- Academic Editor
†These authors contributed equally.
Objective: The aim of this case-control study was to analyze the
association between sirtuin 1 (SIRT1) single
nucleotide polymorphism (SNP) and the risk of acute kidney injury (AKI) in Han
Chinese patients with cirrhosis and to explore its potential mechanism.
Methods: Twenty-nine AKI patients with cirrhosis (AKI group) and 87
non-AKI patients with cirrhosis (control group) were recruited from a Han Chinese
population. SNaPshot sequencing technology was used for the detection of SNPs.
Dual luciferase reporter vectors were constructed and co-transfected into HK-2
human proximal tubular epithelial cells. SIRT1-overexpressing
recombinant plasmids were constructed and co-transfected into HK-2 cells. The
expression of microRNA-599 (miR-599) and SIRT1/peroxisome
proliferator-activated receptor gamma coactivator 1 alpha
(PGC-1
Acute kidney injury (AKI) is a common complication in patients with cirrhosis. In fact, in hospitalized patients with cirrhosis, the incidence of AKI can be as high as 20–40% [1, 2, 3]. The occurrence of AKI in patients with cirrhosis prolongs the hospital stay and increases their risk of multiple organ failure, such that their 30-day and 1-year mortality rates are 10- and 8-fold higher, respectively, than those of cirrhotic patients without AKI [4, 5, 6]. Thus, early detection of and interventions for AKI are important measures to improve patient outcomes [3, 7]. There are several risk factors for AKI patients with cirrhosis, including advanced age, diabetes, and infection [8, 9, 10, 11]. Moreover, candidate gene studies have suggested that some patients have a genetic predisposition to developing AKI in cirrhosis, such as the endothelial nitric oxide synthase gene (eNOS) G894T single nucleotide polymorphism (SNP), the vasopressin 1a receptor gene (AVPR1A) promoter region rs113481894 SNP, or an angiotensin-converting enzyme gene insertion or deletion, as these are associated with the risk of hepatorenal syndrome [12, 13, 14].
Silent mating type information regulator 2 homolog 1 (SIRT1) is a
highly conserved nicotinamide adenine dinucleotide-dependent protein deacetylase
[15], which has been shown to have nephroprotective effects in AKI models induced
by multiple etiologies such as ischemia-reperfusion injury, sepsis, and
nephrotoxic drugs [16, 17, 18, 19, 20]. For example, we showed that in the early stages of a
rat model of AKI in cirrhosis, the expression of SIRT1 and peroxisome
proliferator-activated receptor gamma coactivator 1 alpha
(PGC-1
Currently, the role of SIRT1 SNPs in AKI patients with cirrhosis remains unknown. Accordingly, in this study, we examined the function of SIRT1, as it is a candidate susceptibility gene for AKI in cirrhosis. Our aim was to develop a sound theoretical basis for the pathogenesis of AKI in cirrhosis that can be used for early diagnosis and the identification of therapeutic targets.
This study enrolled patients with liver cirrhosis who were admitted to the
Department of Hepatology, Ningbo No.2 Hospital (Zhejiang, China) from October
2020 to January 2021. The inclusion criteria were being Han Chinese, aged
The data collected on the general condition of the participants comprised their age, sex, primary disease, comorbidities, medical history, family history, having complications of cirrhosis (infection/spontaneous peritonitis, gastrointestinal (GI) bleeding, ascites, and hepatic encephalopathy), body mass index (BMI), systolic blood pressure, and diastolic blood pressure. The laboratory indicators of the participants that were recorded included SCr concentration (baseline SCr concentration, i.e., at admission, and highest SCr concentration), blood urea nitrogen (BUN) concentration, albumin concentration, total bilirubin concentration, alanine aminotransferase (ALT) concentration, aspartate aminotransferase (AST) concentration, glucose concentration, total cholesterol concentration, low-density lipoprotein concentration, triglyceride concentration, blood sodium concentration, C-reactive protein (CRP) concentration, hemoglobin concentration, platelet count, prothrombin time, and international normalized ratio. The simplified Modification of Diet in Renal Disease formula was used to determine the estimated glomerular filtration rate (eGFR).
