- Academic Editors
Background: RUNX2 (Runt-related transcription factor 2) acts as a key regulator in the odontogenic differentiation of human dental pulp stem cells (hDPSCs). Moreover, the inclusion of exon 5 is important for RUNX2 function. Our previous study showed that Y-Box Binding Protein 1 (YBX1) promoted RUNX2 exon 5 inclusion and mineralization of hDPSCs. However, the regulatory mechanism of RUNX2 exon 5 alternative splicing needed further exploration. Methods: The expression level of heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) during the odontogenic differentiation of hDPSCs was analyzed by RT-PCR and Western blot. The roles of hnRNP A1 in the alternative splicing of RUNX2 exon 5 and the odontogenic differentiation of dental mesenchymal cells were analyzed by gain- and loss-of-function experiments. Results: Surprisingly, we found an alternative splicing factor, hnRNP A1, which had an opposite role to YBX1 in regulating RUNX2 exon 5 inclusion and odontogenic differentiation of hDPSCs. Through gain- and loss-of-function assay, we found that hnRNP A1 suppressed the inclusion of RUNX2 exon 5, resulting in the inhibition of odontoblastic differentiation. The overexpression of hnRNP A1 can inhibit the expression of ALP (alkaline phosphatase) and OCN (osteocalcin), and the formation of mineralized nodules during the odontogenic differentiation of both hDPSCs and mouse dental papilla cells (mDPCs), whereas the opposite results were obtained with an hnRNP A1 knockdown preparation. Conclusions: The present study indicated that hnRNP A1 suppressed RUNX2 exon 5 inclusion and reduced the odontogenic differentiation ability of hDPSCs and mDPCs.
RUNX2 (Runt-related transcription factor 2), a transcription factor containing
the Runt structural domain, is a master regulator in osteogenesis and tooth
development [1, 2]. During the development of the mouse tooth, Runx2 is highly
expressed in the dental papilla and follicle mesenchyme at embryonic day 14 [3].
In addition, Runx2 mediates the epithelial-mesenchymal interactions during the
bud-to-cap-stage transition of tooth development [4]. These results suggested
that Runx2 is essential for tooth development. RUNX2 encodes a highly conserved
transcription factor, containing an essential Runt domain, which is involved in
DNA binding [5], heterodimerization [6], and nuclear location [7]. RUNX2 exon 5
encodes a nuclear localization signal (NLS) and partial C-terminus of the Runt
domain [8]. Two shorter isoforms of RUNX2 without exon 5 (RUNX2
Splicing factor heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is one of the most plentiful and extensively expressed nuclear proteins [11]. The hnRNP A1 protein encompasses an N-terminal region with two closely-related RRM domains (RRM1 and RRM2), and a glycine-rich C-terminal region with an RGG box domain relating to RNA binding, and an M9 domain relating to nuclear targeting [12, 13]. HnRNP A1 is a multifunctional protein that participates in mRNA biogenesis, such as transcription, alternative splicing, and stability [13, 14]. As a classical modulator of alternative splicing, hnRNP A1 has been extensively studied [13]. Recently, several studies have elucidated the biological roles of hnRNP A1 in the differentiation of some stem cells [15, 16], but the functions of hnRNP A1 in differentiation of hDPSCs remain unknown.
The hDPSCs are characterized by a rapid proliferation rate, self-renewal capability, and high accessibility to multi-directional differentiation (e.g., odontogenic, osteogenic, chondrogenic, and neurogenic) [17, 18]. Many studies have demonstrated that hDPSCs possesses the potential to form dentin/pulp-like structure, as well as to compensate for defects in dentin and bone formation [19, 20]. But the regulatory mechanisms of hDPSC differentiation are not fully understood.
In the present study, we attempted to analyze the function of hnRNP A1 in RUNX2 exon 5 alternative splicing and odontogenic differentiation of hDPSCs and mouse dental papilla cells (mDPCs).
The collection of human dental pulp tissues was approved by the Ethics Committee of the School and Hospital of Stomatology at Wuhan University (2019-A47). Informed consent was obtained from every participant. The use of animal tissues was approved by the Animal Welfare and Ethics Committee of the School and Hospital of Stomatology at Wuhan University (S07922090F).
Human dental pulp tissues were isolated from healthy and non-functional third molars donated by patients aged 18–26 years old. In brief, healthy dental pulp tissues were collected and cut into tiny pieces in a sterile environment, and then digested with 3 mg/mL collagenase type I (#SCR103, Sigma-Aldrich, Saint Louis, MO, USA) for 1 h at 37 °C. The cells were cultured in minimal essential medium, alpha modification (#SH30625.01, Hyclone, Marlborough, MA, USA), which contains 10% fetal bovine serum (#10099141, Gibco, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin (#15240-062, Gibco, Carlsbad, CA, USA). The surface stemness markers of hDPSCs were determined by flow cytometry. In brief, hDPSCs (3rd passage) were digested with trypsin and rinsed two times with phosphate-buffered saline (PBS) (#SH30256.01, Hyclone, Marlborough, MA, USA). Then, cells were resuspended with flow cytometry staining buffer (#S1001, MultiSciences, Hangzhou, China), followed by incubation with the following fluorescent dye-conjugated antibodies for 30 minutes: isotype control-anti-CD34 (#400207); anti-CD34 (#343603); isotype control-anti-CD45 (#400109); anti-CD45 (#368507); isotype control-anti-CD29, CD90, and CD146 (#400111); anti-CD29 (#303003); anti-CD90 (#328109); and anti-CD146 (#361005) (all antibodies were obtained from Biolegend, San Diego, CA, USA). The stained cells were analyzed by a CytoFLEX Flow Cytometer (#C09748, Beckman, Indianapolis, IN, USA). CD34 and CD45 are hematopoietic stem cells and leukocyte markers, whereas CD29, CD90, and CD146, are mesenchymal stem cell (MSC) markers. Analysis of data was performed with CytExpert software (Beckman, Indianapolis, IN, USA).
