserted into a POPC
(1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) bilayer and (ii) PLLP inserted
into a lipid raft environment consisting of an equimolar mixture of ganglioside
and cholesterol [30, 31] as previously described [32, 33, 34]. Both environments were
solubilized with TIP3P water (the TIP3P water model as implemented in CHARMM
specifies a 3-site rigid water molecule with charges and Lennard-Jones parameters
assigned to each of the 3 atoms) [35] and neutralized with Na
As a preliminary analysis, we submitted the extracellular domains of human PLLP
to our dedicated algorithm for the prediction of ganglioside binding domains
[23]. This algorithm has been previously validated with numerous endogenous and
microbial proteins [38]. The largest extracellular loop of human PLLP (loop 1)
fulfills the algorithm with the RGTRPYNQR motif [39]. A synthetic peptide
encompassing this predicted ganglioside binding domain was thus tested for its
ganglioside binding properties. In these experiments, a ganglioside cluster was
reconstituted as a monolayer at the air-water interface, and the interaction of
the loop 1-derived peptide with this ganglioside monolayer was measured by
surface pressure measurements [28]. Although consisting of a single lipid
leaflet, lipid monolayer systems, including reconstituted lipid rafts coupled
with surface pressure measurements, have been validated as valuable models for
the study of membrane-protein interactions [40, 41, 42, 43, 44]. Most importantly, lipid
raft-like microdomains have been observed in ganglioside monolayers by
fluorescence imaging [45], consistent with the use of these systems for studying
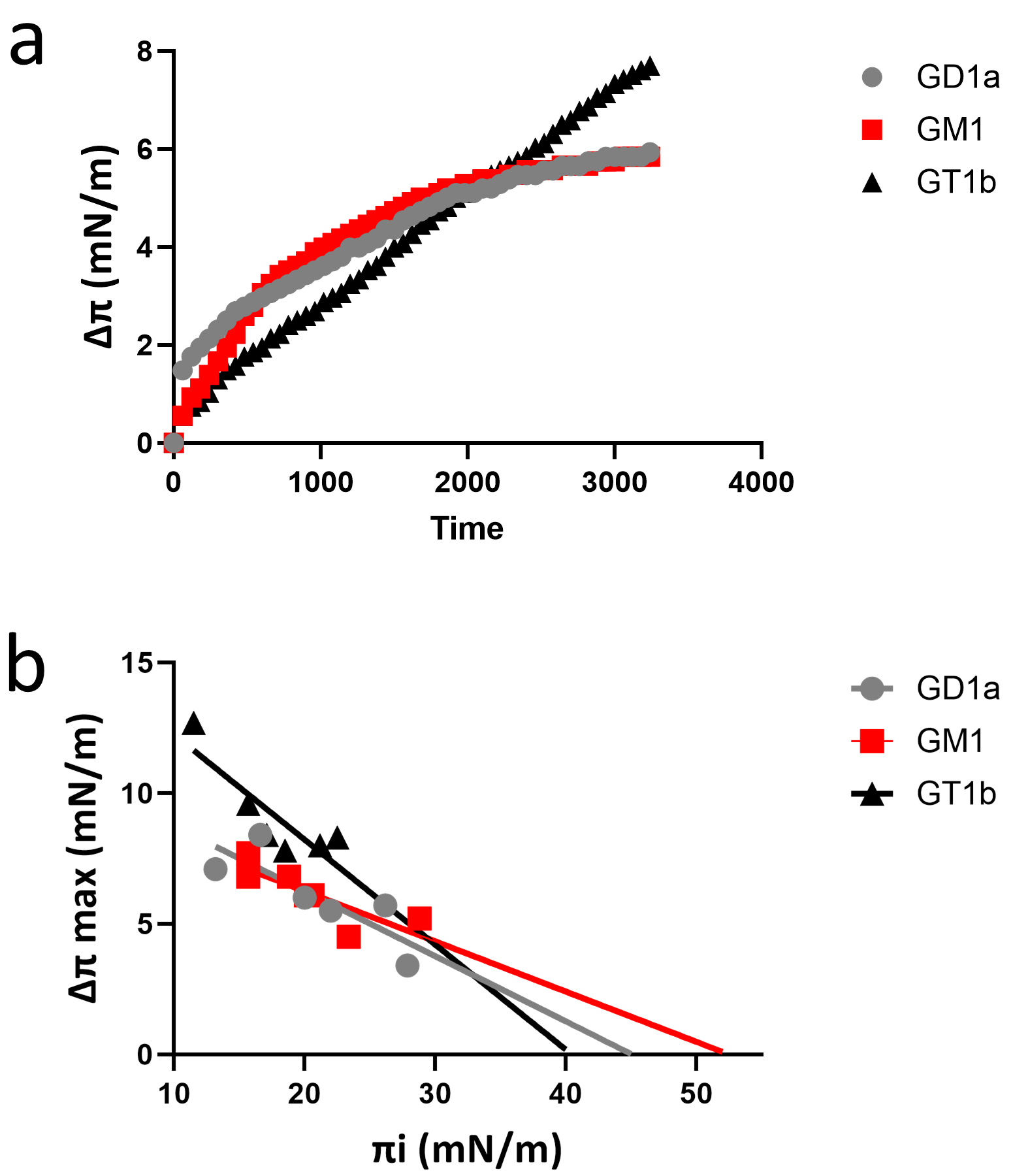
protein binding to lipid rafts [46]. As shown in Fig. 1, the synthetic loop 1
recognized the three gangliosides tested: GM1 (the main ganglioside of myelin)
[47], GD1a, and GT1b (both known to interact with myelin-associated glycoprotein)
[48]. The kinetics of interaction were similar with all three gangliosides (Fig. 1a). However, the data in Fig. 1b indicate that the interaction of the synthetic
loop 1 still occurs with GM1 at higher surface pressure values (
 Fig. 1.
Fig. 1.A synthetic peptide derived from loop 1 of human plasmolipin interacts with gangliosides. (a) Kinetics of peptide SLRGTRPYNQRA interaction with gangliosides GM1, GD1a, and GT1b. (b) Interaction of peptide SLRGTRPYNQRA with ganglioside monolayers prepared at various values of the initial pressure.
