- Academic Editor
Background: As a chronic degenerative disorder of the central nervous
system that affects both motor and non-motor systems, Parkinson’s disease (PD) is
very complex, and explanations and models are needed to better understand how
dopaminergic neurons are affected and microglia are activated. Methods:
A theoretical protein-RNA recognition code that assumes that the second letter in
codons is compatible with specific amino acids involved in protein-RNA
recognition was used to search for compatibility of human
Parkinson’s disease (PD) is classified as the second most common age-related progressive neurodegenerative disease; it is manifested by both motor and non-motor symptoms [1]. Motor symptoms are a classic finding in PD and include resting tremor, bradykinesia, postural instability, and rigidity. Non-motor symptoms of PD have become better identified over time and include cognitive decline, depression, anxiety, anosmia, and dysautonomia [1]. The precise cause of PD is still unknown, but a combination of genetic and environmental factors are thought to play a role [2]. Epidemiological studies have shown that increasing age, male sex (50% higher risk than females), and exposure to pesticides are risk factors, whereas coffee and alcohol drinking appear to be protective [1, 2].
In PD, the pathological feature is depigmentation of the substantia nigra and
locus coeruleus, with loss of neurons in the pars compacta of the substantia
nigra [1]. In PD, the presence of intracellular aggregates/fibrils of
Under physiologically normal conditions, “soluble”
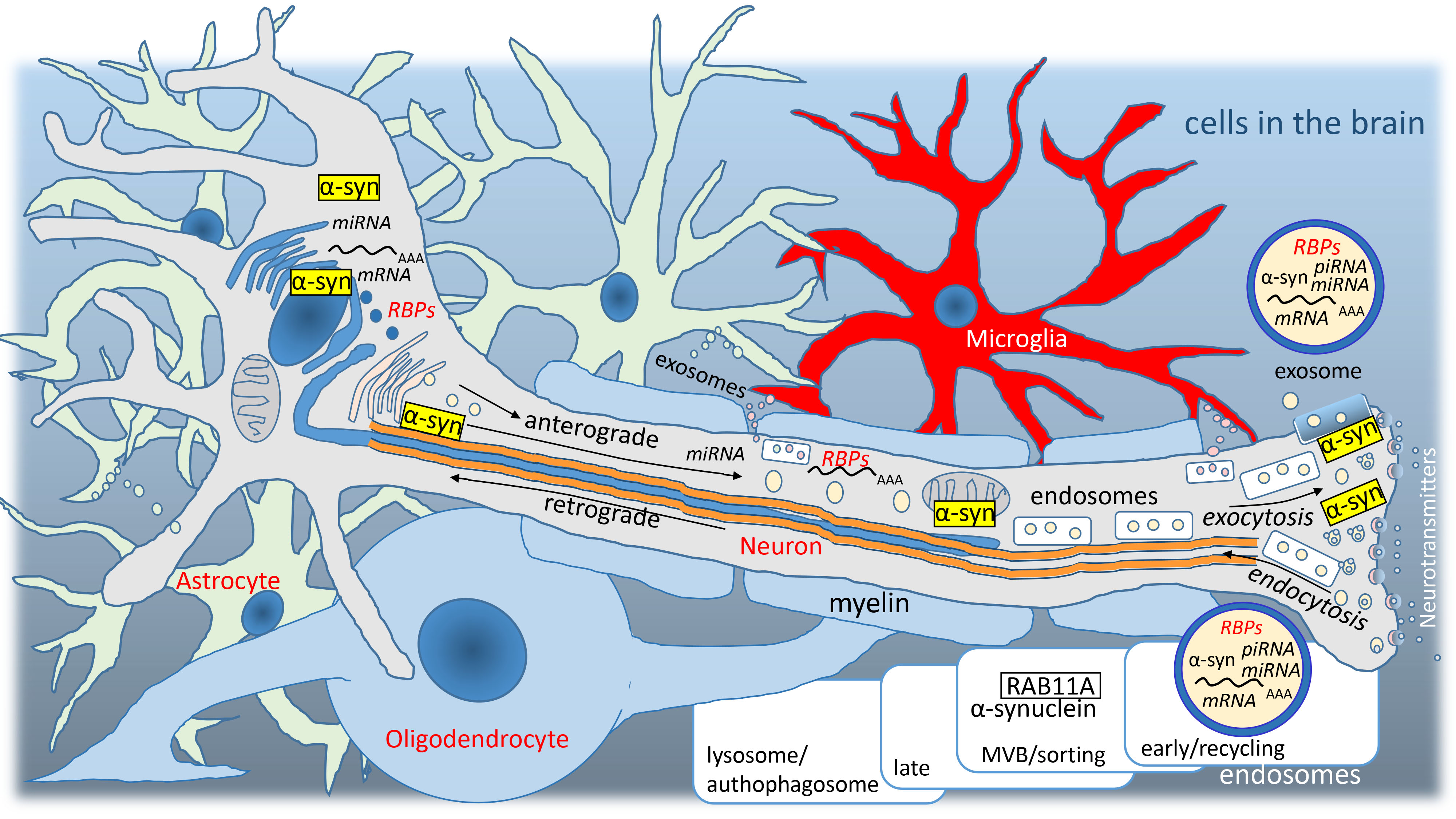 Fig. 1.
