1 Key Laboratory of Microbial Pathogenesis and Interventions of Fujian Province University, the Key Laboratory of Innate Immune Biology of Fujian Province, Biomedical Research Center of South China, Key Laboratory of OptoElectronic Science and Technology for Medicine of the Ministry of Education, College of Life Sciences, Fujian Normal University, Fuzhou, 350117, China
2 State Key Laboratory of Freshwater Ecology and Biotechnology, Key Laboratory of Aquaculture Disease Control, Institute of Hydrobiology, Chinese Academy of Sciences, 430072 Wuhan, Hubei, China
3 College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, 100049 Beijing, China
4 State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Biotechnology, 100071 Beijing, China
Abstract
Glycosylation is one of the most common post-translational modifications of proteins across all kingdoms of life. Diverse monosaccharides and polysaccharides can be attached to a range of amino acid residues generating N-glycosylation, O-glycosylation, C-glycosylation, S-glycosylation, as well as P-glycosylation. The functions of the eukaryotic glycosylation system during protein folding in the endoplasmic reticulum (ER) and Golgi are well-studied. Increasing evidence in the recent decade has demonstrated the presence of oligosaccharyltransferases (OSTs) in bacteria and archaea. In particular, the oligosaccharyltransferase (PglB) of Campylobacter jejuni and oligosaccharyltransferase (PglL) enzyme of Neisseria meningitidis are the most characterized OSTs that catalyze bacterial N-linked glycosylation and O-linked glycosylation, respectively. Glycoprotein administered as glycoconjugate vaccines have been shown to be effective prophylactic to protect against numerous pathogenic bacteria. The chemical synthesis of glycoproteins is complex and expensive, which limits its application to the development of glycoconjugate vaccines. However, studies have demonstrated that the biosynthesis of glycoproteins is realizable by transferring PglB, a plasmid encoding a substrate protein, or PglL, a plasmid encoding genes for glycan synthesis to Escherichia coli. This strategy can be applied to the development of glycoconjugate vaccines using engineered host E. coli. This review summarizes the structure and mechanism of action of the bacterial OSTs, PglB and PglL, and discusses their potential application to glycoconjugate vaccine design.
Keywords
- oligosaccharyltransferases
- PglB
- PglL
- glycoconjugate vaccine
Oligosaccharyltransferases (OSTs) catalyze the transfer of oligosaccharides to
substrate proteins via a glycosidic
bond by a process termed protein glycosylation, which is necessary for protein
trafficking, folding, stability, cellular signaling, and
self/non-self-interaction [1]. N-linked and O-linked glycosylations are the most
well-characterized types of protein glycosylation in eukaryotes [2]. N-linked
glycosylation is catalyzed by OSTs that attach a lipid-linked oligosaccharide
(LLO) to the acceptor asparagine of the Asn-X-Thr/Ser motif, where X represents
any amino acid except proline [3, 4]. Mammalian OSTs that catalyze N-linked
glycosylation reactions are multi-subunit complexes comprising DAD1 (Ost2p),
N33/Tusc3 or MagT1 (Ost3p and Ost6p), OST48 (Wbp1p), ribophorin I (Ost1p),
ribophorin II (Swp1p), and STT3A or STT3B (Stt3p) [5]. STT3A-OSTs catalyze the
co-translational glycosylation of proteins as they emerge from the translocon
into the lumen of the endoplasmic reticulum (ER), while STT3B-OSTs catalyze
post-translational glycosylation after polypeptide synthesis is complete [6].
O-linked glycosylation reactions use more saccharide donors (GalNAc, Fuc, GlcNAc,
Man, Glc, Xyl, and Gal) and substrate residue receptors (serine, threonine,
tyrosine, hydroxylysine, and hydroxyproline) than other glycosylation reactions.
O-linked glycosylation is initiated by distinct glycosyltransferases (GTs) [2, 7]. GTs comprise an N-terminal catalytic domain for binding and glycosylating
substrates, and a C-terminal lectin domain for binding GalNAc [8]. For instance,
UDP-GalNAc, a sugar donor for O-linked glycosylation, is cleaved by GTs into
UDP-
Multiple bacterial protein glycosylation mechanisms have been reported in recent
years in bacterial species from the most diverse genera and demonstrated that
protein glycosylation affects flagellar formation, motility, virulence, and
adhesion to host cells [10, 11]. PglB, a major OST for N-linked glycosylation,
has been found in Campylobacter jejuni, Wolinella succinogenes,
and Desulfovibrio desulfuricans [12, 13]. The deletion of protein
glycosylation locus (pgl) gene clusters has been shown to
weaken bacterial virulence, adherence, and invasion in human intestinal cells, in
addition to impairing temperature sensitivity and antibiotic resistance [14, 15].
PglL, a ubiquitous O-OST for O-linked glycosylation [16], is found in several
bacterial species, including Ralstonia solanacearum, Neisseria
meningitidis, Pseudomonas aeruginosa, and Neisseria gonorrhoeae. PglL
is capable of catalyzing the O-linked glycosylation of pilin proteins, and the
lack of PglL
Therefore, the glycosylation systems of pathogenic bacteria are key factors for regulating bacterial invasion and adherence by modulating the interactions between glycans and host cells; however, these processes also depend on the level of host immunity to pathogenic bacteria. In the last few decades, novel glycoprotein vaccines have been designed by transferring a foreign bacterial glycosylation system to bacteria lacking glycosylation machinery, mostly E. coli. This approach enhances antibody production and the protective effect of vaccines against numerous antigenic species [20, 21]. This review discusses and highlights the structures and mechanisms of bacterial N-linked and O-linked glycosylation systems, with particular emphasis on the PglB and PglL enzymes. More importantly, this paper also discusses that the unique characteristics of OST are expected to become an important tool for the development of new sugar vaccines for glycoengineering in the future.