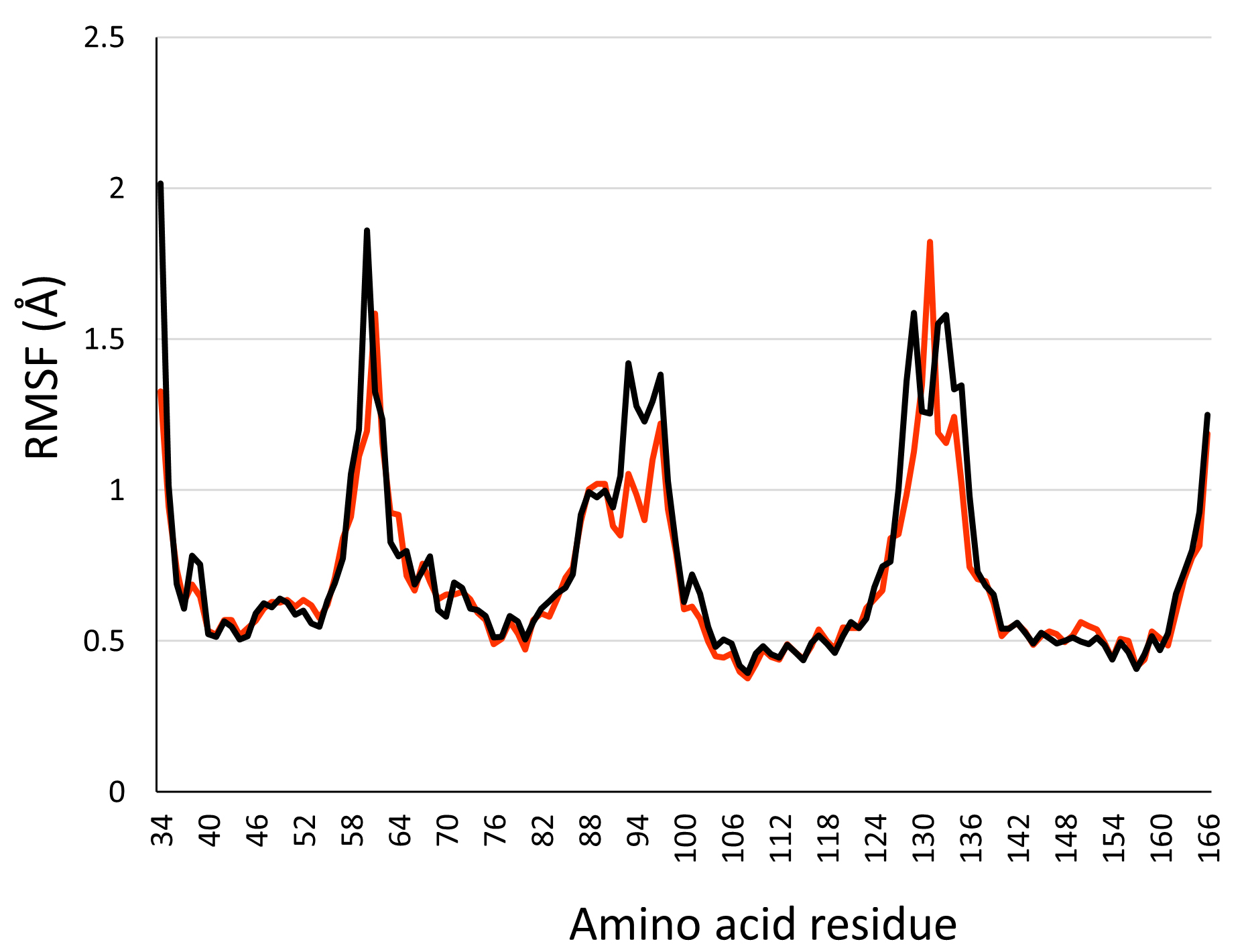
In this study, we present two molecular dynamics simulations which consist of human PLLP inserted into a phosphatidylcholine (POPC) bilayer or a lipid raft environment. To ensure our structure does not go through unstable 3D conformations, we measured its root mean square fluctuation (RMSF). The RMSF is a measure of the degree of flexibility or mobility of atoms or groups of atoms in a protein or other biomolecular systems [36]. The RMSF of the PLLP inserted into a POPC bilayer (black line) or in a lipid raft environment (orange line) are plotted in Fig. 2. The results show that the transmembrane domains of PLLP in either POPC or lipid raft transient around 0.5 Å. In comparison, the flexible loops reach a maximal value of 2 Å. Overall, the small RMSF value of the structured regions of PLLP suggests that our protein is stable over the trajectory.
 Fig. 2.
Fig. 2.RMSF of plasmolipin in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer (black line) and lipid raft environment (orange line).
Lipid rafts are cholesterol-rich plasma membrane microdomains also enriched in
sphingolipids, including sphingomyelin and gangliosides [51]. Among the different
classes of gangliosides expressed by human cells, GM1 is of high interest because
it is the main brain myelin ganglioside [52]. This study aimed to evaluate if
