1. Introduction
In Drosophila, the cryptocephal (crc) gene encodes
the activating transcription factor 4 (ATF4) protein, which belongs to the family
of a basic-leucine zipper transcription factors [1, 2]. ATF4 regulates gene
expression involved in endoplasmic reticulum (ER) stress, amino acid metabolism,
and redox enzymes via a CCAAT-enhancer binding protein-activating transcription
factor response element. Through this transcriptional activity, ATF4 protein is
associated with developmental and disease processes, including anoxia [3],
long-term facilitation [4], stress response [5, 6], apoptosis [7], and cancer [8, 9]. Mutations in the crc (atf4) gene in Drosophila
result in significant lethality during development. Specifically, the hypomorphic
point mutation in crc, known as crc, which involves a
single amino acid change at residue 171 from glutamine to arginine, leads to
delayed larval development and pupal lethality [10, 11, 12, 13].
Integrated stress response (ISR) is a highly conserved homeostatic signaling
pathway that is crucial in controlling translation, amino acid imbalance, and
glucose homeostasis [14, 15, 16]. The common event in this pathway is the
phosphorylation of eukaryotic translation initiation factor 2 on serine 51 of its
alpha subunit (eIF2), which reduces global protein synthesis and induces the
expression of certain genes. GCN2 kinase mediates the ISR signaling pathway as an
amino acid sensor by binding to the uncharged transfer tRNA. The activation of
GCN2 kinase phosphorylates eIF2, leading to ATF4 protein synthesis, and further
triggering ATF4-mediated gene expression to protect cells from amino acid
deprivation. Thus, ATF4 is presumed to be a main downstream component of the ISR.
Previously, we developed an assay tool to detect the in vivo ATF4
translational activity [17]. The study using this reporter indicates that
Drosophila ATF4 protein synthesis increases in response to ER stress and
ISR, and the translational regulatory mechanism of ATF4 is conserved among other
species. Moreover, we demonstrated that the GCN2/ATF4/4E-BP pathway is required
for lifespan extension upon the dietary restriction of amino acids [5, 18]. In
the present study, we analyzed the metabolic status in atf4
mutant flies. These flies appear to use glucose to produce lactate instead of
producing adenosine triphosphate (ATP) through the tricarboxylic acid (TCA) cycle. Microarray analysis of
atf4 mutant flies revealed that ATF4 regulates gene expression
related to enzymes such as hydrolase, acyltransferase, and oxidoreductase, as
well as related to immune response, cell death, and transcription factor.
Further, the overexpression of lipase, which is the transcriptional target of
ATF4, manifests in increased starvation resistance. These results demonstrate
that Drosophila ATF4 regulates gene expression in response to dietary
restriction to resist metabolic stress.
2. Materials and Methods
2.1 Fly Strains
All Drosophila samples were cultivated on standard Bloomington
Drosophila Stock Center cornmeal food containing 1.6% yeast, 0.9% soy flour,
6.7% cornmeal, 1% agar, and 7% light corn syrup at 25 °C. The coding
sequences for CG6283 and CG6295 were obtained via reverse transcription
polymerase chain reaction (RT-PCR) from yw larvae. The HA-tag was added
to the C termini of these coding sequences and subcloned into a pUAST. The
following strains of flies have been previously described:
atf4, atf4 [5], and Act5C [19].
UAS-lacZ flies were obtained from Bloomington Drosophila Stock Center (IN, USA).
For gene induction, 100 µL of a 5 mg/mL solution of RU486 (Sigma, St. Louis, MO,
USA; cat. #M4086) was added on top of food in a vial and dried overnight before
feeding it to the flies.
2.2 Nutrient Restriction on Drosophila Larvae
Larvae were collected approximately 47–49 h after egg laying (AEL) on
apple-juice plates (25% apple juice, 1.25% sucrose, and 2.5% agar) and then
transferred to standard cornmeal food (5.9% glucose, 6.6% cornmeal, 1.2%
baker’s yeast, and 1% agar in water) or to nutrient -restricted medium (5%
sucrose and 1% agar in PBS) for 18 h at 25 °C.