C. jejuni is a foodborne pathogen of humans and animals, and is primarily associated with poultry and domestic animals [22]. The bacterium possesses genes that encode an N-linked glycosylation system, located on the single chromosomal pgl gene cluster [23].
Protein glycosylation has been described in all domains of life. For example, studies have demonstrated that N-linked glycosyltransferases (GTs) are present in prokaryotes and have high similarity to the eukaryotic STT3 family [23, 24]. Furthermore, the discovery of N-linked OSTs in archaebacteria and bacteria increases the understanding of the mechanism of action of OSTs and their potential application [25]. Well-characterized prokaryotic OSTs, oligosaccharyltransferase (AglB) in archaea and PglB in eubacteria, catalyze the transfer of oligosaccharides to the amide moiety of asparagine (N).
The mechanism of N-linked glycosylation is conserved in bacterial species and
has been well-characterized in C. jejuni, which possesses a 17-kb
pgl gene cluster that encodes proteins for synthesizing and transferring
oligosaccharides (Fig. 1A). The process of N-linked glycosylation in C.
jejuni could be divided into two steps, namely oligosaccharide synthesis, and
transfer (Fig. 1B). Oligosaccharide synthesis begins with modification of
UDP-GlcNAc to UDP-2,4-diacetamidoglucosamine (UDP-diNAcBac) by
glycosyltransferases: PglF, PglE, and PglD, followed by partial transfer of
UDP-diNAcBac by PglC onto undecenol diphosphate. The oligosaccharyltransferase
(PglA, PglJ, and PglH enzymes) enzymes subsequently catalyze the formation of a
hexasaccharide by the fusion of five units of GalNAc. LLO synthesis is completed
by PglI, which adds a branched glucose residue to the hexasaccharide [21, 26, 27, 28].
The LLO is then flipped across the inner membrane into the periplasm by PglK
[29, 30], and transferred to the substrate asparagine of the - D/E
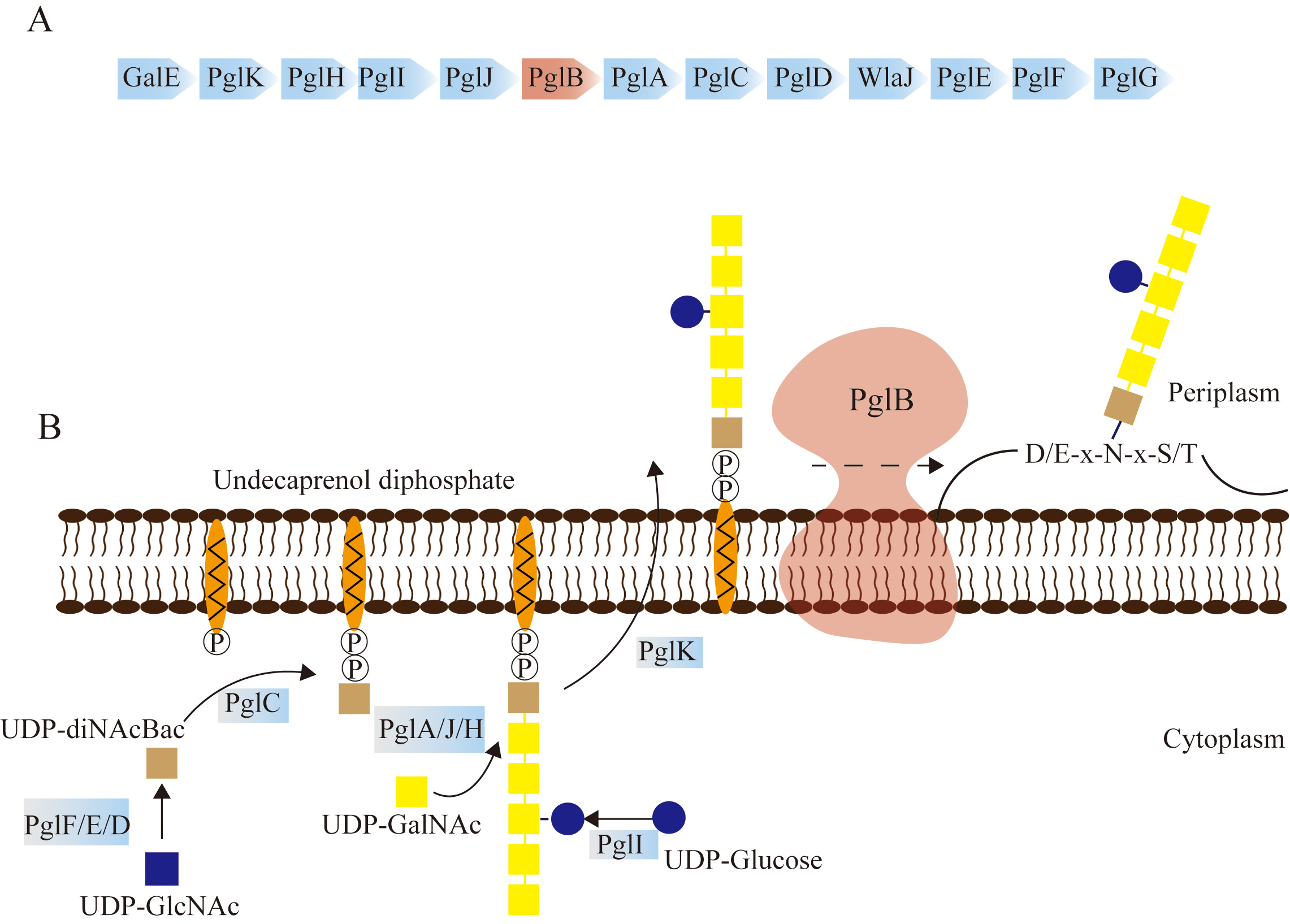 Fig. 1.