lipid raft gangliosides could have a conformational impact on the flexible loops
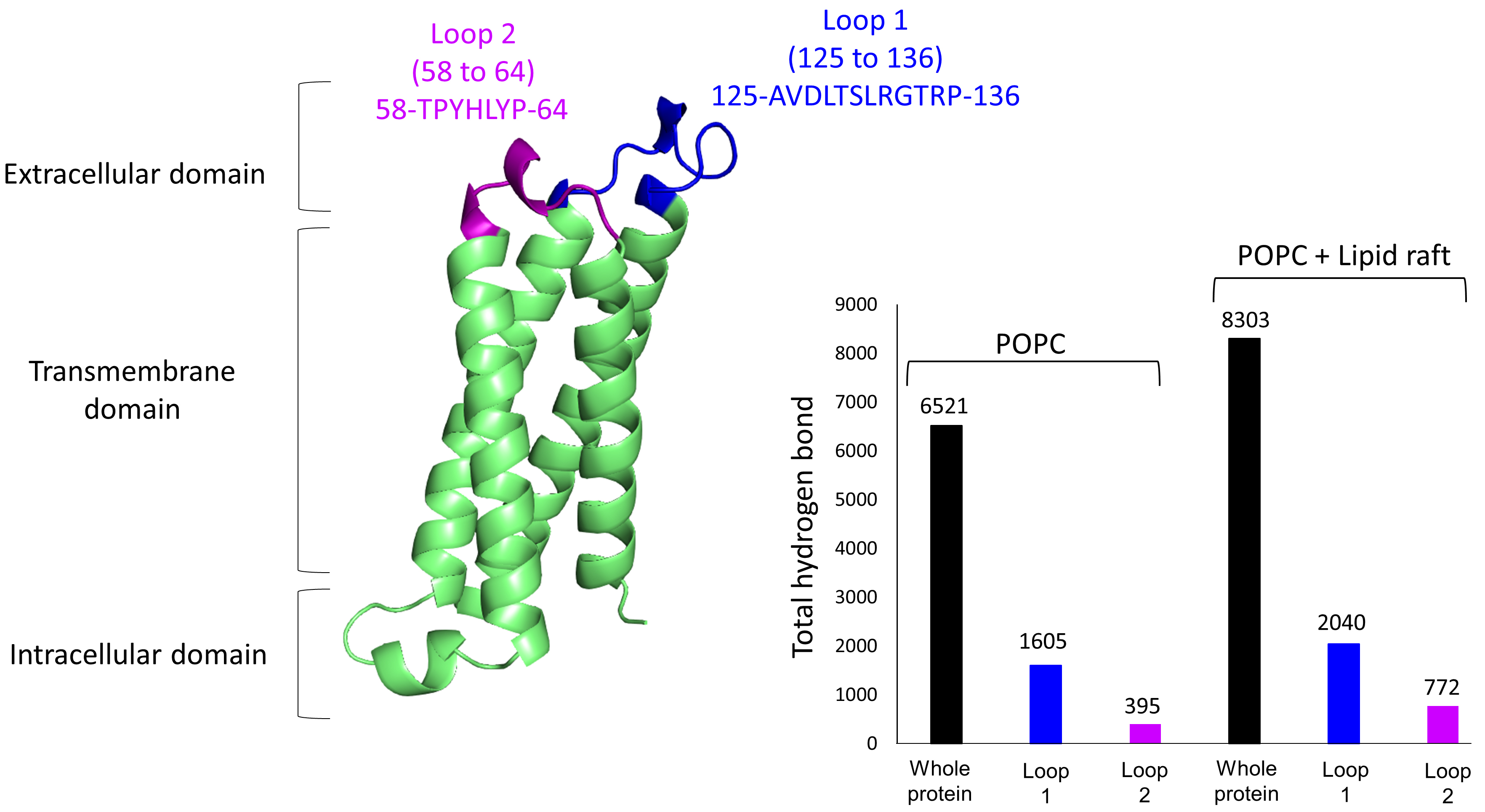
of PLLP. To this end, we first plotted the total number of hydrogen bonds
(H-bonds) formed over time between (i) PLLP and each type of membrane lipid (POPC
bilayer or lipid raft), (ii) loop 1 of PLLP and each membrane or (iii) loop 2 of
PLLP with each membrane. More H-bonds were formed in the lipid raft environment
with a total of 8303 for the whole protein, 2040 for loop 1, and 772 for loop 2,
compared with 6521, 1605, and 395 for the POPC bilayer, respectively (Fig. 3).
The fact that more H-bonds were formed over time in the lipid raft environment
suggests that these bonds are more stable and thus more durable. In the case of
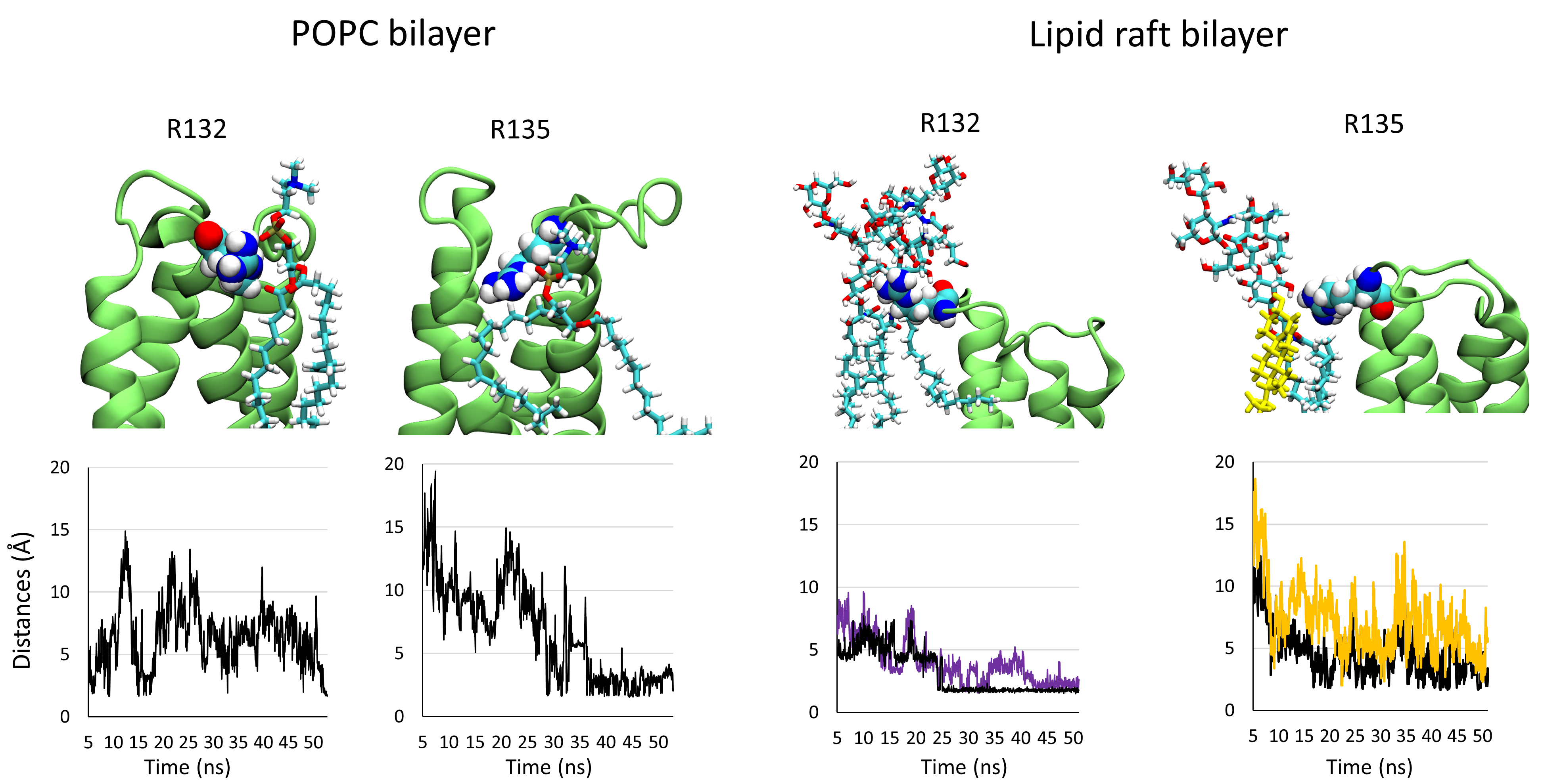
loop 1, the main residues that interact with the membrane component are R132 and
R135. To evaluate the stability of these residues interacting with their lipid
partner, we plotted the distances between them and their partner identified at
the end of the trajectory. We can consider that two atoms enter a non-covalent
interaction between 1.5 and 5 Å. The plots presented in Fig. 4 show that the
distance between the guanidinium group of R132 and an oxygen atom belonging to
the phosphate group of a POPC molecule is very unstable, in contrast with the
intermolecular interactions of the guanidinium group of R132 with the COO
 Fig. 3.