A sample (0.5 mL) of peripheral venous blood was collected from the patients in the morning while they were at rest and had an empty stomachs. The sample was treated with an anticoagulant (ethylenediaminetetraacetic acid) and then stored at –80 °C. After all of the blood samples had been collected, their DNA was extracted using a blood genomic DNA extraction kit and then stored at –20 °C for later use.
The SNP locus information of SIRT1 was obtained, revealing that the functional SNP site of SIRT1 was rs4746720. Thus, the primer sequence was designed as shown in Table 1.
| SNP site | Polymorphism | Sequence 5 |
| rs4746720 | T/C | F: CCAAAGAATGGTATTTTCACTT |
| R: AAGTTAGCTGCCACAGTT |
Note: F, Forward primer; R, Reverse primer.
The kidney proximal tubular epithelial cell line Human Kidney 2 (HK2, cat. ZB188) was purchased from the Shanghai Zhibei Biotechnology Co., Ltd., which has been validated by short tandem repeat profiling and was negative for mycoplasma contamination. Dulbecco’s modified Eagle’s medium/F-12 modification containing 10% fetal
bovine serum was added to recovered human proximal tubular epithelial (HK-2)
cells, which were then cultured in an incubator at 37 °C under an
atmosphere of 5% CO
LipofectamineTM3000 liposomal transfection reagent was used to co-transfect the
pmir-GLO-SIRT1-3
Overexpressed recombinant plasmids pcDNA3.1-SIRT1-T and
pcDNA3.1-SIRT1-C bearing different alleles of SIRT1 rs4746720
were constructed, and HK-2 cells were co-transfected with an miR-599
mimic, an miR-599 inhibitor, or an NC, respectively. Total cellular RNA
was extracted using TRIzol reagent, and the concentration and purity of the
extracted RNA were determined. RNA was reverse transcribed into complementary DNA
(cDNA), which was detected by qPCR using an miR-599 upstream primer
(5
Phenylmethylsulfonyl fluoride was added to lyse and extract total proteins, and
the bicinchoninic acid assay was used to determine the total protein
concentration. Subsequently, proteins were separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and then electrotransferred to a
polyvinylidene difluoride membrane. Then the membrane was blocked in blocking
solution for 1 h, followed by incubation overnight at 4 °C with primary
antibodies against SIRT1, PGC-1
Statistical Package for Social Sciences (SPSS) version 26.0 (IBM SPSS
Statistics, Armonk, NY, USA) was used for the data analysis. The normally distributed
measurements are expressed as the means
Of the 116 participants, 29 (20 males) were assigned to the AKI group and had an
average age of 59.0
| AKI group (n = 29) | Control group (n = 87) | p | |
| Age (year) | 59.0 |
60.1 |
0.677 |
| Male (n, %) | 20 (69.0) | 58 (66.7) | 0.819 |
| BMI (Kg/m |
23.7 |
23.9 |
0.805 |
| Mean arterial pressure (mmHg) | 87.9 |
89.9 |
0.577 |
| Diabetes (n, %) | 5 (17.2) | 29 (33.3) | 0.099 |
| Hypertension (n, %) | 6 (20.7) | 20 (23.0) | 0.797 |
| Infection (n, %) | 18 (62.1) | 23 (26.4) | 0.001* |
| Ascites (n, %) | 22 (75.9) | 50 (57.5) | 0.077 |
| Hepatic Encephalopathy (n, %) | 9 (31.0) | 21 (24.1) | 0.463 |
| Gastrointestinal Bleeding (n, %) | 9 (31.0) | 19 (21.8) | 0.316 |
| Albumin (g/L) | 28.8 |
30.2 |
0.297 |
| Total bilirubin (µmol/L) | 50.8 (30.1, 168.9) | 37.0 (21.9, 72.1) | 0.037* |
| ALT (U/L) | 43.0 (23.0, 103.0) | 32.0 (19.0, 46.0) | 0.038* |
| AST (U/L) | 79.0 (48.5, 204.5) | 56.0 (32.0, 81.0) | 0.004* |
| Serum sodium (mmol/L) | 136.6 |
137.2 |
0.614 |
| CRP (mg/L) | 18.1 (7.5, 45.1) | 4.8 (1.9, 17.5) | |
| PT (s) | 17.9 (15.9, 21.4) | 17.1 (14.6, 19.6) | 0.152 |
| INR | 1.5 (1.4, 2.0) | 1.5 (1.3, 1.7) | 0.118 |
| Baseline SCr (µmol/L) | 66 (52.3, 80.7) | 60.8 (52.5, 75.0) | 0.296 |
| eGFR (mL/min/1.73 m |
56.0 |
101.9 |
|
| MELD score | 20.8 |
11.9 |
Note: BMI, Body Mass Index; ALT, alanine aminotransferase; AST, aspartate
aminotransferase; CRP, C-reactive protein; PT, prothrombin time; INR,
International Normalized Ratio; SCr, serum creatinine; eGFR, estimation of
glomerular filtration rate; MELD, Model of end-stage liver disease; AKI, acute kidney injury; *p
SNaPshot gene sequencing technology was used for SIRT1 genotyping. This revealed that SIRT1 rs4746720 sites exhibited CC, CT, or TT genotypes (Fig. 1).