2.3 Real-Time RT-PCR
Total RNA was isolated using TRIzol (Invitrogen, 15596018, Waltham, MA, USA),
and 100 ng of RNA was transcribed with ReverTra Ace qPCR RT kit (TOYOBO Co.,
Osaka, JAPAN). The real-time RT-PCR was run for 40 cycles using the
TOPreal qPCR 2X PreMIX (SYBR Green with high ROX, enzynomics, Seoul,
Republic of KOREA) and a LightCycler 480 Real-Time PCR system (Roche, Rotkreuz,
Switzerland). The primer sequences are listed in Supplementary Table 1.
2.4 In Situ Hybridization
The full-length CG6283 cDNA was subcloned into pBluescript SK+. The T3 and T7
promoters of the pBluescript SK+ were used to generate DIG-labeled riboprobes for
in situ hybridization using standard protocols [DIG RNA Labeling Kit
(SP6/T7), 11175025910, Roche].
2.5 Microarray Analysis
Microarray experiments were performed using GeneChip® Drosophila
Genome 2.0 Array (Applied Biosystems, Foster City, CA, USA). Total RNA
from the flies was isolated using the Trizol reagent (Invitrogen, Waltham, MA,
USA). Thereafter, cDNA was amplified from a 100-ng aliquot of total RNA from each
sample using the GeneChip WT (whole transcript) amplification kit, as described
by the manufacturer (Affymetrix, Santa Clara, CA, USA). The sense cDNA was then
fragmented and biotin-labeled with terminal deoxynucleotidyl transferase using
the GeneChip WT Terminal labeling kit (Thermo Fisher Scientific, Waltham, MA,
USA). Approximately, 5.5 µg of the labeled DNA target was hybridized to the
Affymetrix GeneChip Array at 45 °C for 16 h. Next, the hybridized arrays
were washed and stained on a GeneChip Fluidics Station 450 and scanned on a GeneChip System (GCS)
3000Dx v.2 Scanner (Affymetrix). Array data export processing and analysis was
performed using Affymetrix® GeneChip Command
Console® Software. Specifically, the fold change (fc) was
determined as follows: First, lagfc was calculated as the difference
between the normalized values of the Test and Control samples (logfc =
Normalized value of Test – Normalized value of Control). Second, the calculated
logfc value was computed as 2 raised to the power of logfc (fc =
2) to convert it into a linear scale. If the calculated
fc value fell within the range of 0 to 1, it was interpreted as downregulated. If
the value was 1, it was considered upregulated. To effectively represent
values between 0 and 1, the fc value was transformed into its negative reciprocal
(-1/fc) (Macrogen, Seoul, Korea).
2.6 Metabolic Profiling
Fifty larvae of each genotype were collected at approximately 47–49 h AEL and
homogenized using TissueLyzer (Qiagen, Hilden, Germany) with MeOH. Internal
standard solutions (malonyl-C CoA, 5 µM Gln-d) were
added to the samples. Thereafter, the samples were centrifuged at 15,700
g for 10 min (Eppendorf Centrifuge 5415R). The precipitate was stored
for further measurement of the protein amount by the Bradford assay. For the
supernatant, the aqueous phase after liquid–liquid extraction was collected and
used for subsequent analysis. Metabolites were analyzed via liquid chromatography
with tandem mass spectrometry (LC-MS/MS) [1290 HPLC (Agilent)-Qtrap 5500
(ABSciex)]. For metabolites related to energy metabolism, Synergi Fusion RP 50
2 mm was used. Here, 5 mM CHCOONH in HO and in MeOH
served as mobile phases A and B, respectively. The separation gradient was as
follows: hold at 0% B for 5 min, 0%–90% B for 2 min, hold at 90% for 8 min,
90%–0% B for 1 min, and then hold at 0% B for 9 min. The LC flow was 70
µL/min, except for 140 µL/min between 7–15 min, at 23 °C.