Fig. 3.3D structure of plasmolipin. Extracellular loops 1 and 2 are highlighted in blue and purple, respectively. The structure is accompanied by a histogram showing the total number of hydrogen bonds (H-bonds) formed between the protein and its membrane environment over time for the whole structure (black bar), loop 1 (blue bar), and loop 2 (purple bar).
 Fig. 4.
Fig. 4.Snapshots showing the interaction of residues R132 and R135 belonging to loop 1 in interaction with their lipid/glycolipid partner after 50 ns of simulation. The plots below each panel show the variation of the distances between both partners over the trajectory.
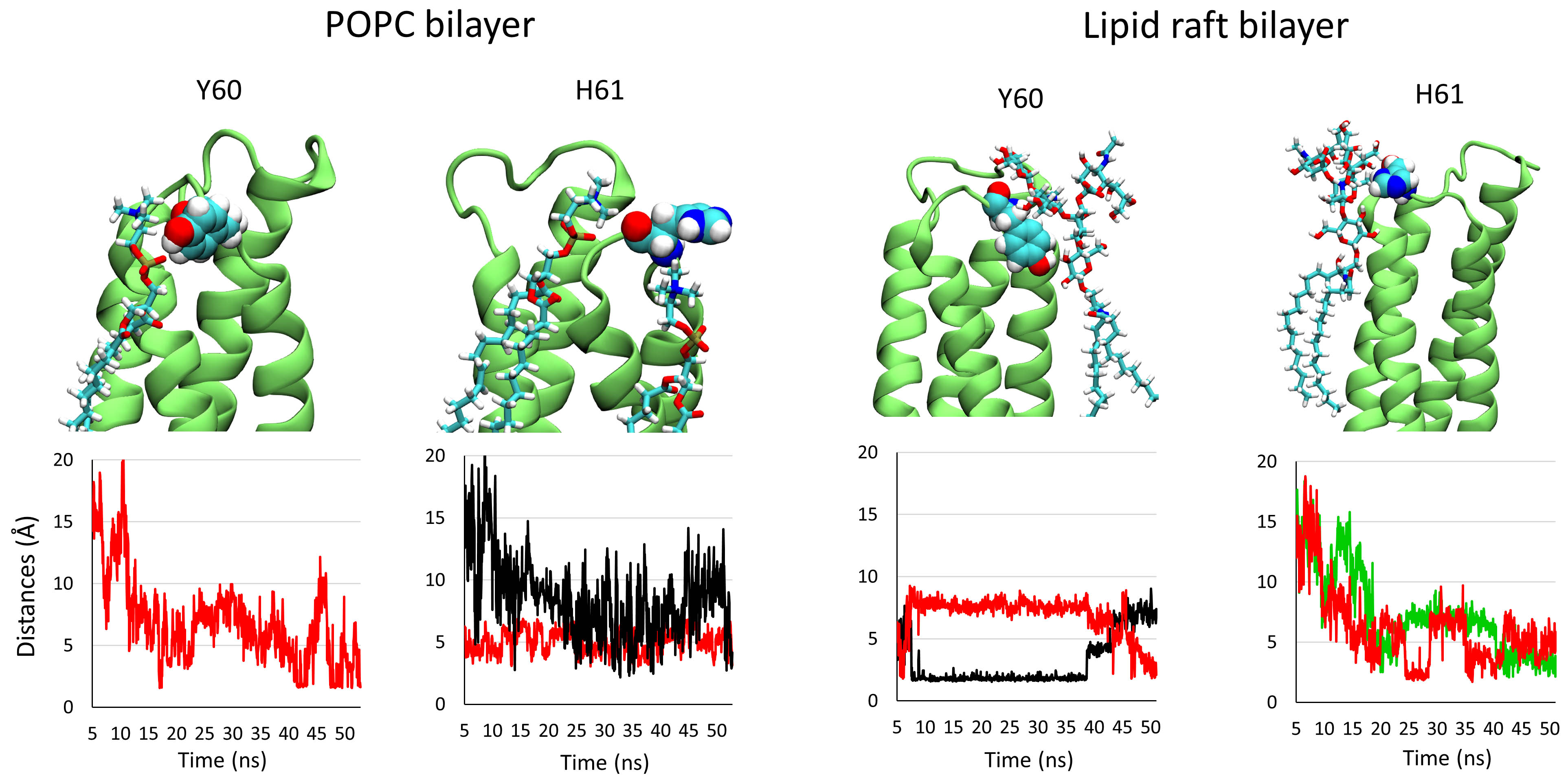
Similar measurements were completed for the extracellular loop 2. The main
residues of loop 2 that interact with the membrane components are Y60 and H61.
The distances of the intermolecular interactions between these residues and their
final partner are plotted in Fig. 5. The distance between the OH group of Y60 and
an oxygen atom of the phosphate group of a POPC molecule is mostly superior to 5
Å, indicating that the intermolecular interaction between both chemical
groups is not stable over the trajectory. It is the same for H61, which was found
to interact with two molecules of POPC via van der Waals (black line) and
CH-
 Fig. 5.
Fig. 5.Snapshots showing the interaction of residues Y60 and H61 belonging to loop 2 in interaction with their lipid/glycolipid partner after 50 ns of simulation. The plots below each panel show the variation of the distances between both partners over the trajectory.