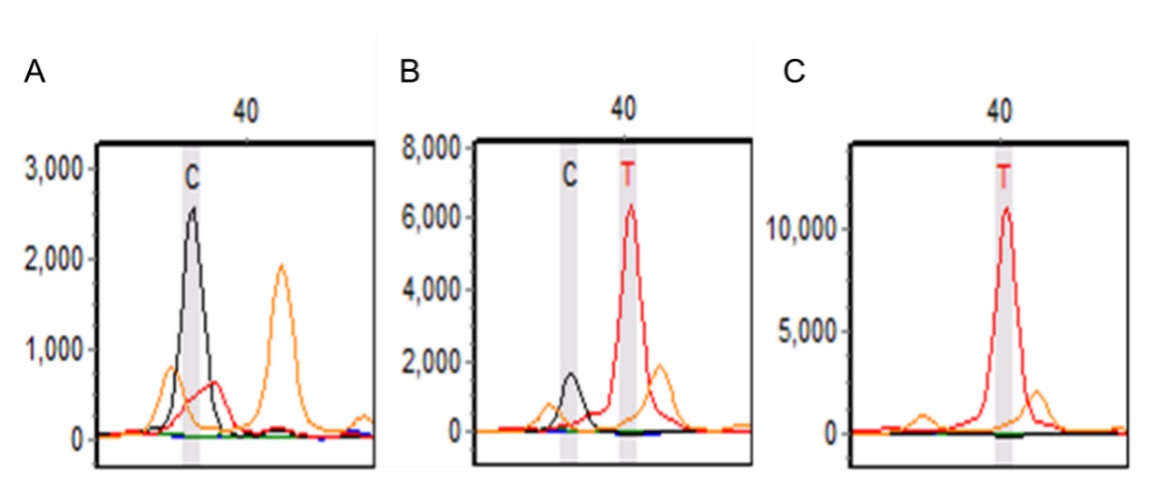 Fig. 1.
Fig. 1.The genotyping results of SIRT1 rs4746720 polymorphism in the AKI group and the control group. (A) CC homozygous. (B) CT heterozygotes. (C) TT homozygous. Note: SIRT1 rs4746720 extends forward, and the product is consistent with polymorphism.