Mice were raised in a specific pathogen-free facility with a 12:12-h day/night cycle. Primary mDPCs were isolated from the first mandibular molars of C57BL/6 mice at postnatal day 0.5 (PN 0.5) and digested by 0.25% trypsin (#SH30042.02, Hyclone, Marlborough, MA, USA) for 10 min, and primary cells were fusiform or triangular under phase contrast microscope (Supplementary Fig. 1). CAL 27 cells, HEK 293 cells and HEK 293T cells were authenticated using short tandem repeat (STR) profiling analysis. Mycoplasma testing has been done for the cell lines used. The mDPCs, CAL 27 cells, HEK 293 cells, and HEK 293T cells, were grown in Dulbecco’s modified Eagle medium (DMEM) (#SH30022.01, Hyclone, Marlborough, MA, USA), which contains 10% fetal bovine serum.
To induce differentiation, hDPSCs and mDPCs were cultured in differentiation
medium which comprised basal medium, 10 nM dexamethasone (#T5648, Sigma-Aldrich,
Saint Louis, MO, USA), 50 µg/mL ascorbic acid (#A4544, Sigma-Aldrich,
Saint Louis, MO, USA) and 10 mM
T7-tagged human hnRNP A1 overexpression plasmid was made for our previous
publication [21]. The mouse hnRNP A1 gene was amplified using
primers 5
A minigene of human RUNX2 with genomic DNA sequence from exon 4 to exon 6
(including exon 4, exon 5, exon 6, and partial intron) was amplified from the CAL
27 genome by the following primers: 5
The sequences of hnRNP A1 siRNAs are as follows:
5
Total cellular protein samples were lysed with 2
Total RNA was extracted using the AxyPrep miniprep kit (#AP-MN-MS-RNA-250,
Axygen, Union City, CA, USA). RNA was reversely transcribed to cDNA by using
Maxima H Minus Transcriptase (#EP0752, Thermo Fisher Scientific, Carlsbad, CA,
USA), and amplified with Green Taq Mix (#P131, Vazyme, Nanjing, China) according
to the manufacturer’s protocols. Sequences of primers were as follows:
5
After being cultured in differential medium for 7 days, the cells were rinsed three times with PBS and fixed with 4% paraformaldehyde for 15 min. Then, cells were washed with PBS for three times and alkaline phosphatase staining was performed with a BCIP/NBT ALP Color Development Kit (#C3206, Beyotime, Shanghai, China) according to the manufacturer’s instructions. The ALP activity of the harvested proteins was measured with an ALP activity kit (#A059-2-2, Jiancheng, Nanjing, China), and the absorbance values at 520 nm were measured. ALP activity relative to that of the control group was calculated after normalization with the total protein level.
After hDPSCs were cultured in differentiation medium for 14 days or mDPCs were cultured in differentiation medium for 7 days, cells were rinsed three times with PBS and fixed with 4% paraformaldehyde for 15 min. Then, cells were washed with PBS for three times. Calcium deposition was stained with ARS (#ALIR10001, Cyagen, Suzhou, China). Furthermore, mineralized nodules were dissolved in 10% cetylpyridinium chloride (#C129534, aladdin, Shanghai, China). The concentration of calcium was calculated by measuring the absorbance value at 562 nm.
All statistical analyses were performed using GraphPad Prism 8.0 (San Diego, CA, USA). Student’s t tests were used to analyze the statistical differences between two groups. One-way analysis of variance was performed for the comparison among three groups.
We explored the expression profile of hnRNP A1 during the mineralization induction of hDPSCs. Primary hDPSCs were isolated from extracted third molars, and the surface markers were analyzed. The results showed that these cells were positive for the MSC markers, including CD29, CD90, and CD146, although they were negative for the hematopoietic stem cells and leukocyte markers like CD34 and CD45 (Fig. 1A). In the current study, hnRNP A1 expression showed reduced at RNA (Fig. 1B) and protein (Fig. 1C) levels after hDPSC mineralization induction for 7 days. The expression levels of ALP and OCN were applied to represent the mineralized state of hDPSCs (Fig. 1B).