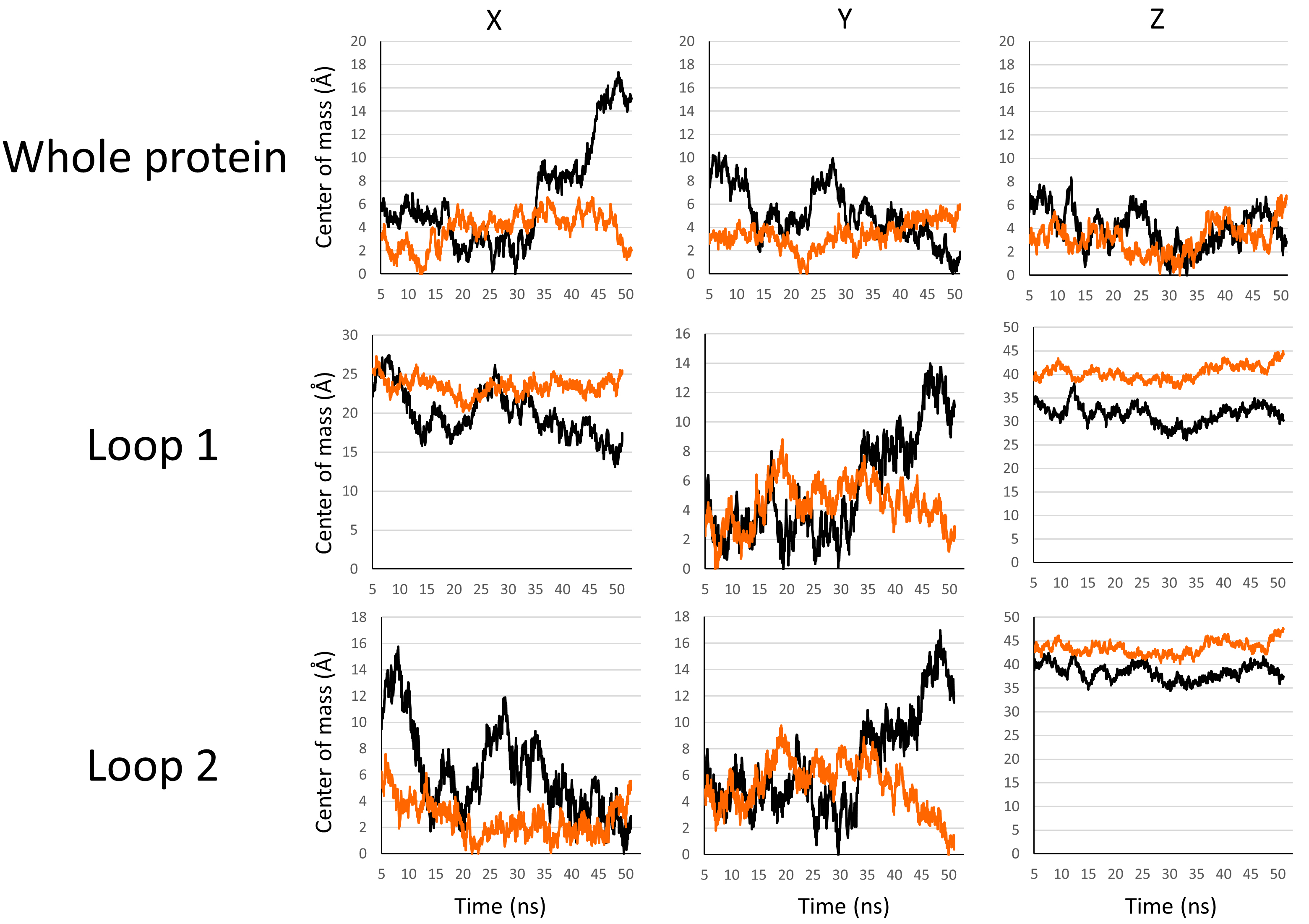
To gain more accuracy in the evaluation of the differences in stability of the PLLP loops in the two different lipidic environments, we studied the confinement of the mass center of the backbone of these structures along the x, y, and z-axis. The more the mass center of a structure is confined, the less the conformation of this structure varies. Thus, this parameter indicates the conformational freedom of the protein surrounded by membrane lipids. In lipid rafts, the confinement of a protein is expected to be stronger in the central area of the raft, where cholesterol and sphingolipids are densely packed, compared with the more flexible peripheral zones [54]. The mass centers for the backbone of the full protein, the backbone of loop 1, and the backbone of loop 2 in the POPC bilayer (black lines) and the lipid raft (orange lines) are plotted in Fig. 6. The data show that for the full protein, loop 1 or loop 2, the mass center of all these structures fluctuates less along the x, y, and z axis in the lipid raft environment. Our in silico data suggest that the different structures of the protein are more stable in the presence of gangliosides and cholesterol molecules, which appear to be key membrane components with which PLLP can durably interact over time.
 Fig. 6.
Fig. 6.Set of plots showing the variations of the mass center of the backbone of the whole protein (first line), loop 1 (second line), and loop 2 (third line) along the x, y, and z axis in a POPC bilayer (black lines) or lipid raft (orange lines).
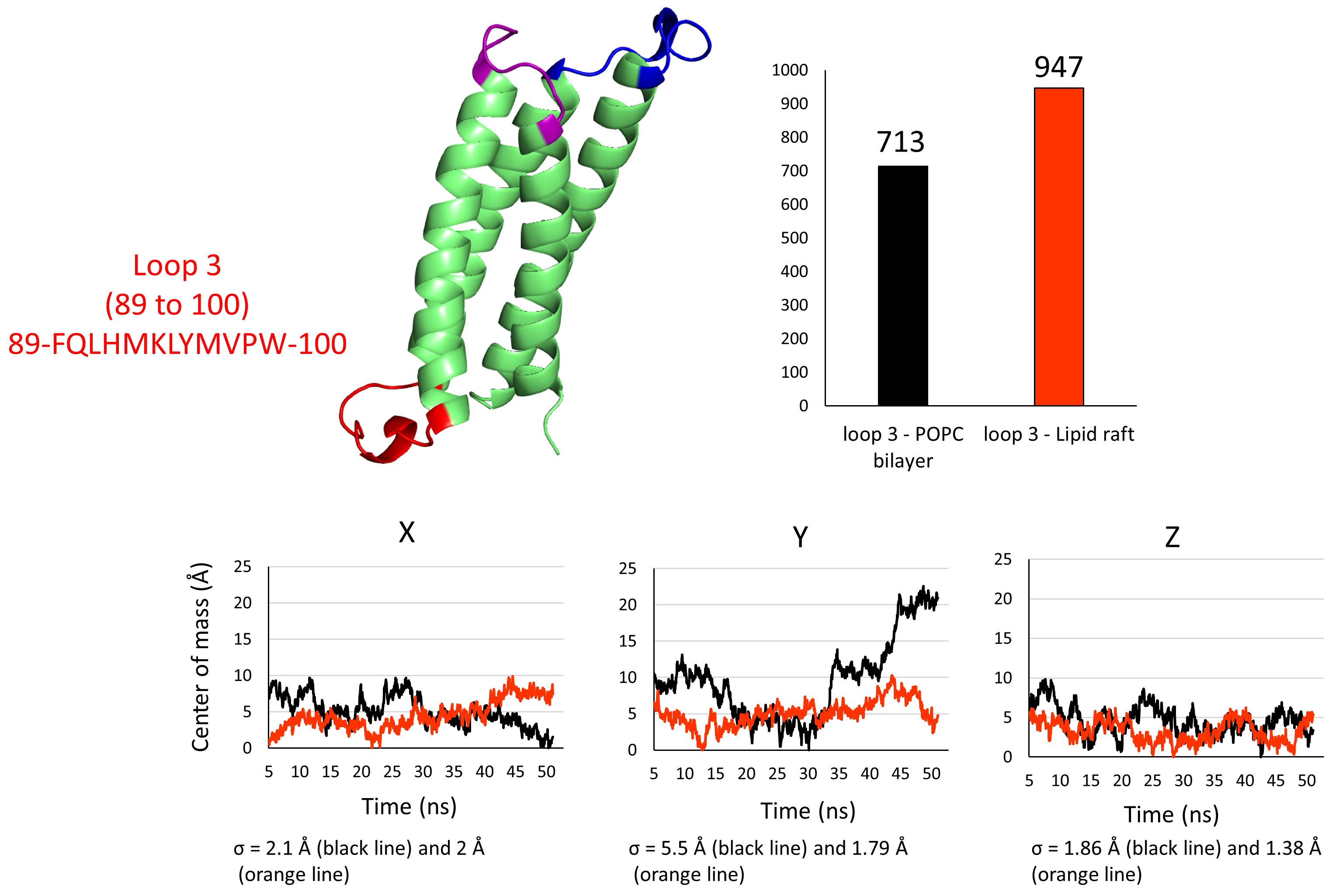
Next, we speculated if the lipid raft, which is a microdomain formed by the clustering of gangliosides and cholesterol in the upper leaflet of the plasma membrane, has an impact on the conformation of the intracellular loop of PLLP that we coined “loop 3” (Fig. 7). In any case, the only partners loop 3 has in both systems are POPC molecules. We performed the same measurements as for loops 1 and 2 to confirm. The histogram presented