The goodness-of-fit
| SNP site | Genotype | Observed value | Expected value | p | |
| rs4746720 | TT | 30 (34.5) | 27 (31.0) | 1.66 | 0.437 |
| CT | 37 (42.5) | 17 (19.5) | |||
| CC | 20 (23.0) | 43 (49.4) |
Note: p
There was no significant between-group difference in the frequency distribution
of the SIRT1 rs4746720 TT, CT, and CC genotypes (
| Heredity model | Genotype/allele | AKI group (n = 29) | Control group (n = 87) | OR (95% CI) | p |
| Co-dominant | TT | 12 (41.4) | 30 (34.5) | 1.34 (0.43–4.21) | 0.611 |
| CT | 11 (37.9) | 37 (42.5) | 0.98 (0.32–3.07) | 0.978 | |
| CC | 6 (20.7) | 20 (23.0) | 1 (reference) | ||
| Allele | T | 35 (60.3) | 97 (55.7) | 1.21 (0.66–2.23) | 0.534 |
| C | 23 (39.7) | 77 (44.3) | 1 (reference) | ||
| Dominant genes | TT+ CT | 23 (79.3) | 67 (77.0) | 1.14 (0.41–3.21) | 0.802 |
| CC | 6 (20.7) | 20 (23.0) | 1 (reference) | ||
| Recessive genes | TT | 12 (41.4) | 30 (34.5) | 1.36 (0.57–3.24) | 0.490 |
| CC+ CT | 17 (58.6) | 57 (65.5) | 1 (reference) | ||
| Hyperdominant genes | TT+CC | 18 (62.1) | 50 (57.5) | 1.22 (0.52–2.91) | 0.647 |
| CT | 11 (37.9) | 37 (42.5) | 1 (reference) |
Note: OR and p are parameters after adjusting age and gender.
Stratified analyses based on age, sex, and complications of cirrhosis
(infection, ascites, GI bleeding, and hepatic encephalopathy) were conducted to
study the effect of genetic factors on the risk of AKI in patients with
cirrhosis. In the subgroup with hepatic encephalopathy, SIRT1 rs4746720
SNPs were significantly linked with the development of AKI, and patients with the
T allele had a six times greater risk of AKI than those with the C allele (OR =
6.00, 95% CI = 1.22–29.48; p = 0.027). However, in subgroups based on
other factors, SIRT1 rs4746720 SNPs were not significantly associated
with AKI in cirrhosis (p
| Variable | Genotype/allele | AKI group (n = 29) | Control group (n = 87) | OR (95% CI) | p | |
| Age (years) | TT vs. CC | 8/3 | 13/12 | 2.46 (0.53–11.50) | 0.252 | |
| CT vs. CC | 5/3 | 20/12 | 1.00 (0.20–4.96) | 1.000 | ||
| T vs. C | 21/11 | 46/44 | 1.83 (0.79–4.22) | 0.156 | ||
| TT vs. CC | 4/3 | 17/8 | 0.63 (0.11–3.49) | 0.595 | ||
| CT vs. CC | 6/3 | 17/8 | 0.94 (0.19–4.76) | 0.942 | ||