Fig. 1.Mechanism of N-linked glycosylation mediated by the PglB Oligosaccharyltransferase (OST) of C. jejuni. (A) The 17 kb protein glycosylation locus pgl of C. jejuni. N-linked glycosylation in C. jejuni is cooperatively accomplished by the pgl gene cluster. PglB is the core factor for the glycosylation process. (B) Schematic diagram depicting N-linked glycosylation in C. jejuni. The first step of glycosylation involves oligosaccharide synthesis in the cytoplasm, which is mediated by glycosyltransferases (GTs), including PglF/E/D, PglC, PglA/J/H, PglI, and PglK. The transfer of oligosaccharides to the periplasm is carried out by PglK. Subsequently, PglB transfers the hexasaccharide to the asparagine residue of the substrate with the sequence D/ExNxS/T.
The mechanism of bacterial N-linked glycosylation by OSTs has been investigated
in detail by determining the structures of the AglB enzyme of
Archaeoglobus fulgidus and the PglB enzymes of N. gonorrhoeae,
C. jejuni, and C. lari. The full-length structures of AglB and PglB
comprise an N-terminal transmembrane domain and a C-terminal periplasmic domain
(Fig. 2A). The C-terminal domain of the AglB protein of Ar. fulgidus and
the PglB enzyme of C. jejuni share a high structural
similarity, with both proteins containing a central core (CC) domain and an
inserted
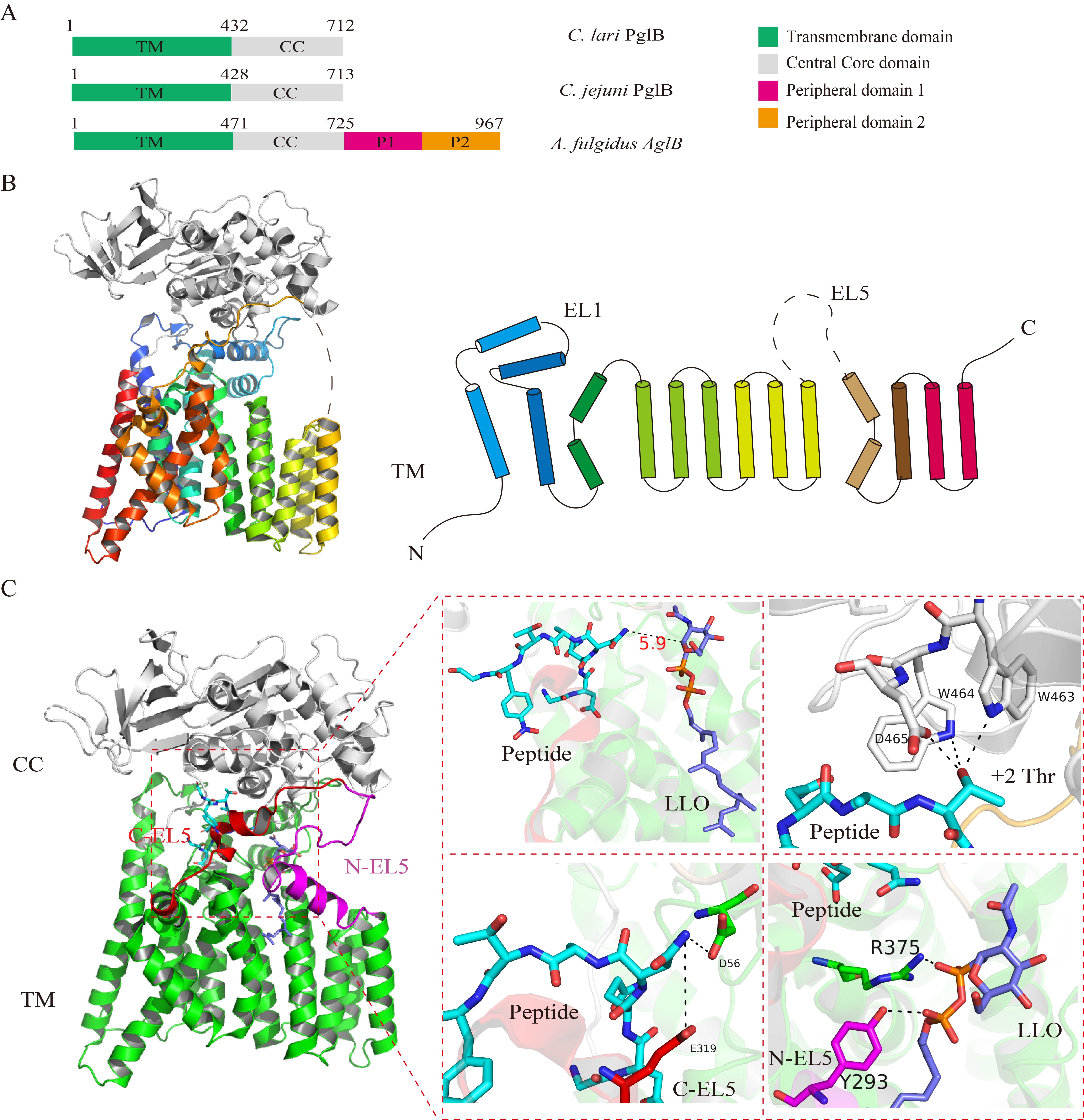 Fig. 2.