For fatty acyl CoAs, a Zorbax 300 Extend-C18 column (2.1 150 mm) was
used. Mobile phase A comprised acetonitrile (ACN)–HO (10:90) with 15 mM
NHOH, and mobile phase B comprised ACN containing 15 mM NHOH. The
separation gradient was as follows: hold at 0% B for 3 min, 0%–50% B for 2
min, 50%–80% B for 5 min, 80%–0% B for 0.1 min, and then hold at 0% B for
4.9 min. The LC flow was 200 µL/min, and the column was kept at
25 °C. Multiple reaction monitoring was employed for analysis. The
quantitative value of each metabolite was normalized to the total protein amount.
2.7 Starvation Assay on Adults
Twenty female flies (5 days old) of each genotype were transferred to vials
containing 1% agar in PBS. The flies were supplied fresh food every 12 h and
maintained at 25 °C; deaths were recorded at 96 h after starvation.
3. Results
3.1 Metabolomic Analysis Revealed the Metabolic Status in
atf4 Mutant Larvae
To investigate the potential involvement of ATF4 in metabolic homeostasis, we
assessed the nutrient reserves in the atf4 mutant larvae.
Specifically, we measured the level of intermediates of major metabolic pathways,
including glycolysis, the TCA cycle, and the pentose phosphate pathway, as well
as coenzymes related to fatty acid metabolism. As shown in Fig. 1, there were
notable differences in the levels of intermediates in the major metabolic
pathways between the yw (atf4, control) flies and the atf4 (atf4) mutant flies
(Fig. 1A–C). Overall, the relative levels of metabolites were higher in the
atf4 mutant flies compared to the control
flies. However, the level of ATP, primarily produced during the TCA cycle, was
significantly decreased in the atf4 larvae
(Fig. 1B). Interestingly, the lactate level, the end product of glycolysis, was
higher in the atf4 larvae than in the control