| T vs. C | 14/12 | 51/33 | 0.76 (0.31–1.83) | 0.534 | ||
| Gender | male | TT vs. CC | 8/4 | 17/15 | 1.77 (0.44–7.06) | 0.422 |
| CT vs. CC | 8/4 | 26/15 | 1.15 (0.30–4.49) | 0.836 | ||
| T vs. C | 24/16 | 60/56 | 1.40 (0.68–2.91) | 0.366 | ||
| female | TT vs. CC | 4/2 | 13/5 | 0.77 (0.11–5.61) | 0.796 | |
| CT vs. CC | 3/2 | 11/5 | 0.68 (0.09–5.45) | 0.718 | ||
| T vs. C | 11/7 | 37/21 | 0.89 (0.30–2.65) | 0.837 | ||
| Infection | no | TT vs. CC | 4/3 | 20/15 | 1.00 (0.19–5.15) | 1.000 |
| CT vs. CC | 4/3 | 29/15 | 0.69 (0.14–3.49) | 0.653 | ||
| T vs. C | 12/10 | 69/59 | 1.03 (0.41–2.55) | 0.956 | ||
| yes | TT vs. CC | 8/3 | 10/5 | 1.33 (0.24–7.35) | 0.741 | |
| CT vs. CC | 7/3 | 8/5 | 1.46 (0.25–8.43) | 0.673 | ||
| T vs. C | 23/13 | 28/18 | 1.14 (0.46–2.80) | 0.780 | ||
| Ascites | no | TT vs. CC | 3/1 | 15/9 | 1.80 (0.16–20.03) | 0.633 |
| CT vs. CC | 3/1 | 13/9 | 2.08 (0.19–23.30) | 0.553 | ||
| T vs. C | 9/5 | 43/31 | 1.30 (0.40–4.25) | 0.666 | ||
| yes | TT vs. CC | 9/5 | 15/11 | 1.32 (0.35–5.05) | 0.685 | |
| CT vs. CC | 8/5 | 24/11 | 0.73 (0.20–2.76) | 0.647 | ||
| T vs. C | 26/18 | 54/46 | 1.23 (0.60–2.52) | 0.571 | ||
| Hepatic encephalopathy | no | TT vs. CC | 5/6 | 22/15 | 0.57 (0.15–2.21) | 0.414 |
| CT vs. CC | 9/6 | 29/15 | 0.78 (0.23–2.59) | 0.680 | ||
| T vs. C | 19/21 | 73/59 | 0.73 (0.36–1.49) | 0.387 | ||
| yes | TT vs. CC | 7/0 | 8/5 | - | - | |
| CT vs. CC | 2/0 | 8/5 | - | - | ||
| T vs. C | 16/2 | 24/18 | 6.00 (1.22–29.48) | 0.027* | ||
| Gastrointestinal bleeding | no | TT vs. CC | 9/4 | 26/13 | 1.13 (0.29–4.35) | 0.865 |
| CT vs. CC | 7/4 | 29/13 | 0.78 (0.20–3.16) | 0.732 | ||
| T vs. C | 25/15 | 81/55 | 1.13 (0.55–2.34) | 0.738 | ||
| yes | TT vs. CC | 3/2 | 4/7 | 2.63 (0.30–23.00) | 0.383 | |
| CT vs. CC | 4/2 | 8/7 | 1.75 (0.24–12.64) | 0.579 | ||
| T vs. C | 10/8 | 16/22 | 1.72 (0.56–5.33) | 0.348 |
Note:
The relationship between genotype and liver and kidney function indexes was
analyzed in each group. In the AKI group, patients with the SIRT1
rs4746720 TT genotype had significantly higher SCr and BUN concentrations and
significantly lower eGFRs than those with the CC+CT genotypes (p
| Variable | AKI group | p | Control group | p | ||
| TT | CC+CT | TT | CC+CT | |||