Fig. 2.The structure of the PglB enzyme of C. jejuni and the
ternary complex of PglB bound to a substrate DQNATF peptide and an lipid-linked oligosaccharide (LLO) analog.
(A) Schematic diagram of the PglB enzymes of C. lari and
C. jejuni and the AglB protein of Ar.
fulgidus. All the enzymes possess a transmembrane domain and a periplasmic
domain, while the AglB protein of Ar. fulgidus comprises two
more peripheral domains, P1 and P2. (B) Crystal structure of the PglB protein of
C. jejuni (PDB ID: 3RCE). The PglB enzyme of C. jejuni
comprises 13 transmembrane helices, EL1, and EL5. EL1 is a periplasmic helix and
EL5 is a periplasmic loop located between the transmembrane helices. The
periplasmic EL1 and EL5 domains are considered to be important for peptide
recognition and LLO binding. (C) Crystal structure of the ternary complex of
C. jejuni PglB with a substrate DQNATF peptide and an LLO analog (PDB
ID: 5OGL). The substrate peptide and the LLO analog in the ternary complex are
situated opposite to each other in the platform formed by the transmembrane and
periplasmic domains, and the distance between the asparagine residue and LLO
analog is 6 Å. The WWD motif in the periplasmic domain is important for
peptide binding, and forms three hydrogen bonds with the
Interestingly, the long external EL5 loop is partially ordered, and 25 residues
of the EL5 loop are disordered in the electron density map [34]. However, a study
demonstrated that EL5 contains a conserved tyrosine residue (Tyr293), and the
Y293A mutation leads to an almost complete loss of catalytic activity, confirming
that the aromatic side chain of Y293 is essential for catalysis [35]. Moreover,
LLO binding induces dynamic changes in the EL5 loop, resulting in movements in
the transmembrane domain, which progressively induces the formation of an open
conformation of EL5 [36]. In order to obtain further insights into the mechanism
of LLO transfer and the function of the EL5 loop, Napiórkowska et
al. [37] solved the ternary complex of PglB bound to an acceptor DQNATF peptide
and a synthetic nonhydrolyzable (
Metal ions are essential for enzymatic activity, and it has been reported that
Zn
O-linked glycosyltransferases (GTs) transfer a GlcNAc moiety from UDP-GlcNAc to the hydroxyl oxygen of a serine or threonine residue [41, 42]. O-linked glycosylation has been reported to be important for fundamental cellular processes, including transcription, signal transduction, metabolism, subcellular localization, and immune response [43]. It has been observed that the pathway for the O-glycosylation of proteins in B. cepacia affects the growth of the bacterium. Non-glycosylated proteins have been shown to not fully utilize multiple carbon sources, and are easy to be broken down [44]. Furthermore, both O-antigen ligases (WaaL ligases) and O-OSTs harbor an evolutionarily conserved Wzy_C domain, but differ concerning the lipid A-core and amino acid residues for glycan reception (Fig. 3A) [45, 46]. The mechanism of action and function of the O-linked GTs, PilO from P. aeruginosa, and PglL of N. meningitidis are well studied. Both enzymes share very little sequence identity in their glycan recognition region and acceptor proteins.