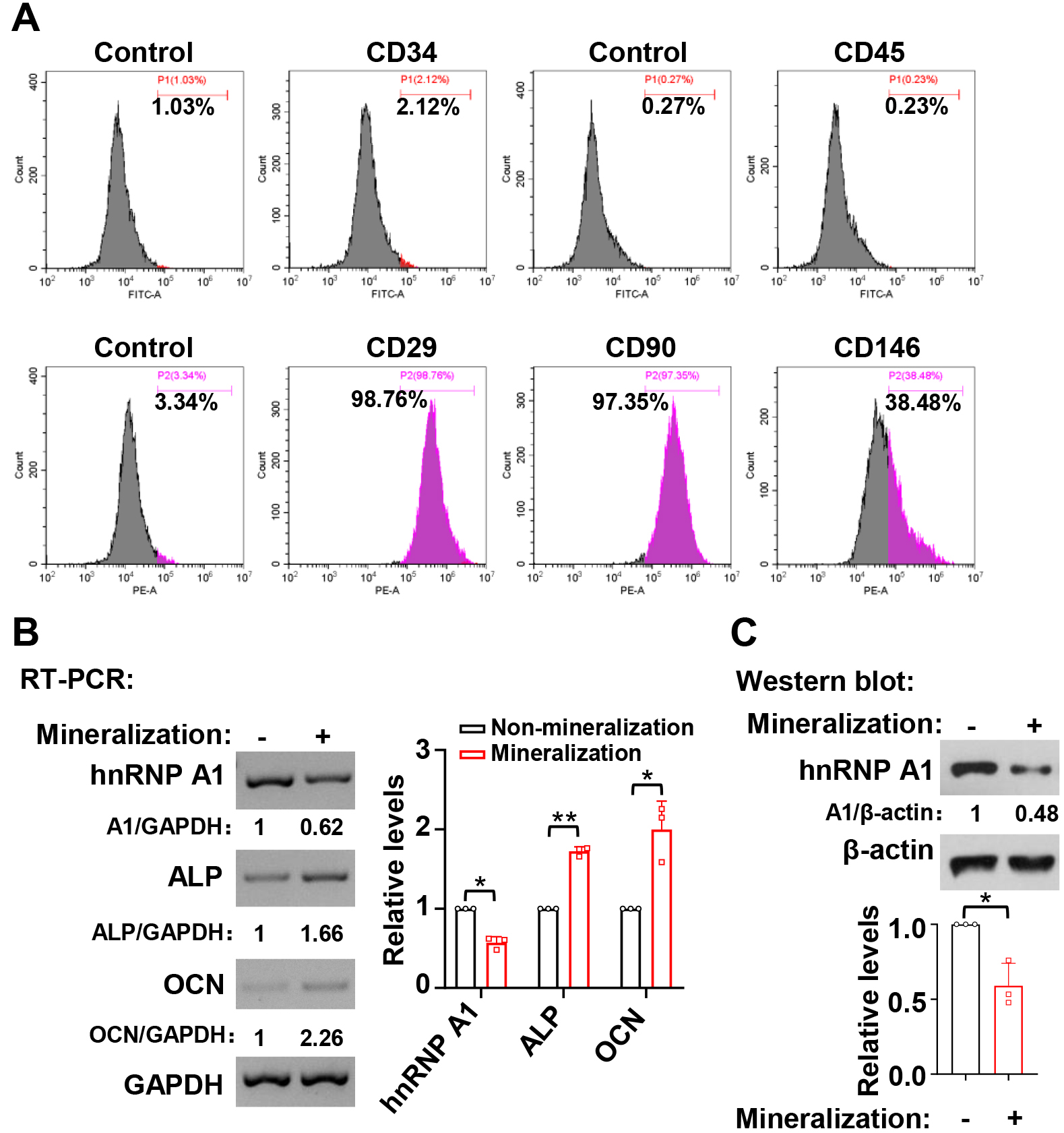 Fig. 1.
Fig. 1.HnRNP A1 expression decreased during the mineralization
induction in hDPSCs. (A) The expression of cell surface markers on hDPSCs was
analyzed by flow cytometric. (B) RNA expression levels of ALP and OCN increased
while the levels of hnRNP A1 decreased after seven days of mineralization
induction. Histogram showed quantification for the relative RNA expression levels
of hnRNP A1, ALP, and OCN, n = 3. GAPDH served as a loading control. (C) The
corresponding protein expression levels of hnRNP A1 decreased after seven days of
mineralization in hDPSCs. Histogram showed quantification for the relative
protein expression levels of hnRNP A1, n = 3.
Our previous study demonstrated that the inclusion of RUNX2 exon 5 is enhanced during the odontogenic differentiation of hDPSCs [10]. To explore whether hnRNP A1 participates in RUNX2 exon 5 alternative splicing, we applied an online RNA binding sequence database, SpliceAid. We found that RUNX2 exon 5 has four potential binding sites for hnRNP A1 (Fig. 2A). We further explored the role of hnRNP A1 in the alternative splicing of RUNX2 exon 5 by gain- and loss-of-function experiments. We found that overexpression of hnRNP A1 significantly suppressed RUNX2 exon 5 inclusion in hDPSCs (Fig. 2B,C). In contrast, in the hnRNP A1 knockdown, there was a markedly increased RUNX2 exon 5 inclusion (Fig. 2D,E). To confirm the inhibition of hnRNP A1 on the inclusion of RUNX2 exon 5, we co-transfected HEK 293 cells with RUNX2 exon 5 minigene and hnRNP A1 expression plasmid. Consistently, hnRNP A1 overexpression notably suppressed RUNX2 exon 5 inclusion compared with the control group in the minigene system (Fig. 2F). These results suggested that hnRNP A1 suppressed the inclusion of RUNX2 exon 5.