| SCr (µmol/L) | 171.5 |
111.8 |
0.026* | 68.0 |
72.0 |
0.255 |
| eGFR (mL/min/1.73 m |
43.1 |
65.1 |
0.009* | 104.4 |
100.6 |
0.564 |
| BUN (mmol/L) | 15.7 |
9.5 |
0.015* | 5.2 |
6.0 |
0.256 |
| Albumin (g/L) | 27.6 |
29.6 |
0.350 | 30.5 |
30.0 |
0.748 |
| Total bilirubin (µmol/L) | 138.7 |
117.7 |
0.723 | 75.2 |
56.0 |
0.229 |
| ALT (U/L) | 48.7 |
88.8 |
0.191 | 59.6 |
47.5 |
0.454 |
| AST (U/L) | 79.0 |
225.1 |
0.073 | 84.9 |
65.0 |
0.189 |
| MELD Score | 23.5 |
18.8 |
0.193 | 12.2 |
11.7 |
0.685 |
Note: SCr, serum creatinine; eGFR, estimation of glomerular filtration rate;
BUN, urea nitrogen; ALT, alanine aminotransferase; AST, aspartate
aminotransferase; MELD, Model of end-stage liver disease;
The bioinformatics software programs TargetScan and miRanda were used to predict
the target gene of microRNA-599 (miRNA-599). To verify the effect of
SIRT1 rs4746720 SNP binding with miR-599, whole-gene
SIRT1–3
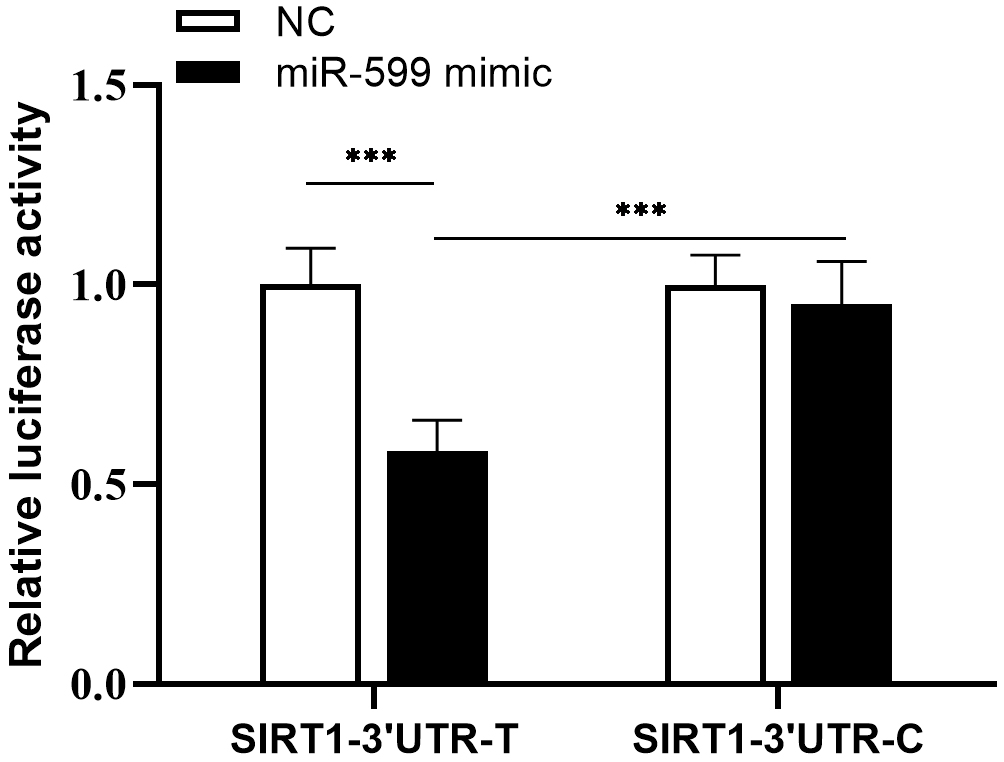 Fig. 2.
Fig. 2.
Results of dual luciferase reporter genes interaction between
SIRT1 rs4746720 and miR-599 (
To further study the effect of the SIRT1 rs4746720-mediated effect of
miR-599 on SIRT1 expression, overexpression plasmids
pcDNA3.1-SIRT1-T/C, which contained different alleles (i.e., T or C) of
SIRT1 rs4746720, were constructed and verified by gene sequencing. Next,
pcDNA3.1-SIRT1-T/C plasmids and pcDNA3.1 (empty plasmid) were used to
transfect HK-2 cells with NC, an miR-599 mimic, or an miR-599
inhibitor, respectively. Subsequently, the relative expression of
miR-599, SIRT1 mRNA, and protein in cells was determined by
qPCR and Western blotting, respectively. The results were as follows: (1)
Compared with transfection with NC, transfection with the miR-599 mimic
caused the expression of miR-599 in the SIRT1-T overexpression
group and the empty plasmid group to increase by 41% and 58%, respectively
(p
 Fig. 3.
Fig. 3.Expression of miR-599 and SIRT1 in HK-2 cells
after co-transfection with overexpression plasmid and miR-599. (A)
Relative expression of miR-599. (B) Relative expression of
SIRT1 mRNA. (C) Relative expression of SIRT1; *p
The effect of the SIRT1 rs4746720 SNP-mediated interaction of
miR-599 with SIRT1 on the
PGC-1
 Fig. 4.