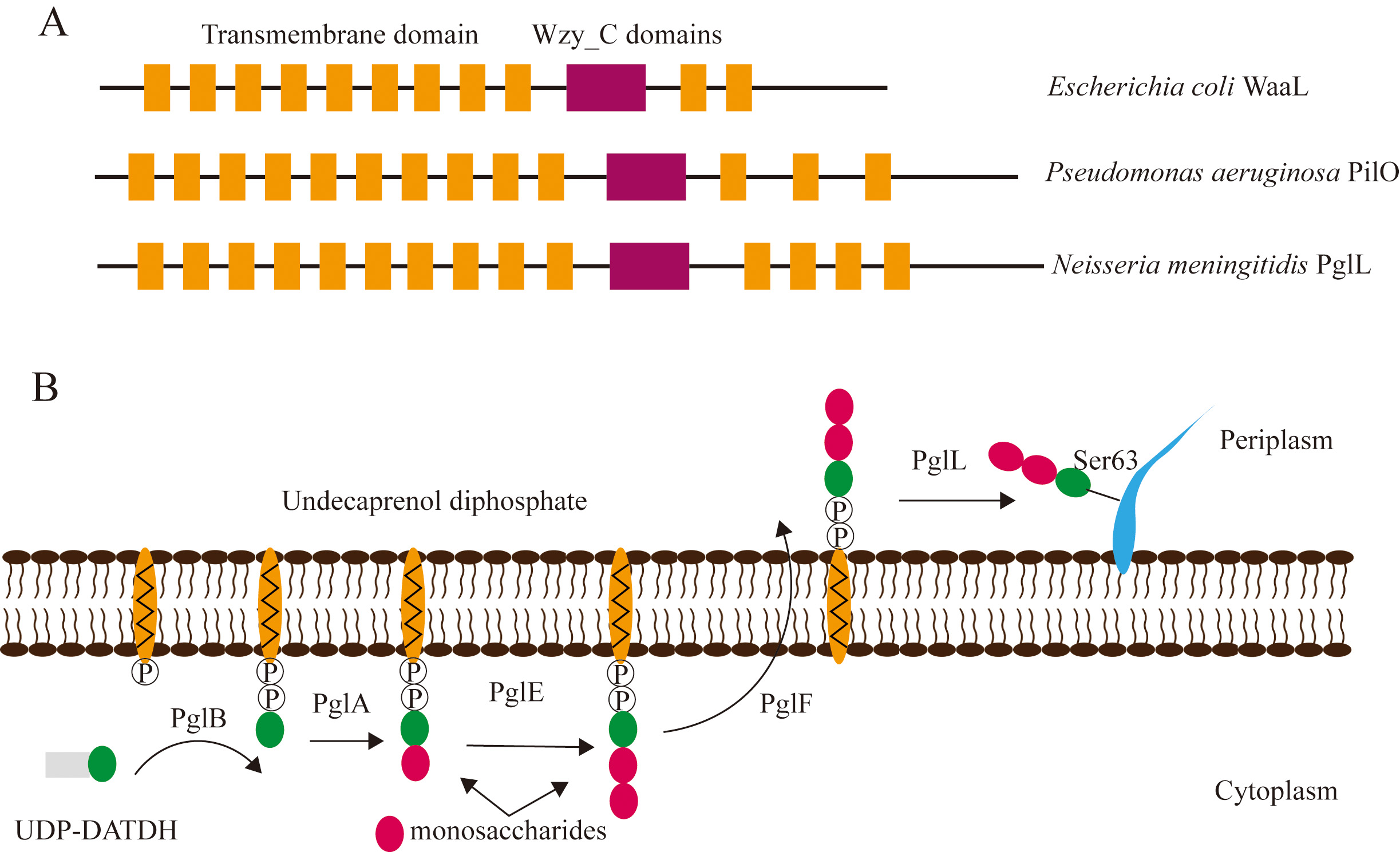 Fig. 3.
Fig. 3.Structure of enzymes and mechanism of bacterial O-linked glycosylation. (A) Structure of the WaaL protein of E. coli, PilO enzyme of P. aeruginosa, and the PglL protein of N. meningitidis. The Wzy_C domain is shared among the three species of bacteria. (B) Schematic diagram depicting the mechanism of O-linked glycosylation in N. meningitidis. The PglB, PglA, and PglE enzymes transfer UDP-DATDH and monosaccharides to undecaprenol diphosphate, and PglF flips the oligosaccharides from the cytoplasm to the periplasm. The LLO is finally transferred by PglL to a serine residue of the substrate.
The PilO enzyme of P. aeruginosa comprises a transmembrane domain that
anchors to the periplasmic membrane, and a periplasmic core domain that catalyzes
the transfer of glycans to pilin via its Wzy_C domain (Fig. 3A) [47]. Analysis
of the transmembrane helix of PilO revealed that the transmembrane region
includes residues 1-266, and the residues include nine transmembrane helices.
Residues 267-461 of PilO have some amino acids that are less hydrophilic and form
four transmembrane domains, in which there are two periplasmic loops and two
cytoplasmic loops. A subsequent BLAST search of the Wzy_C structural domain of
PilO (residues 275-325) revealed that residues 281-301 are conserved and have no
homologous sequences. In order to investigate whether this region affects
glycosylation, mutations were introduced at different positions. Residues 281-287
(
N. meningitidis is a gram-negative bacterium that causes meningitis and
is a common commensal in the human respiratory tract. The genes and phenotype of
N. meningitidis have been reported to undergo in vivo
alteration within a very short time to evade the host immune system and
enhance virulence [50]. The O-linked protein glycosylation system affects
polysaccharide diversity owing to selective gene expression. It has been reported
that ST-11 strains of N. meningitidis express 7 to 21 different glycans,
resulting from the diversity and variation of pgl genes [51]. The PglL
enzyme of N. meningitidis contains multiple consecutive transmembrane
domains and two periplasmic domains (Fig. 3A). The PglL enzyme has been
demonstrated to be responsible for attaching UndPP-glycan to pilin (PilE), and
Ser63 is one of the glycosylation sites on PilE [52, 53] (Fig. 3B). The
glycosyltransferase (GT) activity of PglB transfers
2,4-diacetamido-2,4,6-trideoxyhexose (DATDH) to undecaprenol diphosphate [49, 54]. The PglA and PglE GTs subsequently add two monosaccharide units to the side
chain of the sugar moiety. The trisaccharide is finally flipped by the PglF GT
and transferred to the serine residue of its substrate by PglL (Fig. 3B). The
final recognition motif of PglL on the exotoxin A (EPA) protein of
Pseudomonas aeruginosa was initially identified as
However, full-length structures of the PglL/PilO enzymes or PglL/PilO enzymes bound to the substrate peptide complex have not been solved to date. These structures are necessary for further studies on elucidating the mechanism of peptide binding and glycan transfer by PglL/PilO.