in Fig. 7 reveals that more H-bonds were formed between loop 3 and the POPC molecules in the lipid raft system. This result suggests that these bonds are more stable in the lipid raft environment, which may therefore help to stabilize the conformational landscape of loop 3. The plots presenting the variation of the center of mass of the backbone of loop 3 in both environments along the x, y, and z axis suggest that the conformation of loop 3 varies less along the y-axis with a standard deviation of 1.79 Å in the lipid raft system against 5.5 Å in the POPC bilayer. It varies slightly less along the z-axis with a standard deviation of 1.38 Å in the lipid raft system against 1.86 Å in the POPC bilayer. The difference in variation of both systems along the x-axis is unimportant, with a standard deviation of 2 Å in the lipid raft against 2.1 Å in the POPC bilayer. Together, our computational results suggest that the intracellular flexible loop of PLLP, which is not in direct contact with the lipid raft components of the exofacial membrane leaflet, is stabilized by the global lipid raft environment.
 Fig. 7.
Fig. 7.3D structure of plasmolipin. Loops 1, 2, and 3 are highlighted in blue, purple, and red, respectively. The structure is accompanied by a histogram showing the total number of hydrogen bonds (H-bonds) formed between loop 3 and the lipids in the POPC bilayer (black bar) or loop 3 and the lipids in a lipid raft environment (orange bar). The colors for the variations of the mass center of the backbone are the same as in Fig. 6 (black lines for the POPC bilayer, orange lines for the lipid raft).
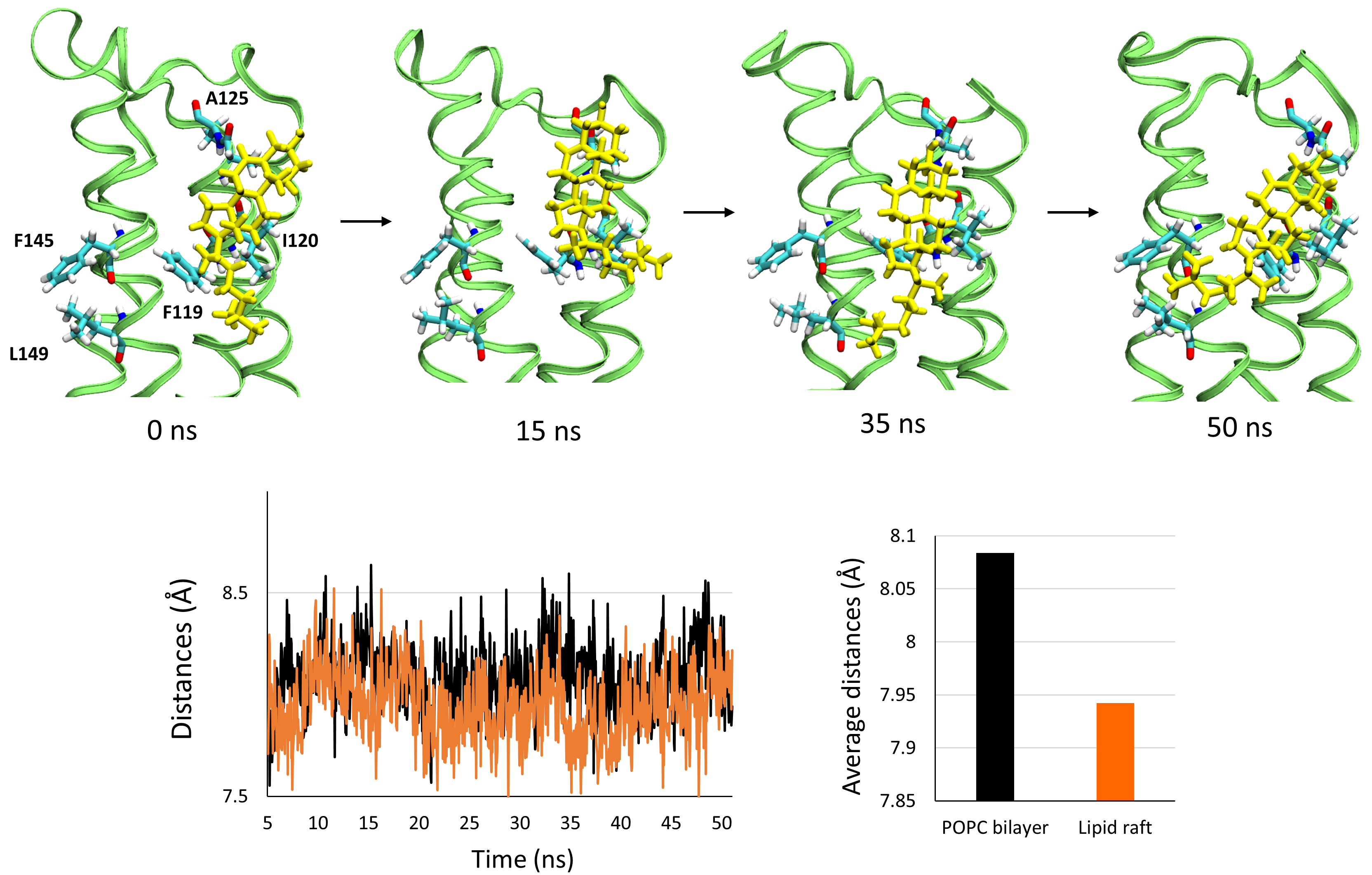
Finally, we observed that one cholesterol molecule specifically interacts with
PLLP. The interaction is depicted in Fig. 8, in which we present a snapshot of
the cholesterol- PLLP complex at 0, 15, 35, and 50 ns. First, the cholesterol
molecule interacts with the residues F119, I120, and A125, which belong to the
3rd transmembrane domain (TM3), via van der Waals and CH-
 Fig. 8.