Fig. 4.Expression of
PGC-1
The rapid development of genomics technology has enabled research revealing the important role of genetic factors in the occurrence and development of AKI [21, 22, 23, 24]. However, there have been few studies on patients with cirrhosis and genetic susceptibility to develop AKI. Seckin et al. [12] reported that the GT/TT genotypes and T mutant allele of the eNOs G894T SNP might be risk factors for the development of hepatorenal syndrome, as patients with the GT or TT genotype had a 5.0 or 5.8 times greater risk, respectively, of developing hepatorenal syndrome than those with the GG genotype. Wang et al. [13] studied 60 patients with cirrhosis and found that those with the T allele of the promoter of AVPR1A rs113481894 had a 2.23 times higher risk of developing type I hepatorenal syndrome than those with the C allele of the promoter of AVPR1A rs113481894. These candidate genes code for proteins involved in classical splanchnic-vasodilation-related processes. In recent years, further research has increased our understanding of the pathogenesis of AKI in cirrhosis [25, 26]. In addition to traditional hemodynamic mechanisms, non-hemodynamic microvascular toxicities (caused by, for example, endotoxins or bile acids) may be directly related to the development of AKI [10, 27]. The synergistic effect of toxic factors and microvascular dysfunction increases damage to proximal tubular epithelial cells, mediates the downregulation of mitochondrial function, and triggers intrarenal activation of the renin-angiotensin-aldosterone system, resulting in a decreased GFR [28].
SIRT1 is a highly conserved protein deacetylase that is widely
expressed in renal tissue. In ischemia-reperfusion injury, sepsis, and AKI caused
by cisplatin, SIRT1 acts on many transcription factors to regulate
members of downstream pathways, such as PGC-1
SIRT1 rs4746720 is associated with aging, type 2 diabetes, diabetic
nephropathy, and other diseases [36, 37, 38, 39]. In the current study, we found that the
TT genotype and T allele frequencies of the SIRT1 rs4746720 locus in the
AKI group were higher than those in the control group (41.4% vs.
34.5%, 60.3% vs. 55.7%), and the risk of AKI in cirrhosis was
increased in patients with the T allele; however, the latter difference did not
reach statistical significance (p = 0.534). Stratified analyses revealed
that the T allele led to a greater risk of developing AKI in cirrhosis with
hepatic encephalopathy. Analyses of the liver and kidney function indicators of
the AKI group with respect to genotype demonstrated that the SCr and BUN
concentrations of patients with the TT genotype were significantly higher, and
their eGFRs were significantly lower than those with the CC+CT genotype
(p
Bioinformatics software was used to predict the miRNAs that may bind to
rs4746720 in the 3
The above-described results, combined with those from other studies, indicate
that the T allele of SIRT1 rs4746720 may mediate the ability of
miR-599 to target and inhibit the expression of SIRT1 and its
downstream PGC-1
This was a single-center retrospective case-control study with a small sample size that only included a Han Chinese population with cirrhosis. No relationship between SIRT1 rs4746720 and the development of AKI was observed in cirrhotic patients. Thus, our conclusions need to be validated in future centralized studies with larger sample sizes. Moreover, in the current study, the serum levels of SIRT1 and miR-599 in patients in the AKI and control groups were considered to fully represent the effect of SIRT1rs4746720 SNP-mediated miR-599 on AKI patients with cirrhosis at the population level. Furthermore, no in vivo functional verification experiments were performed in this study. Therefore, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 technology could be used to construct a knock-in SIRT1 rs4746720 mutant mouse model of AKI in cirrhosis. This would enable verification of the mechanism of the susceptibility of SIRT1 and its downstream pathways to SIRT1 rs4746720 SNP-mediated miR-599 control.
The SIRT1 rs4746720 SNP might be linked with AKI in cirrhotic patients,
and the T allele increased the risk of AKI in those with hepatic encephalopathy.
In AKI patients with cirrhosis, the renal function of patients with the TT
genotype was significantly lower than that of patients with the CC+CT genotype.
The SIRT1 rs4746720 SNP mediated miR-599 binding to
SIRT1, thereby affecting the expression of SIRT1 and its
downstream effectors, PGC-1
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by FZ, YC, YX, and QL. The first draft of the manuscript was written by FZ and YC, and it was revised by QL. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
The research protocol was approved by the Medical Ethics Committee of Ningbo NO.2 hospital (YJ-KYSB-NBEY-2018-002-01), and all of the participants provided signed informed consent.
Not applicable.
This study was funded by the Project of NINGBO Leading Medical & Health Discipline (Project No. 2022-S03), China, and Medical Scientific Research Foundation of Zhejiang Province (Project No. 2019KY181).
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