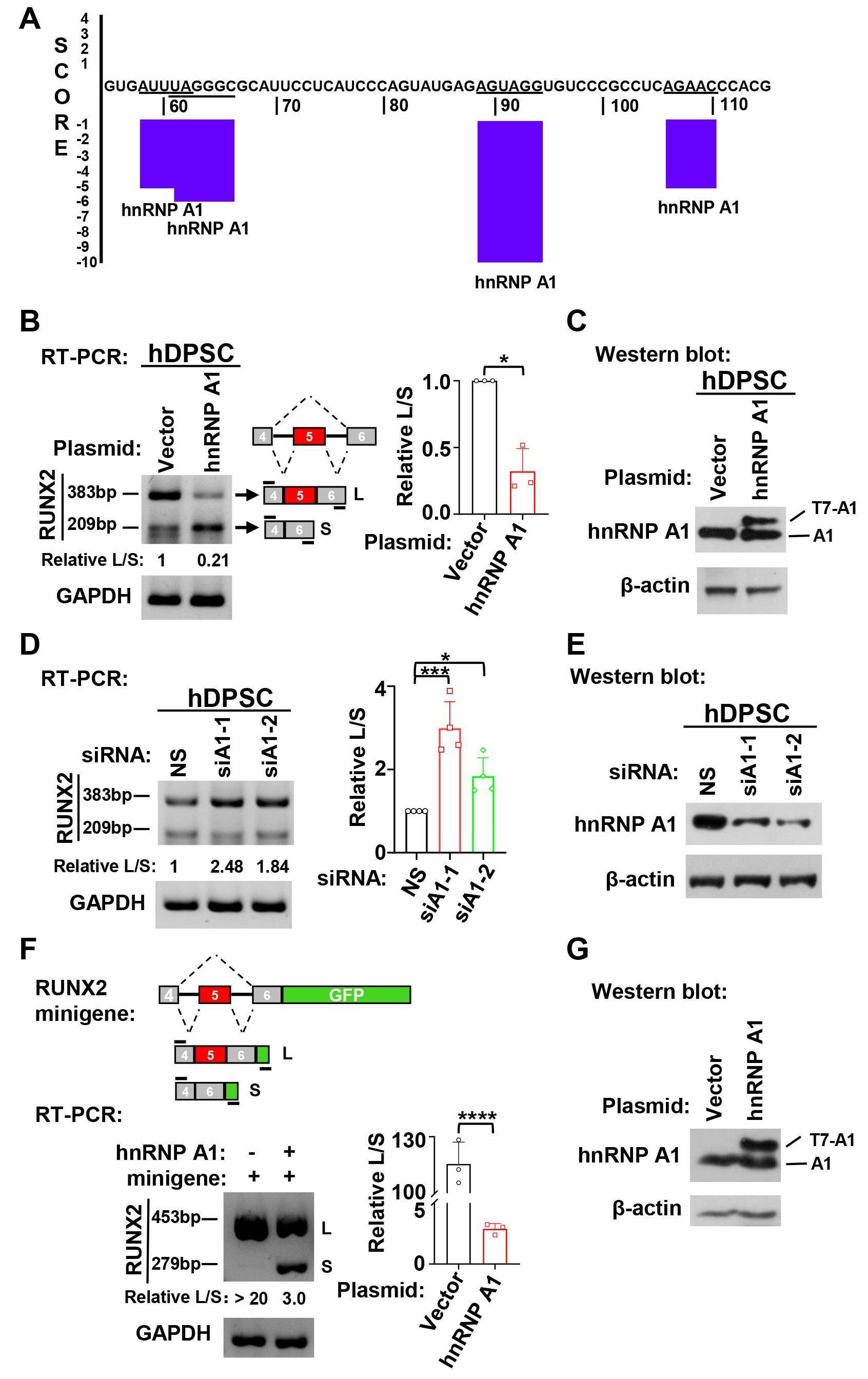 Fig. 2.
Fig. 2.HnRNP A1 suppressed the inclusion of RUNX2 exon 5. (A) HnRNP A1
binding sites in human RUNX2 exon 5 RNA sequence were predicted by SpliceAid
online program. Negative score represents suppressing exon inclusion. (B)
Overexpression of hnRNP A1 suppressed RUNX2 exon 5 inclusion in hDPSCs. The ratio
of inclusion versus exclusion of RUNX2 exon 5 in hDPSCs was represented by
histogram, n = 3. GAPDH served as a loading control. (C) Overexpression of
exogenous T7-tagged hnRNP A1 in hDPSCs was confirmed by Western blot.