larvae, although not significantly (Fig. 1A). In contrast, the amount of coenzyme
and diacylglycerol did not differ between the two groups. These results indicate
that the ATF4 is involved in energy metabolism including glycolysis and this
perturbation in energy balance may contribute to the physiology of the
atf4 mutant flies.
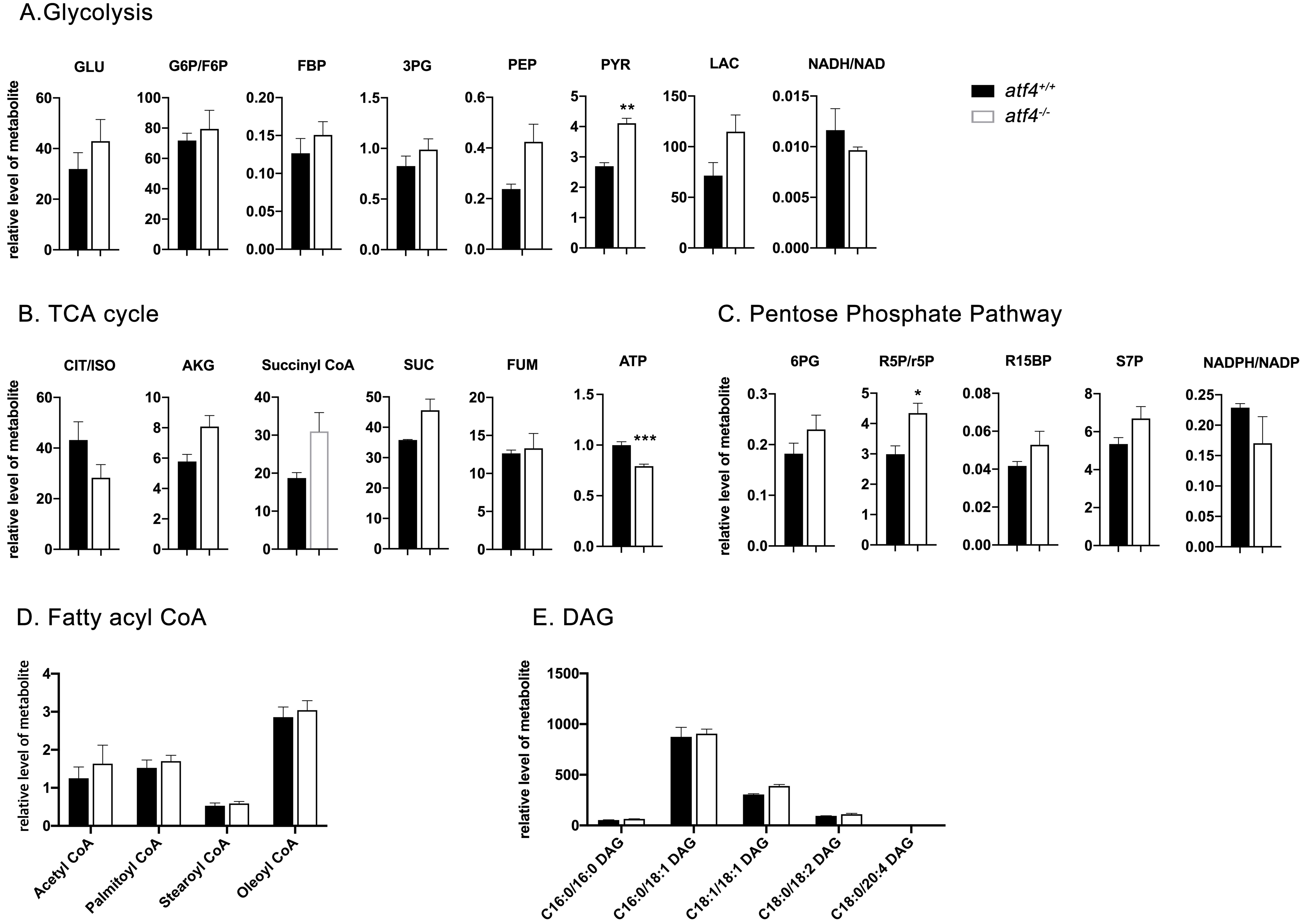 Fig. 1.
Fig. 1.
Metabolic reprogramming in atf4 mutants.
Control (yw) and atf4 (atf4) mutant larvae were collected at
approximately 47–49 h after egg laying. The metabolites were monitored via
liquid chromatography-tandem mass spectrometry (LC-MS/MS). (A) The relative
amount of the intermediates of glycolysis. The values of each metabolite were
normalized to the total protein level. (B) The relative levels of the
intermediates of the tricarboxylic acid (TCA) cycle. (C) The relative amount of the intermediates of
the pentose phosphate pathway (PPP). (D,E) The levels of fatty acyl CoA and
diacylglycerol (DAG). These experiments were conducted in triplicate. Data are
presented as mean standard error of the mean (SEM). p-values were determined using Student’s
t-test. *p 0.05, **p 0.01, and ***p
0.001. Abbreviation: GLU, Glucose; G6P, Glucose-6-phosphate; F6P,
Fructose-6-phosphate; FBP, Fructose-1,6-bisphosphate; 3PG, 3-phosphoglycerate;
PEP, Phosphoenolpyruvate; PYR, Pyruvate; LAC, Lactate; NADH, reduced nicotinamide
adenine dinucleotide; NAD, Nicotinamide Adenine Dinucleotide; CIT, Citrate; ISO,
Iso citrate; AKG, alpha-ketoglutarate; SUC, Succinate; FUM, Fumarate; ATP,
Adenosine triphosphate; 6PG, 6-phosphogluconate; R5P, Ribulose-5-phosphate; r5P,
Ribose-5-phosphate; R15BP, Ribose-1,5-bisphosphate; S7P,
Sedoheptulose-7-phosphate; NADPH, reduced Nicotinamide Adenine Dinucleotide
Phosphate; NADP, Nicotinamide Adenine Dinucleotide Phosphate; DAG,
diacylglycerol.