V. cholerae is a gram-negative pathogen causing cholera, a water-borne
diarrheal disease in animals and humans [57]. Bioinformatics analysis identified
the VC0393 gene, which encodes O-OTases in the O1 E1 strain of V.
cholerae, and is referred to as PglL
O-linked glycosylation systems have also been detected in gram-positive
bacteria, including L. monocytogenes, Clostridium
difficile, Staphylococcus aureus, and Streptococcus
agalactiae [13, 60]. The flagellin protein of L. monocytogenes is
modified by the addition of O-linked N-acetylglucosamine (GlcNAc) moieties at
serine and threonine residues, which is regulated by the GmaR GT [61, 62]. GmaR
is a bifunctional protein in which one hand is a temperature sensor that controls
the temperature-dependent transcription of flagellar motility genes by
interacting with MogR via conformational changes [63, 64]; the other hand is a
GT-2 family GT that glycosylates the flagellin protein FlaA. Unlike the PilO or
PglL enzymes that transfer oligosaccharides to the substrate peptides, GmaR only
transfers a mono-GlcNAc to serine and threonine residues of FlgA. The secondary
structure of GmaR comprises an N-terminal domain for GT activity, a C-terminal
domain that binds MogR for temperature sensing, and a linker region. Small-angle
X-ray scattering (SAXS) analysis revealed that the apo GmaR protein adopts a
multidomain shape, and the binding of Mg
Bacterial lipopolysaccharide (LPS) comprises a conserved lipid A moiety, a core
oligosaccharide chain, and a variable polysaccharide chain [65]. Variable
polysaccharide chains are crucial for antigenicity, and polysaccharides are ideal
vaccine targets to enhance the immune protection of vaccines [66]. Therefore, the
development of novel vaccines using glycans conjugated to protein is an efficient
strategy for enhancing the immunoprotective effect of the vaccine. Glycoconjugate
vaccines have attracted considerable attention for their novel vaccine design
during the past three decades since glycoproteins can enhance immunogenicity and
induce T-cell-dependent immune responses against numerous pathogens, including
Salmonella Typhi, Streptococcus pneumoniae, and
Neisseria meningitidis [67, 68]. Current chemical conjugation methods
allow the continuous production of recombinant vaccines, especially against
pathogenic targets, but the production of glycoconjugate vaccines is complex and
expensive. For example, a chemical glycoconjugate vaccine against F.
tularensis requires purification of the receptor CRM
To overcome the challenges of glycoconjugate vaccine production, a one-step purification of glycoconjugate vaccines produced by a bioengineered bacterial host, especially using the PglB or PglL oligosaccharyltransferases (OSTs), has been employed in bacterial vaccine design. E. coli lacks the pathways for synthesizing N-linked polysaccharide proteins; consequently, glycoengineering with E. coli generally involves the introduction of three components: a glycan plasmid for the production of the lipid-linked polysaccharide, a plasmid for the carrier protein, and a plasmid encoding an OST (Fig. 4) [70].
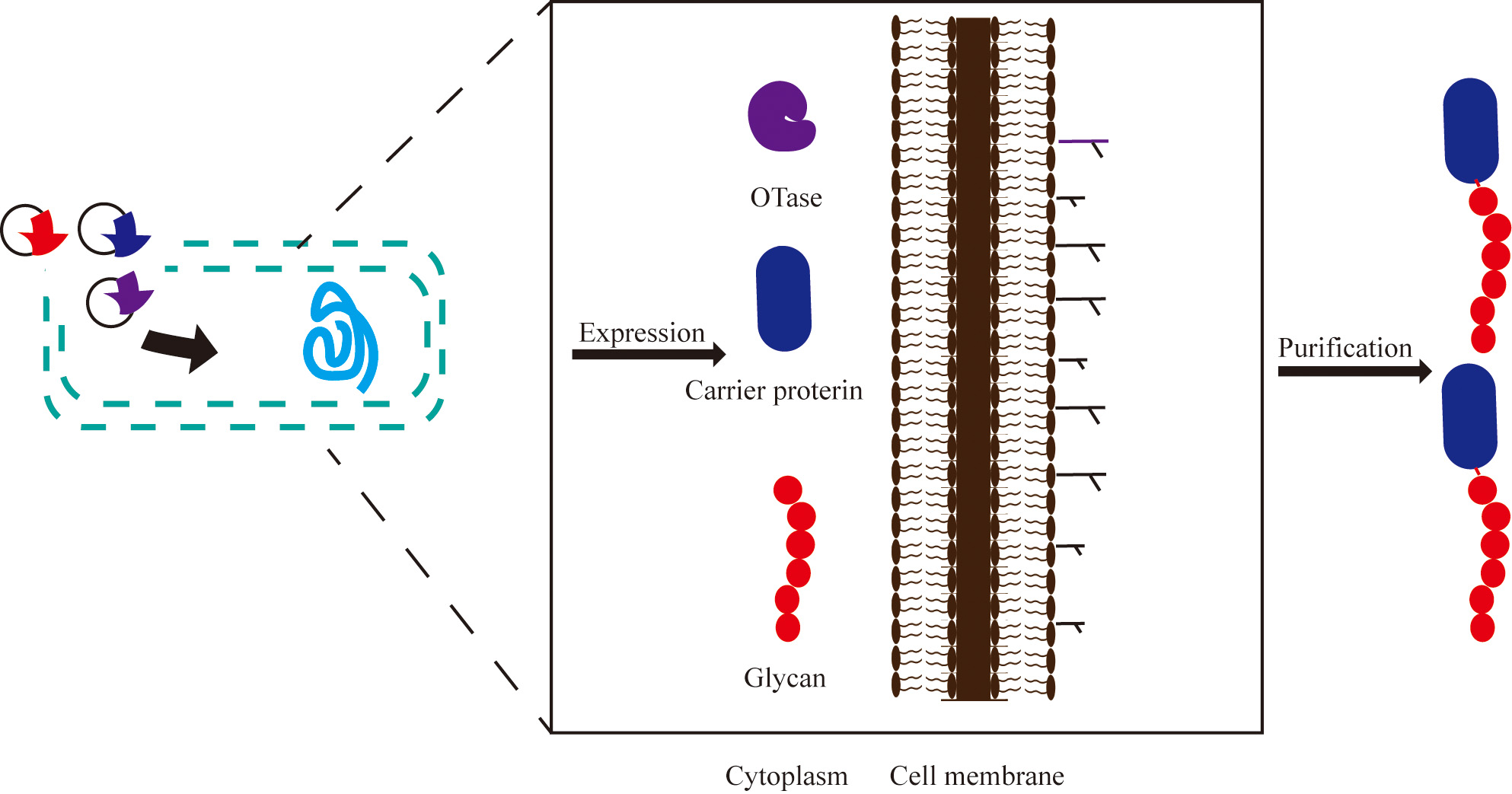 Fig. 4.