Next, the function of hnRNP A1 in the odontogenic differentiation of hDPSCs was investigated. The expression levels of ALP and OCN were reduced after overexpression of hnRNP A1 in the mineralization induced hDPSCs (Fig. 3A). Meanwhile, ALP activity (Fig. 3B) and the formation of mineralized nodules were reduced in hDPSCs with overexpressed hnRNP A1 (Fig. 3C). Conversely, knockdown of hnRNP A1 led to a great increase in the expression of ALP and OCN in mineralized hDPSCs (Fig. 3D). ALP activity (Fig. 3E) and the formation of mineralized nodules (Fig. 3F) were also promoted in hDPSCs with hnRNP A1 knockdown. These results demonstrated that hnRNP A1 inhibited the mineralization of hDPSCs.
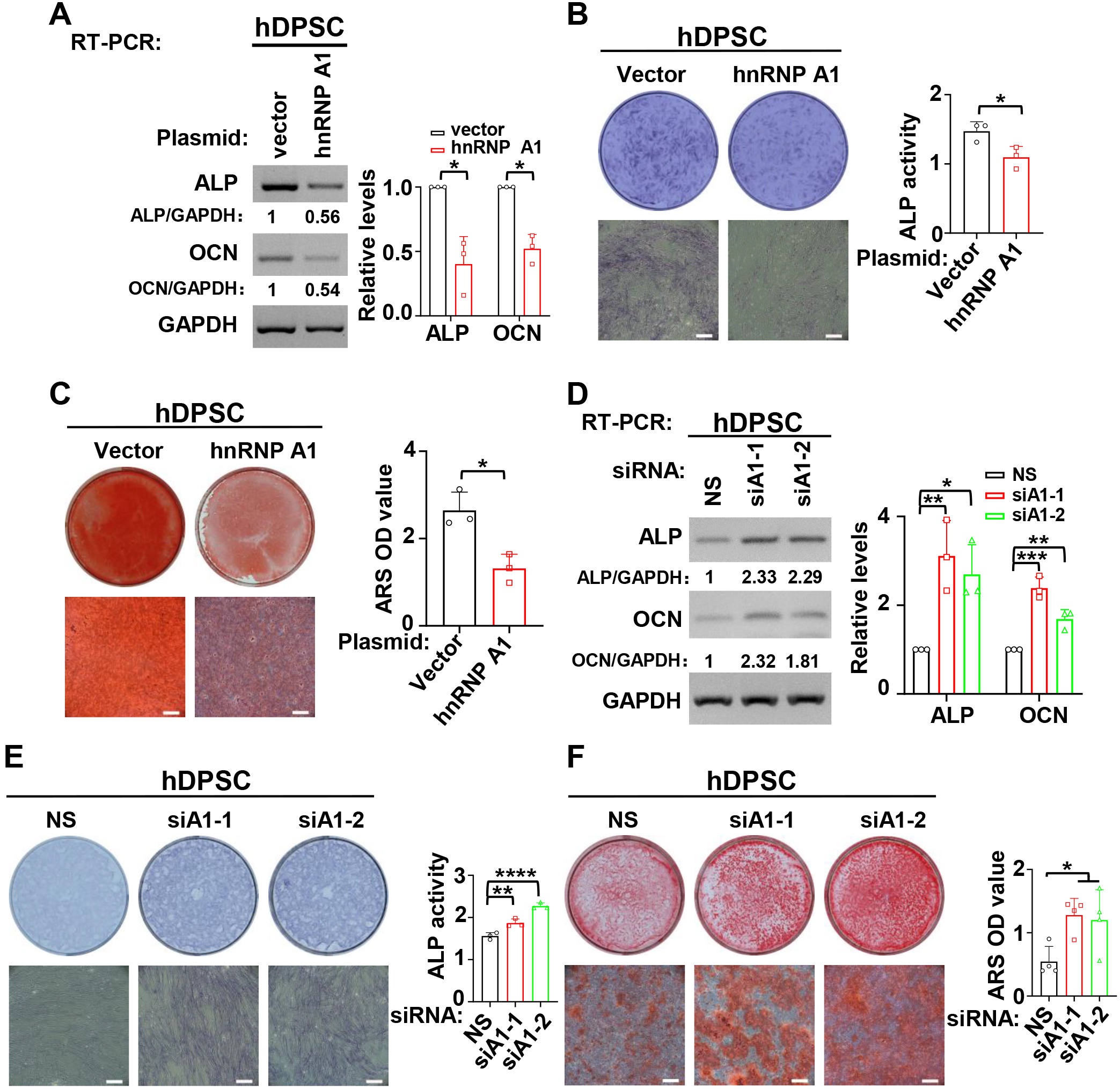 Fig. 3.
Fig. 3.HnRNP A1 inhibited the differentiation of hDPSCs.