3.2 ATF4 Regulates Gene Expression Upon Nutrient Restriction
Considering that ATF4 is known to respond to nutrient restriction in various
species [5, 20, 21], we conducted a genome-wide expression profiling of control
(atf4) and atf4 larvae subjected to dietary
restriction for 18 hours using GeneChip® Drosophila Genome 2.0
Array (see Material and Methods). The microarray analysis was performed three
times using independent biological replicates, and we compared the gene
expression between the wild type (yw, atf4) and
atf4 mutant larvae subjected to 18 hours of starvation
(Supplementary Fig. 1 and Supplementary Table 2). The
differentially expressed genes (the value of atf4 starved/the
value of atf4 starved) with p 0.05 and a fold
change 2 were selected for subsequent gene ontology analysis. Based on these
criteria, we identified 141 genes that were downregulated in atf4mutant larvae compared to the control (Supplementary Table 3), and 132
genes that were upregulated (Supplementary Table 4). Out of the 141
downregulated genes, 110 genes could be grouped into significantly enriched
functional categories, which included lipase activity, oxidoreductase activity,
acyltransferase, immune response, cell death, and transcription factor functions
(Table 1 and Supplementary Table 3).
Table 1.The analysis of genes regulated by ATF4 in response to nutrient
restriction.
| Function |
Probe ID |
Drosophila gene |
Human othologs |
Fold |
Function |
Probe ID |
Drosophila gene |
Human othologs |
Fold |
| Lipase activity |
1635045 |
CG6271 (triglyceride lipase) |
|
–8.61 |
Acyltransferase |
1625235 |
CG13325 |
|
–4.22 |
|
1629367 |
CG15534 (sphingomyelin phosphodiesterase) |
SMPD1 |
–6.14 |
|
1626764 |
CG10182 |
|
–4.20 |
|
1623775 |
CG31089 (triglyceride lipase) |
LIPA (lipase, family member) |
–5.02 |
|
1631234 |
CG18173 |
PIGW |
–2.83 |
|
1636343 |
CG6283 (triglyceride lipase) |
|
–2.81 |
|
1632163 |
CG8481 |
NAT6 |
–2.42 |
|
1635868 |
CG6295 (serine hydrolase) |
|
–2.76 |
|
1629934 |
CG14219 |
|
–2.10 |
|
1633709 |
CG2772 (triglyceride lipase) |
LIPA (lipase, family member) |
–2.51 |
Hydrolase activity |
1635812 |
CG16965 |
ATHL1 |
–3.33 |
|
1632120 |
CG15533 (sphingomyelin phosphodiesterase) |
SMPD1 |
–2.30 |
|
1637357 |
CG9463 |
|
–3.23 |
| Oxidoreductase activity |
1640566 |
Cyp4p2 |
cytochrome P450 |
–62.81 |
|
1637602 |
CG32801 /// Edem1 |
EDEM1 |
–3.02 |
| 1625436 |
Uro |
|
–4.16 |
|
1639401 |
Mal-A1 |
|
–2.78 |
|
1634623 |
Cyp6a14 |
|
–3.88 |
Immune response |
1626319 |
CG33470 /// IM10 |
|
–13.32 |
|
1623000 |
CG33093 |
|
–3.82 |
|
1627986 |
PGRP-SC1a /// PGRP-SC1b |
PGLYRP |
–5.26 |
|
1622906 |
Sod3 |
SOD1, CCS |
–3.82 |
|
1627613 |
Mtk |
|
–4.95 |
|
1633471 |
Prx2540-2 |
|
–3.73 |
|
1636490 |
PGRP-SB1 |
PGLYRP |
–3.15 |
|
1626401 |
Cyp6a2 |
|
–3.53 |
Cell death |
1630010 |
pnt |
ETS1, ETS2 |
–3.75 |
|
1626503 |
CG2254 /// DsecGM11216 |
DHRS3, HSD17B11, RDH10, SDR16C5 |
–3.45 |
|
1625981 |
rab3-GEF |
MADD |
–3.40 |
|
1624101 |
Cyp6a23 |
|
–3.23 |
|
1627463 |
Damm |
Caspase-like domain |