Fig. 4.Engineering bacteria to produce glycoproteins. OTase-dependent glycosylation is used as an example. Oligosaccharyltransferase, carrier protein, and polysaccharide are expressed in the host and assembled in the periplasm. Purple: plasmid encoding OST; Blue: plasmid with carrier protein sequence; Red: a glycan plasmid for the production of the lipid-linked polysaccharide.
PglB
| Pathogen/Antigens | Glycosyltransferase | Linked | Substrate protein | Efficacy results | Clinical development phase |
| Shigella flexneri 2a /polysaccharide component of the Shigella flexneri 2a O-antigen | PglB |
N-linked | exotoxin protein A of Pseudomonas aeruginosa (EPA) | Access to vaccine safety and tolerability data. | Phase 1 (Complete) |
| Shigella Flexyn2a /polysaccharide component of the Shigella flexneri 2a O-antigen | PglB |
N-linked | exotoxin protein A of Pseudomonas aeruginosa (EPA) | Testing the ability of vaccines to induce an immune response. | Phase 2 (Complete) |
| Shigella /polysaccharide component of the Shigella flexneri 2a O-antigen | PglB |
N-linked | exotoxin protein A of Pseudomonas aeruginosa (EPA) | To demonstrate the safety and reactogenicity of the vaccine alone or in combination with an adjuvant (aluminum hydroxide). | Phase 1 (Complete) |
| Shigella /polysaccharide component of the Shigella O1 lipopolysaccharide | PglB |
N-linked | exotoxin protein A of Pseudomonas aeruginosa (EPA) | First-in-human data on safety and immunogenicity in infants was obtained. | Phase 1 |
| Phase 2 (Complete) | |||||
| E. coli Tetravalent /Oligosaccharide | PglB |
N-linked | exotoxin protein A of Pseudomonas aeruginosa (EPA) | This study aims to assess the safety and efficacy of a Tetravalent E. coli vaccine in inducing an immune response and reducing the incidence of urinary tract infections. | Phase 1 (Complete) |
| Klebsiella Pneumoniae Tetravalent /O-polysaccharide (OPS) | PglL |
O-linked | cholera toxin B subunit (rCTB) | Obtain first-in-human data on its safety and immunogenicity in healthy adults. | Phase 1 |
| Phase 2 (Complete) | |||||
| New Pneumococcal /Capsular polysaccharides (CPS) | PglS | O-linked | exotoxin protein A of Pseudomonas aeruginosa (EPA) | Assess the safety and immunogenicity of a bioconjugate investigational vaccine compared to the control group (Pneumovax23). | Phase 1 (Complete) |
Although PglB
Compared to N-linked glycosylation, there is a greater flexibility of glycan structures and protein acceptors in O-linked glycosylation. Faridmoayer et al. [49] demonstrated that in engineered E. coli and Salmonella cells, PglL can transfer any sugar carried by undecaprenyl pyrophosphate (UndPP) to the pilin substrate. Considering whether the lipid carrier was a limiting factor for PglL, the authors established the authors established a glycosylation system in vitro using purified PglL, pilin, and chemically synthesized lipid farnesyl pyrophosphate (FarPP) with pentasaccharides. The results indicated that synthetic lipid carriers could also transfer polysaccharides to the periplasm and localize polysaccharides around PglL. PglL is also broadly selective for vectors. The flexibility of PglL is likely to play a role in the production of future glycoconjugate vaccines [44]. Therefore, it has a higher application potential in the development of new conjugate vaccines. Salmonella enteritidis (SE) is a zoonotic pathogen. Li M et al. [82] constructed the PglL plasmid, recombinant Pseudomonas aeruginosa exotoxin A (rEPA), and cholera toxin B subunit (CTB) plasmids into S. enteritidis strains. It was detected that when PglL was expressed, rEPA and CTB could undergo glycosylation. The results provide the basis for the SE glycoprotein biosynthesis [82]. A conjugate vaccine against S. enterica serovar Paratyphi A produced by the co-expression of PglL and the CTB4573 carrier protein in vivo evokes a protective and specific immune response against the bacterium [83]. A polysaccharide conjugate vaccine against Klebsiella pneumoniae serotype O2 strain was developed by transfecting a K. pneumoniae waaL mutant with a vector encoding PglL and a recombinant cholera toxin B subunit, and the vaccine conferred effective protection in a murine model (Table 1) [84, 85]. Brucellosis is a common human and animal infectious disease worldwide, endangering human health. There is currently no effective vaccine against this disease. Huang et al. [86] designed a vaccine by replacing the Brucella O-antigen polysaccharide (OPS) with the low-pathogenic Yersinia enterocolitica serotype O:9 (YeO9) OPS via an O-linked glycosylation system. As a result, specific immunity was successfully induced, effectively defending against brucellosis and avoiding large-scale cultivation of pathogenic strains [86]. In order to further mass-produce biological conjugate vaccines against B. abortus, Li S et al. [87] established a new vaccine engineering production process in 2023. The OPS gene cluster was divided into five separate fragments and then reassembled, and subsequently expressed in E. coli. The final product is glycosylated by PglL. A series of experiments proved that the vaccine can induce a strong immune response, which induces the production of specific antibodies [87]. Other glycosyltransferases in the O-linked glycosylation system have also been used in vaccine development in recent years. For example TfpM from Moraxella osloensis. Knoot CJ et al. [88] found that TfpM-derived bioconjugates can trigger specific antibody IgG production in mice and are immunogenic. Because a variety of long-chain polysaccharides can be transferred to the substrate protein carrier, the shortest recognition glycosylation sequence is identified. This provides new options for the production of new sugar-conjugated vaccines in the future [88].