Fig. 8.Interaction of a cholesterol molecule with the surface of the transmembrane domains of plasmolipin at 0, 15, 35, and 50 ns. The cholesterol is depicted as yellow sticks, the structure of plasmolipin as ribbons colored in lime, and the amino acids interacting with cholesterol as color labelled sticks (white for hydrogens, cyan for carbons, blue for nitrogen, and red for oxygen). The snapshots are accompanied by a plot showing the variations in the distance between the center of mass of the backbone of the transmembrane domains TM3 and TM4 throughout the simulation in the POPC bilayer (black line) and the lipid raft (orange line). The histogram shows the value of the average distance between the center of mass of the backbone of the transmembrane domains TM3 and TM4.
PLLP is a special case of a membrane protein associated with rafts but has only a small surface area available to anchor itself to the lipids of these microdomains. PLLP is a major component of myelin [3]. It is also involved in signal transduction, which implies that its extracellular domains must be able to transmit conformational information to the other side of the plasma membrane. One of the major results of our study is the identification, in one of the two extracellular loops of PLLP, of a functional ganglioside binding motif displaying a high specificity for the ganglioside GM1. This motif corresponds to a canonical domain of interaction with gangliosides, which we identified 20 years ago [39]. It has long been part of the list of potential ganglioside-binding domains identified in brain proteins [56]. The experimental confirmation of this domain’s functionality is required to correlate its binding capacities to raft gangliosides with the 3D structure of the protein in its membrane environment. Our team’s recent progress in obtaining realistic 3D structures of membrane proteins in various plasma membrane environments has made the task possible, which motivated the present study [27, 33, 57, 58, 59].
The data obtained in silico showed that human PLLP interacts with ganglioside GM1 in a reconstituted lipid raft system. The interaction is driven by the two extracellular loops of PLLP, which undergo a specific structuration that does not occur in a POPC system. The largest loop (loop 1) displays a canonical ganglioside binding motif whose ganglioside binding properties can be demonstrated with a synthetic peptide in a ganglioside monolayer assay. Considering the results, we can conclude that this ganglioside binding motif does not have a pre-established structure. On the contrary, this extracellular loop is globally disordered. In fact, it is precisely the role of gangliosides to induce the structuring of this loop when the protein reaches a raft. In this respect, our data may explain why knocked-out mice lacking major brain gangliosides show decreased central myelination and axonal degeneration [18]. The key point is that in the liquid disordered (Ld) phase of the plasma membrane containing chiefly glycerophospholipids such as POPC [51], the disordered loops of PLLP are not sufficiently constrained to acquire a functional fold. In contrast, the liquid-ordered (Lo) phase of lipid rafts contain gangliosides [51] which act as membrane chaperones for PLLP. In the raft environment, PLLP can acquire a functional structure that is mandatory for the biogenesis of the myelin sheath. Hence, the lack of gangliosides leads to a non-functional PLLP, explaining the demyelination phenotype of ganglioside-negative mice.
The localization of PLLP in lipid rafts is also consistent with its function in signal transduction [10]. Our in silico study suggests that the acquisition of a functional 3D structure of PLLP is first initiated by the binding of the proteolipid to raft gangliosides. Then, this conformational information propagates through the plasma membrane, relayed by a cholesterol molecule controlling the relative position of the TM domains. This conformational wave then reaches the intracellular (cytosolic) area of the protein, which reacts by adopting and stabilizing a particular structure.