–3.13 |
|
1633401 |
Cyp12d1-d /// Cyp12d1-p |
|
–3.00 |
Transcription factor activity |
1626392 |
Mef2 |
MEF2 |
–6.59 |
|
1639892 |
Sodh-1 |
|
–2.88 |
|
1641365 |
jim |
|
–3.17 |
|
1629745 |
CG6439 |
IDH3G |
–2.75 |
|
1625195 |
Cyp6v1 /// shn |
|
–2.62 |
|
1635110 |
Cyp6a13 |
|
–2.72 |
Peptidase activity |
1634477 |
CG42335 |
ERAP1-like C-terminal domain |
–8.57 |
|
1630244 |
CG31809 /// CG31810 |
HSDL1, HSD17B3 |
–2.43 |
|
1627156 |
CG33225 |
GZMB (granzyme B), CTSG (cathepsin G) |
–3.69 |
|
1631452 |
CG8665 |
ALDH1L1 |
–2.41 |
|
1635453 |
Bace |
|
–3.64 |
|
1630359 |
CG31810 |
HSDL1, HSD17B3 |
–2.16 |
|
1623493 |
CG12717 |
|
–2.32 |
|
1624159 |
Cyp9h1 |
CYP3A4 |
–2.07 |
|
1635398 |
CG10587 /// Scp2 |
|
–2.22 |
|
|
|
|
|
|
1639391 |
CG17109 |
PM20D1 |
–2.19 |
# Fold: the value of atf4 starved/the value of atf4 starved.
To validate the microarray results, we performed quantitative RT-PCR. As shown
in Fig. 2, the expression of lipase genes (CG6271, CG15533, CG15534, and CG31089)
increased in response to 18 hours of larval starvation. However, this increase in
gene expression due to nutrient restriction was suppressed in
atf4 mutants, indicating that these lipases are induced by ATF4
in response to nutrient restriction. Similarly, the expression of
CG11893 and Cyp4p2, which exhibit transferase and
oxidoreductase activities, respectively, was also increased by starvation, and
their regulation was dependent on ATF4. In contrast, the expression of
CG33039 (with oxidoreductase activity), CG31436 (with
transferase activity), and nol (involved in neuroblast proliferation) were
regulated by ATF4 independently of the nutrient status.
 Fig. 2.
Fig. 2.
Relative expression levels of the selected transcripts
identified as the transcriptional targets of ATF4. Control (yw) and
atf4 larvae at 48 h after egg laying were collected and
subjected to a restricted diet. After 18 h, total mRNA was extracted and used for
quantitative reverse transcription polymerase chain reaction (RT-PCR). The values
were normalized to the Rp49 data. (A) Lipase activity. (B) Transferase
activity. (C) Oxidoreductase activity. (D) Neuroblast proliferation. The data
represent the mean and SEM from at least four independent experiments.
p-values were determined using Student’s t-test. *p
0.05, **p 0.01, and ***p 0.001.
3.3 Increased Expression of Lipase, the Transcriptional Target of
ATF4, Enhances Resistance to Starvation
To investigate the localization of lipase gene expression regulated by ATF4, we
performed in situ hybridization. Among the lipases in
Drosophila, CG6283 is classified as a gastric lipase, but is
not well characterized. Under fed conditions, we detected mRNA expression of
CG6283 in the midguts of both atf4mutant flies and the
wild-type flies (Fig. 3A). However, upon starvation, the mRNA levels of CG6283
significantly increased. Specifically, we
observed an induction of CG6283 in the gastric caeca (arrow in Fig. 3A), which is
involved in enhancing digestive enzyme secretion and nutrient absorption.