(A–C) During the process of mineralization induction, overexpression of hnRNP A1
decreased the expression of ALP and OCN (A), ALP activity (B), and the formation
of mineralized nodules (C). Alizarin red staining (ARS) was applied to stain
mineralized nodules after hDPSCs were cultured in differentiation medium for 14
days. Mineralized nodules were collected with 10% cetylpyridinium chloride and
the absorbance values at 562 nm were measured for quantification of mineralized
nodules. Histogram respectively showed quantification for the expression of ALP
and OCN, ALP activity or the formation of mineralized nodules, n = 3. GAPDH
served as a loading control. Scale bar = 500 µm. (D–E) HnRNP A1 knockdown
increased the expression of ALP and OCN (D), ALP activity (E) and the formation
of mineralized nodules (F). ARS was applied to stain mineralized nodules after
hDPSCs cultured in differentiation medium for 14 days. Histogram respectively
showed quantification for the expression of ALP and OCN (n = 3), ALP activity (n
= 3) or the formation of mineralized nodules (n = 4). GAPDH served as a loading
control. Scale bar = 500 µm. *: p
According to our previous study, RUNX2 exon 5 is highly conserved in human and mouse except for one nucleotide replacement [10]. Therefore, we speculated that hnRNP A1 may also suppress mouse Runx2 exon 5 inclusion and odontogenic differentiation in mDPCs. As we speculated, hnRNP A1 plays an identical role both in hDPSCs and mDPCs in Runx2 exon 5 inclusion and odontogenic differentiation. Overexpression of hnRNP A1 significantly suppressed the inclusion of Runx2 exon 5 (Fig. 4A), and hnRNP A1 knockdown markedly promoted the inclusion of Runx2 exon 5 (Fig. 4E). We also found that overexpression of hnRNP A1 inhibited the expression of Alp and Ocn in the differentiation of mDPCs (Fig. 4C). In addition, the formation of mineralized nodules was reduced in hnRNP A1 overexpressed mDPCs compared with the control group (Fig. 4D). On the contrary, knockdown of hnRNP A1 led to increased expression of Alp and Ocn and increased mineralized nodules (Fig. 4G,H). These findings demonstrated that hnRNP A1 contributed to the skipping of RUNX2 exon 5 and repressed odontogenic differentiation of mDPCs.
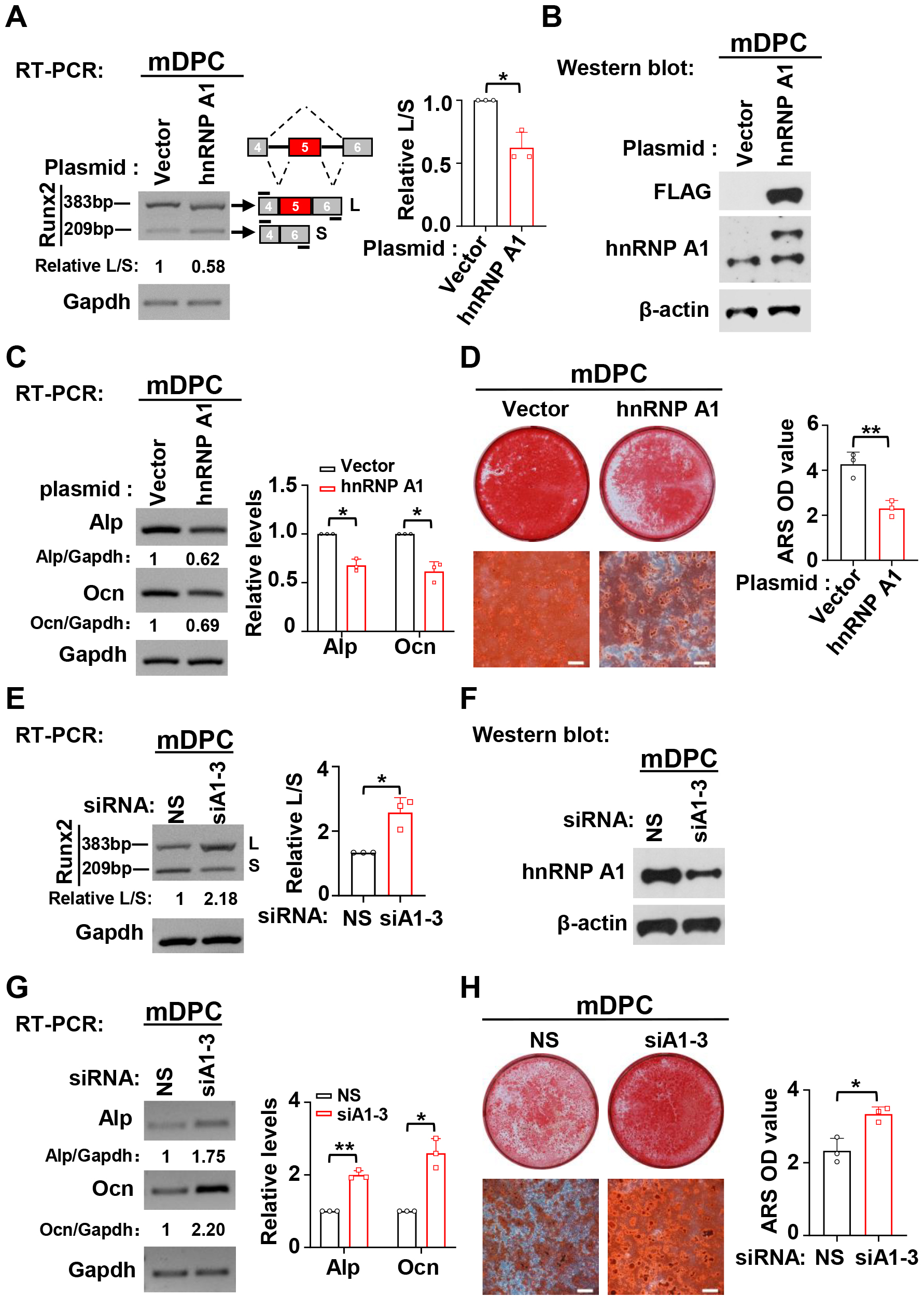 Fig. 4.