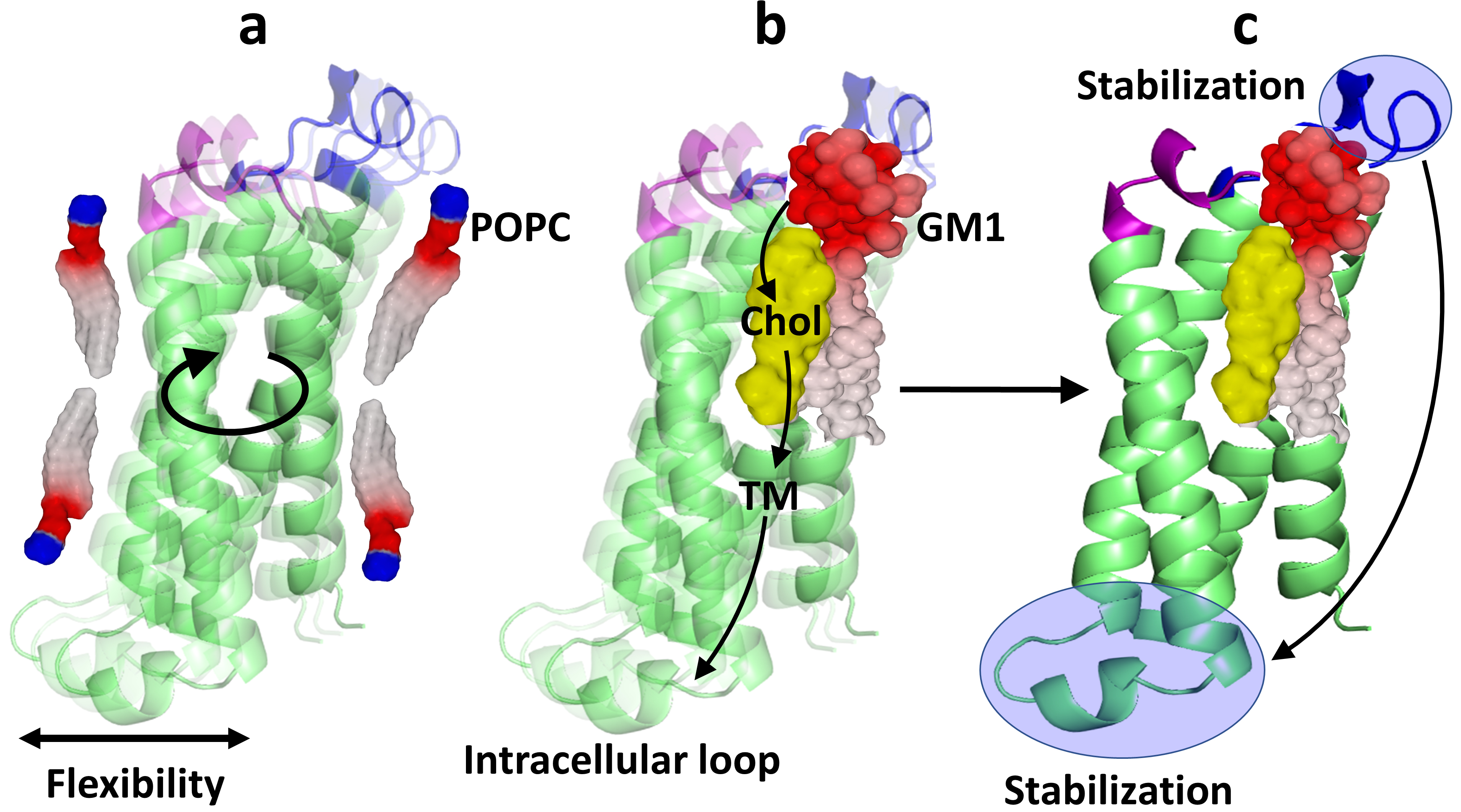
The concept of conformational coupling across the plasma membrane during receptor activation has been recognized for a long time [60]. The propagation of a conformational wave has also been proposed to explain how the transient receptor potential vanilloid 1 (TRPV1) ion channel is activated by capsaicin [61]. The data presented here not only confirm the interest in these concepts but explains for the first time how raft lipids may control this complex mechanism. We suggest that a conformational wave represents a long-distance transfer of energy throughout the thickness of the lipid bilayer. This could also be interpreted as a long-range chaperone effect. In the case of PLLP, the wave is triggered by the interaction of the extracellular loops with the headgroups of GM1 gangliosides in the raft environment (Fig. 9). Outside the raft, phosphatidylcholine molecules (POPC) have little impact on PLLP structure which has the freedom to oscillate between several closely related conformers (Fig. 9a). The structuring of the extracellular loops by raft gangliosides then generates coordinated movements of the TM helices (Fig. 9b). The role of cholesterol would be to control the conformational flexibility of the TM domains and determine their tilt during the energy transfer of the wave. In this model, the four-helix bundle structure of PLLP is optimally tuned by cholesterol, according to the conformational information transmitted by raft gangliosides. Overall, the conformational wave generated by the extracellular loops propagates through the TM domains and reaches the intracellular loop, which eventually acquires a stabilized structure (Fig. 9c). By clustering gangliosides and cholesterol, lipid rafts can be viewed as transducer platforms able to propagate such controlled conformational waves throughout the plasma membrane. The tail-to-tail organization of cholesterol molecules [62], which conformationally links the exofacial and intracellular leaflets of the plasma membrane through a mirror code, allowing the same TM domain to accommodate two cholesterol molecules in opposite orientations [63], seems particularly well adapted for propagating these conformational waves. Finally, it is important to note that in addition to the mechanical waves described here, this concept also includes the “swipe card” model based on an electron transfer wave across the TM domains of membrane receptors, as proposed for odorant recognition (see Figs. 2 and 3 in ref. [64]). In this respect, our model may reconcile two distinct scenarios of signal transduction: (i) the classic transmembrane conformational coupling [65], based on the complementary shape theory and thus viewed as a “mechanical” wave, and (ii) the more iconoclast “quantum” wave model that includes a molecular vibration frequency as additional information to explain how odorant molecules activate their receptors [64, 66].
 Fig. 9.
Fig. 9.The conformational wave model. (a) When surrounded by phosphatidylcholine (POPC) molecules, plasmolipin oscillates between several conformers. The intracellular loop, which is not constrained by the plasma membrane, remains flexible. (b) The interaction of the extracellular loops with GM1 induces a conformational change in the extracellular domains of plasmolipin. This initial interaction triggers a conformational wave which is transmitted to the TM domains under the control of cholesterol and finally reaches the intracellular loop. (c) The conformational wave starts in the extracellular loops, propagates throughout the thickness of the plasma membrane, and ends in the intracellular loop. Overall, the stabilization of the extracellular loop by raft gangliosides induces the structuration of the intracellular loop through a ganglioside-cholesterol-dependent mechanism.
The chaperone effect of raft lipids in the 3D folding of membrane proteins is one of the fundamental elements of the theory we have recently developed and called the epigenetic dimension of protein structure [57]. The epigenetic dimension of protein structure, which considers the chaperone-like conformational regulation of raft lipids (gangliosides and cholesterol), is a major limitation of the AlphaFold algorithm [58]. This concept applies both to membrane proteins such as PLLP and to extracellular proteins that trigger toxic effects in brain cells and cause neurodegenerative diseases such as Alzheimer’s disease [67] and Parkinson’s disease [68]. By understanding the molecular mechanisms that control the interaction of these proteins with rafts, we have already begun to design innovative therapeutic agents for the prevention and treatment of neurodegenerative diseases [69, 70]. These new drugs are rationally designed to target raft gangliosides, which play a central role in the changes in raft homeostasis associated with several pathological conditions, including cancer, Alzheimer’s disease, and type 2 diabetes [54].
PLLP is among the potential targets of such therapeutic agents specifically designed to block the interaction of selected proteins with raft gangliosides. The involvement of PLLP in numerous pathologies (Alzheimer’s disease, type 2 diabetes, cancer metastasis, and viral infections) [2] makes this protein a new target of choice to improve our therapeutic arsenal against these diseases. Finally, the results of our study shed light on the molecular mechanisms associating the raft components with the physiological functions of PLLP, in particular signal transduction and myelin formation [2].
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
JF, HC, NY and CDS designed the research study. FA performed the molecular modeling studies. MM and CDS performed the monolayers studies. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity.
Not applicable.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.