Notably, this increase in CG6283 expression was absent in atf4
mutants. To further explore the physiological function of lipase under nutrient
restriction, we induced the expression of CG6283 or CG6295 using
Act5C-gal4, which led to the expression of these genes throughout the
entire body only when RU486 was added to the diet. We subjected the flies to
complete starvation for 4 days and monitored their survival rate. Flies
overexpressing lipases (Act5C CG6283 or Act5C CG6295; 71.4% and 83.3%, respectively) exhibited
longer survival than control flies (Act5C lacZ; 26.7%) (Fig. 3B). In summary, these findings suggest that Drosophila ATF4
plays a critical role in promoting organism survival during nutritional
starvation by inducing the expression of enzymes such as lipases, which provide
essential energy sources for the organism.
 Fig. 3.
Fig. 3.
Lipase gene contributes to the survival of flies from
starvation. (A) the transcripts of CG6283 are at the basal level in the
midgut of wild type (atf4) and atf4 mutants but are significantly enhanced only in the gastric caeca of wild type
by starvation. The induction of CG6283 by starvation was not detected in
atf4 mutants. (B) lipase overexpressing flies are more resistant
to starvation. Five-day-old flies (20 flies in each vial) were cultured in a
complete starvation medium for 4 days. The percentage indicated the number of
surviving flies (n = 3). The data represent the mean and SEM from at least three
independent experiments. p-values were determined using Student’s
t-test. *p 0.05 and **p 0.01.
4. Discussion
In our previous work, we developed an in vivo reporter to detect ATF4
translational activity, which demonstrated that ATF4 responds to ER stress and
ISR [17]. In this study, we performed a metabolomic analysis to investigate the
metabolic status of Drosophila atf4 mutants. Most of
the analyzed metabolites were found to be higher in the atf4
flies compared to the control (yw) flies. Notably, the lactate level was
elevated in the atf4 flies, although not significantly, while
ATP production was reduced.
Lactate is a product of glucose metabolism and is produced in highly glycolytic
tissues, such as the skeletal muscle. It can be converted to pyruvate by lactate
dehydrogenase and utilized in mitochondria in various tissues, including the
liver and kidney. Under certain anaerobic conditions (hypoxia) and hypostasis,
lactate levels can substantially increase (hyperlactatemia), potentially leading
to cell injury. Hyperlactatemia is associated with various diseases, such as
heart disease, severe anemia, and diabetes mellitus [22, 23]. Moreover, the
increase in lactate levels in the atf4 mutants resembles the
Warburg effect, characterized by increased glucose uptake and lactate
accumulation even under aerobic conditions. The Warburg effect is a metabolic
reprogramming observed in cancer cells and is essential for cancer progression.
Cancer cells primarily employ the glucose pyruvate lactate pathway for
their proliferation. However, when they require energy for metastasis, similar to
normal cells, they undergo a metabolic shift to produce ATP in mitochondria [24, 25]. Interestingly, the atf4 mutant flies seem to preferentially
use glucose to produce lactate rather than generating ATP through the TCA cycle,
similar to the Warburg effect. This is supported by the decreased ATP levels and
relatively low or similar levels of TCA cycle intermediates in the
atf4 mutants (Fig. 1B). These metabolic characteristics are
typically observed in proliferating cancer cells but not in differentiating
cells. Although some TCA cycle metabolites, such as AKG and SUC, were increased
in the atf4 mutants, they could potentially be produced through
a salvage pathway. Overall, it appears that the atf4
mutants experience dysregulation of energy metabolism.
Furthermore, these mutants die during the pupal stage [10]. They exhibit certain
developmental defects, such as the absence of head eversion and abnormal
differentiation of the abdomen but show normal eye pigmentation and proper
differentiation of wings and legs. Considering that metamorphosis at the pupal
stage requires substantial energy [26], it is plausible that the dysregulation of
energy metabolism in the atf4 mutants may play a role in their
lethality. However, additional research is necessary to comprehensively
understand this phenomenon.
The findings from numerous previous studies have reported ATF4 as a
transcriptional regulator of genes involved in various cellular processes under
different stress conditions. For instance, ATF4 promotes the expression of genes
related to amino acid import, glutathione biosynthesis, and resistance to
oxidative stress during ER stress in eukaryotes [20]. Another study in
Drosophila S2 cells subjected to ER stress revealed that ATF4 controls
the gene expression of glycolytic enzymes [27].