Another glycosyltransferase, PglS, can transfer many different types of bacterial glycans, including those with glucose at the reducing end. For example, Feldman et al. [89] successfully developed a bivalent K1/K2 Klebsiella pneumoniae biological conjugate vaccine with immunogenicity and effectiveness through PglS. Vaccines protect mice against infection by hypervirulent K. pneumoniae (hvKp). Although in vivo work suggests vaccine efficacy against hvKp in the lung, further studies are needed to assess efficacy against hvKp in other sites including the liver, blood, and meninges (Table 1) [89]. Harding CM et al. [90] utilized PglS to successfully produce sugar-conjugated vaccines with glucose polysaccharides at the reducing end, thereby overcoming the limitations imposed by the structural requirements of polysaccharides. Furthermore, PglS demonstrated the ability to glycosylate various vaccine carrier proteins. Inoculating mice with the polyvalent pneumococcal bioconjugate vaccine resulted in the production of antisera that effectively killed bacteria in vitro [90]. Group B Streptococcus (GBS) is a leading cause of neonatal infections and invasive diseases in nonpregnant adults worldwide. Duke et al. [91] expressed GBS types Ia, Ib, and III in recombinant E. coli and studied the generation of bioconjugate vaccines using PglS conjugation with the carrier protein Pseudomonas aeruginosa exotoxin A (EPA). This technology provides a basis for the subsequent development of multivalent GBS bioconjugate vaccines [91]. These results support our expansion to future sugar-conjugate vaccines.
Prokaryotic glycosylation systems are simpler than eukaryotic systems as protein glycosylation is achieved by a single OST. The transfer of an N-linked or O-linked OST, a substrate protein, and a plasmid for glycans synthesis to E. coli is therefore sufficient for the production of glycoconjugate proteins. To date, the structure and mechanism of N-linked glycosylation by the PglB OST of C. jejuni have been studied in depth and applied to the design of glycoconjugate vaccines against F. tularensis, B. pseudomallei, A. pleuropneumoniae, and S. flexneri. O-linked glycosylation allows a broader variability of substrate amino acid residues, including serine, threonine, and tyrosine. However, the structure and mechanism of LLO binding by the PilO protein of P. aeruginosa and the PglL enzyme of N. meningitidis remain unelucidated, which necessitates further research on bacterial O-linked glycosylation mechanisms and their application. Although sugar-conjugated vaccines have been expressed and produced by engineering bacteria, it is still difficult to address the problems of exogenous plasmids, recombinant expression, and host cell modification. Shibata et al. [92] encountered these problems when expressing the cell wall polysaccharide of Streptococcus mutans in E. coli. For example, the expressed glycan only has the rhamnose backbone and lacks side chain residues. They speculated that another genetic modification may be required for glucose side chain formation in recombinant expression. The deletion experiment of genes involved in the assembly of rhamnose synthesis infers that the genes play a role in the process of rhamnose assembly in a certain order, but the method to modify them in the host body has not been confirmed [92]. These issues still await further research.
Conceptualization, SO, MXC and LZ; methodology, SO, MXC and LZ; software, PL and RL; validation, SO; formal analysis, PL and RL; data curation, PL and RL; writing —original draft preparation, PL and RL; writing review and editing, MXC, and LZ; visualization, PL and RL; supervision, SO, MXC and LZ; project administration, SO, MXC and LZ; fund acquisition, SO, LZ. All the authors have read and approved the final version of the manuscript. All authors contributed to editorial changes in the manuscript.
Not applicable.
Not applicable.
This study was supported by the National Key Research and Development Project of China (grant number: 2021YFC2301403, 2021YFC2102100), the National Natural Science Foundation of China (grant numbers: 82225028, 82172287, 32271507 and U20A20361), and the High-level personnel introduction grant of Fujian Normal University (grant number: Z0210509).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.