Fig. 4.HnRNP A1 suppresses the inclusion of Runx2 exon 5 and
differentiation in mDPCs. (A) Overexpression of hnRNP A1 suppressed the
inclusion of Runx2 exon 5. Bar graph showed the summarized ratio
of inclusion versus exclusion of Runx2 exon 5, n = 3. Gapdh served as a loading
control. (B) Overexpression of exogenous FLAG-tagged hnRNP A1 was confirmed by
Western blot. Actin served as a loading control. (C,D) Overexpression of hnRNP A1
reduced the expression of Alp and Ocn (C) and the formation of mineralized
nodules (D). ARS was applied to stain mineralized nodules after mDPCs cultured in
differentiation medium for 7 days, Scale bar = 500 µm. Histogram showed
quantification for the expression of Alp and Ocn, or the formation of mineralized
nodules, n = 3. Gapdh served as a loading control. (E) Knockdown of hnRNP A1
promoted the inclusion of Runx2 exon 5. Bar graph showed the summarized ratio of
inclusion versus exclusion of Runx2 exon 5, n = 3. Gapdh served as a loading
control. (F) Knockdown efficiency of hnRNP A1 was analyzed by Western blot.
Alternative splicing was considered as a ubiquitous regulatory mechanism of gene expression that generates multiple mRNA species from a single gene [22]. Recent studies have shown that alternative splicing plays a role in MSC differentiation [23, 24]. The hDPSC is a type of human dental-tissue-derived hMSC, that possesses considerable differentiation capacity [18, 25]. A few studies have shown that alternative splicing plays an essential role in the differentiation of hDPSC. For instance, Octamer-binding transcriptional factor 4A (OCT4A), one isoform derived from OCT4 alternative splicing, enhanced the differentiation capability of hDPSC [26, 27]. RUNX2 consists of nine exons, several studies have demonstrated that the inclusion of exon 5 is essential for the function of RUNX2 [9, 28]. Multiple studies have indicated that RUNX2 is essential for odontogenesis [3, 4]. However, few studies focused on the regulatory mechanism of alternative splicing on RUNX2 exon 5. Our previous study showed that YBX1 enhances the inclusion of RUNX2 exon 5 [10]. In the current study, we showed that hnRNP A1 had an opposite function to YBX1 in regulating the inclusion of RUNX2 exon 5. The hnRNP A1 protein belongs to the hnRNP A/B subfamily and is traditionally considered to be a splicing repressor [29]. The DNA sequence of human RUNX2 exon 5 is highly conserved in mouse, with only one nucleotide substitution and the amino acid sequences are identical. In this study, through gain- and loss-of-function experiments, we found that hnRNP A1 suppressed the inclusion of exon 5 in both human and mouse cells, suggesting that the inhibitory effect of hnRNP A1 on RUNX2 exon 5 inclusion is conserved in both species. This finding is complementary to the alternative splicing mechanism of RUNX2 exon 5.
Several studies have shown that the function of hnRNP A1 in stem cell differentiation can be achieved by regulating alternative splicing or transcription [15, 16]. Fang et al. [15] found that the ubiquitination of hnRNP A1 enhanced the exclusion of Arhgap1 exon 2, which ultimately impaired the differentiation of hematopoietic stem/progenitor cells. In another study, Huang et al. [16] found that hnRNP A1 promoted the differentiation of embryonic stem cells into smooth muscle cells (SMC) by directly binding to the promoter of SMC differentiation genes. However, the role of hnRNP A1 in differentiation of hDPSCs was poorly understood. Our current study showed that hnRNP A1 repressed the differentiation of hDPSCs by the reduction of the expression of ALP and OCN and the formation of mineralized nodules. These findings suggested that hnRNP A1 participated in the stem cell differentiation, and that the different roles of hnRNP A1 in differentiation of different stem cells may be related to the diversity of stem cell types.
In conclusion, we elucidated the roles of the RNA binding protein hnRNP A1 in the alternative splicing of RUNX2 exon 5 and the differentiation of hDPSCs and mDPCs. We discovered a novel inhibitory mechanism of RUNX2 exon 5 inclusion and the differentiation of dental mesenchymal cells (Fig. 5).