In our study, we observed that genes categorized as redox/detoxification and
secretion/transmembrane transport were upregulated under nutrient restriction and
that their expression was regulated by ATF4 (Table 1 and Fig. 2A). As
demonstrated in Table 1, the expression of genes with oxidoreductase activity was
significantly reduced in atf4 mutants. When considering the
lower levels of NADH/NAD and NADPH/NADP observed in atf4 mutants
compared to that in control flies (Fig. 1), it suggests that NADH and NADPH could
serve as compensatory reducing agents, given the insufficient reductase activity
in atf4 mutants flies. In contrast, when we subjected larvae to
nutrient restriction, we specifically found that the gene expression related to
lipid catabolism was increased, which was suppressed in atf4
mutants (Table 1 and Fig. 2A). Additionally, we found that gene expression
related to lipid catabolism was specifically increased during nutrient
restriction and suppressed in atf4 mutants (Table 1 and Fig. 2A). These genes,
including CG6271, CG15533, CG31089, and CG6283, showed significant homology to
human LIPH, SMPD1, LIPA, and LIPH, respectively. The human orthologs of these
genes are known to be associated with various metabolic diseases, such as type 2
diabetes mellitus, hypotrichosis 7, lysosomal acid lipase deficiency, and
Niemann-pick disease. According to the modENCODE project
(http://www.modencode.org), these genes are highly expressed in
Drosophila larvae and adults, although their expression levels
differ between the two developmental stages.
In situ hybridization assay indicated that in Drosophila,
CG6283 is specifically expressed in the gut and is induced by nutrient
restriction in the gastric caeca (Fig. 3A). Gastric caeca are finger-like
projections in the gut found in several insects and play a crucial role in
secreting digestive enzymes and facilitating nutrient absorption. Additionally,
gastric caeca contain lysosomes, multivesicular bodies, autophagosomes, and lipid
droplets, all of which are essential for energy metabolism [28]. Based on these
observations, we hypothesized that ATF4 may aid the organism’s survival under
starvation by increasing the expression of various genes, including CG6283 and
CG6295, as illustrated in Fig. 3B. In our previous studies, we demonstrated that
the GCN2/ATF4/4E-BP pathway controls the lifespan of flies under dietary amino
acid restriction by regulating stress-response protein synthesis [5, 18].
Collectively, we believe that ATF4 plays a pivotal role in controlling lifespan
under nutrient restriction by upregulating the gene expression of lipases to
provide the required energy sources.
5. Conclusions
Considering that ATF4 in mammals is involved in metabolic diseases [29, 30, 31],
understanding the function of ATF4 in relation to the regulation of lipase
activity under excess energy conditions would be beneficial for further research.
Availability of Data and Materials
The datasets utilized and/or examined during the present study can be obtained
from the corresponding author upon reasonable request.
Author Contributions
SO, JEP, and MJK designed the research study. SO, JEP, SB, KK and MJK performed
the research. JS provided help and advice on metabolic analysis. SO, JEP, SB, KK,
JS, and MJK analyzed the data. MJK wrote the manuscript. All authors contributed
to editorial changes in the manuscript. All authors read and approved the final
manuscript. All authors have participated sufficiently in the work and agreed to
be accountable for all aspects of the work.
Ethics Approval and Consent to Participate
Not applicable.
Acknowledgment
The authors are grateful to the Bloomington Drosophila Stock Center Center at
Indiana University (NIH P40OD018537) for providing fly strains.
Funding
This research was funded by grants from the National Research Foundation of
Korea, NRF-2022R1A2C1003431, and from the Asan Institute for Life Sciences
(Seoul, Republic of KOREA; 2022IL0010 and 2023IP0121).
Conflict of Interest
The authors declare no conflict of interest.
 Fig. 1.
Fig. 1. Fig. 2.
Fig. 2. Fig. 3.
Fig. 3.