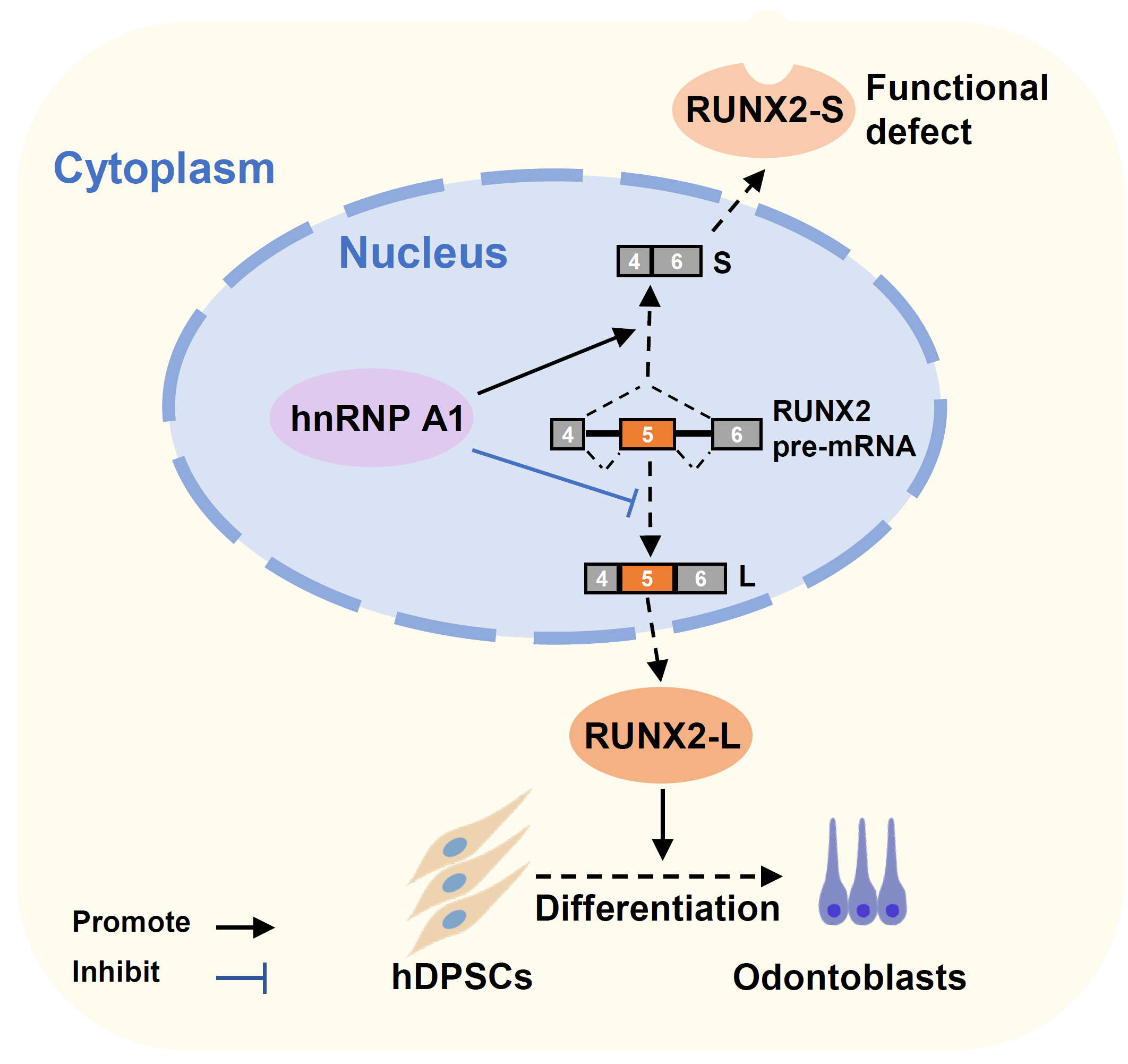 Fig. 5.
Fig. 5.A schematic model of hnRNP A1 inhibits RUNX2 exon 5 inclusion and hDPSCs differentiation. HnRNP A1 represses RUNX2 exon 5 inclusion, decreases the expression of full-length functional RUNX2 protein, and leads to the suppression of hDPSCs’ odontogenic differentiation.
Data supporting the findings of this study are available from the corresponding author upon reasonable request.
JG and RJ designed the research study. YZ and JH performed the research. YZ, JH and LY conducted the statistical analysis of data. YZ, JG and RJ were responsible for writing of manuscript. YZ, JH, LY, RJ and JG contributed to the manuscript review and editing. All authors read and approved the final submitted manuscript.
This study was conducted in accordance with the Declaration of Helsinki. The collection of human dental pulp tissues was approved by the Ethics Committee of the School and Hospital of Stomatology at Wuhan University (2019-A47). Informed consent was obtained from every participant. The use of animal tissues was approved by the Animal Welfare and Ethics Committee of the School and Hospital of Stomatology at Wuhan University (S07922090F).
We would like to acknowledge the contribution to the dental pulp tissue collection of Xiulin Ran and Yujing Zhang, in the Department of Endodontics, School & Hospital of Stomatology, Wuhan University, Wuhan, 430079, China.
This study was supported by National Natural Science Foundation of China (Grant No. 81970933).
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