Fig. 1.Graphic representation of cells in the brain. Nerve mRNAs and
RNA-binding proteins (RBPs) are transferred from the nucleus via the axon
(exosomal pathway, anterograde) and glial RNAs, RBPs and other proteins are
transferred from the extracellular space to the nucleus (endosomal pathway,
retrograde). Dysregulation of
1-L transcription is a very simple method that was introduced recently [13, 14, 15]
that applies a theoretical protein-RNA recognition code [16, 17]. An example can
be given here on the RBP DAZL (deleted in azoospermia-like). DAZL binds the
3
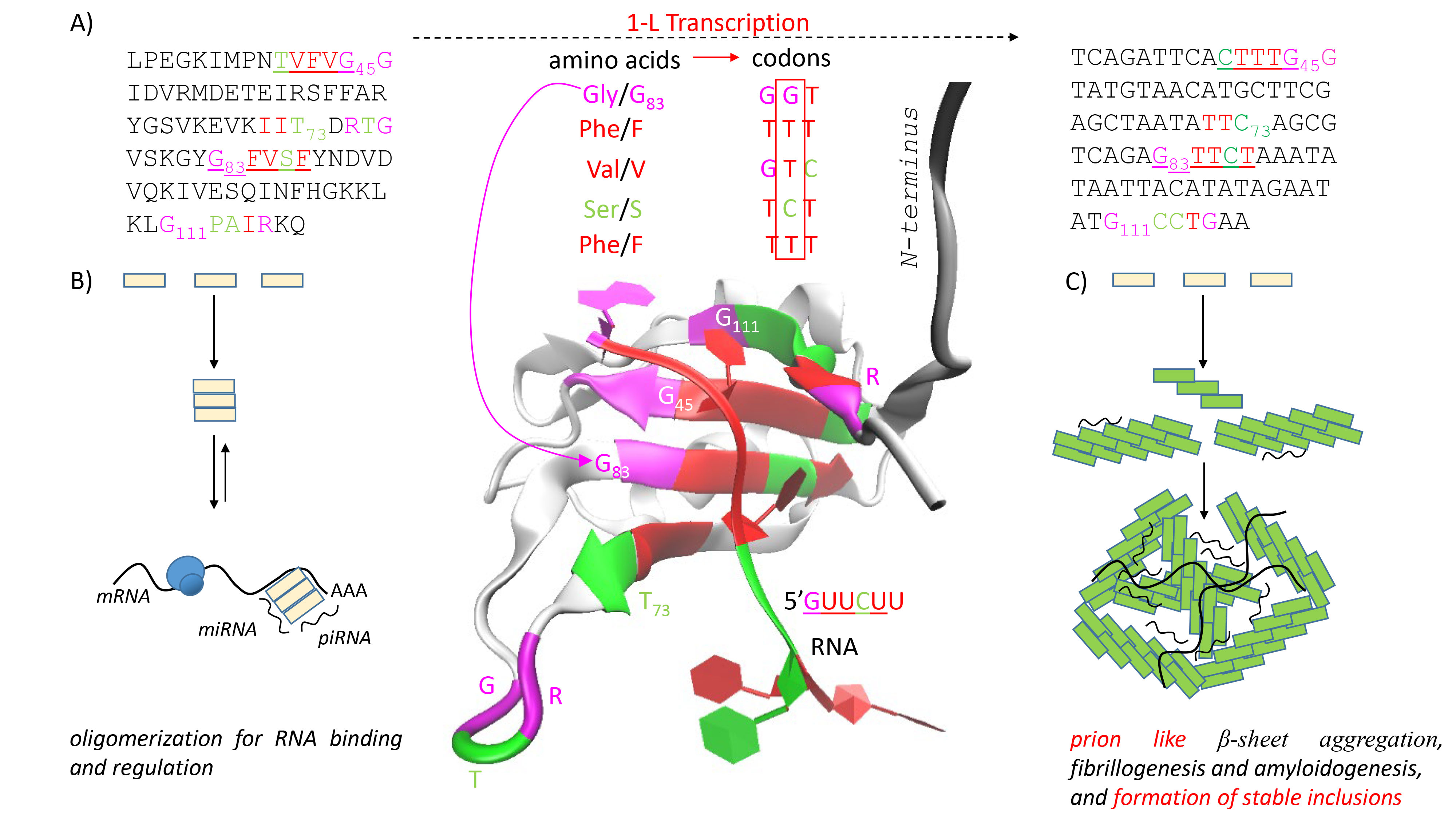 Fig. 2.
Fig. 2.An RNA binding protein (RBP) named deleted-in-azoospermia-like (DAZL). (A) 1-L transcription of DAZL. RBP DAZL binds a GUU(C/U) motifs [18]. The 1-L protein-RNA recognition code means that RBPs use at least one amino acid sequence that is exactly compatible with the recognized RNA nucleotide sequence, 83GFVSF87 for GUUCU and 45GVFVT41 for GUUUC. 1-L transcription uses the second position in the amino acid codon. The structure 2XS2 shows that both sequences are involved in the RNA binding. (B) A model of RBP oligomerization at low concentrations for RNA binding and regulation. (C) A model of prion like fibrillogenesis and amyloidogenesis at high concentrations, followed by RNA binding and formation of stable inclusions.
A single RBP typically recognizes hundreds of transcripts and forms extensive
regulatory networks, so it is not surprising that RBPs are evolutionarily more
conserved than are transcription factors [21]. In the present study,
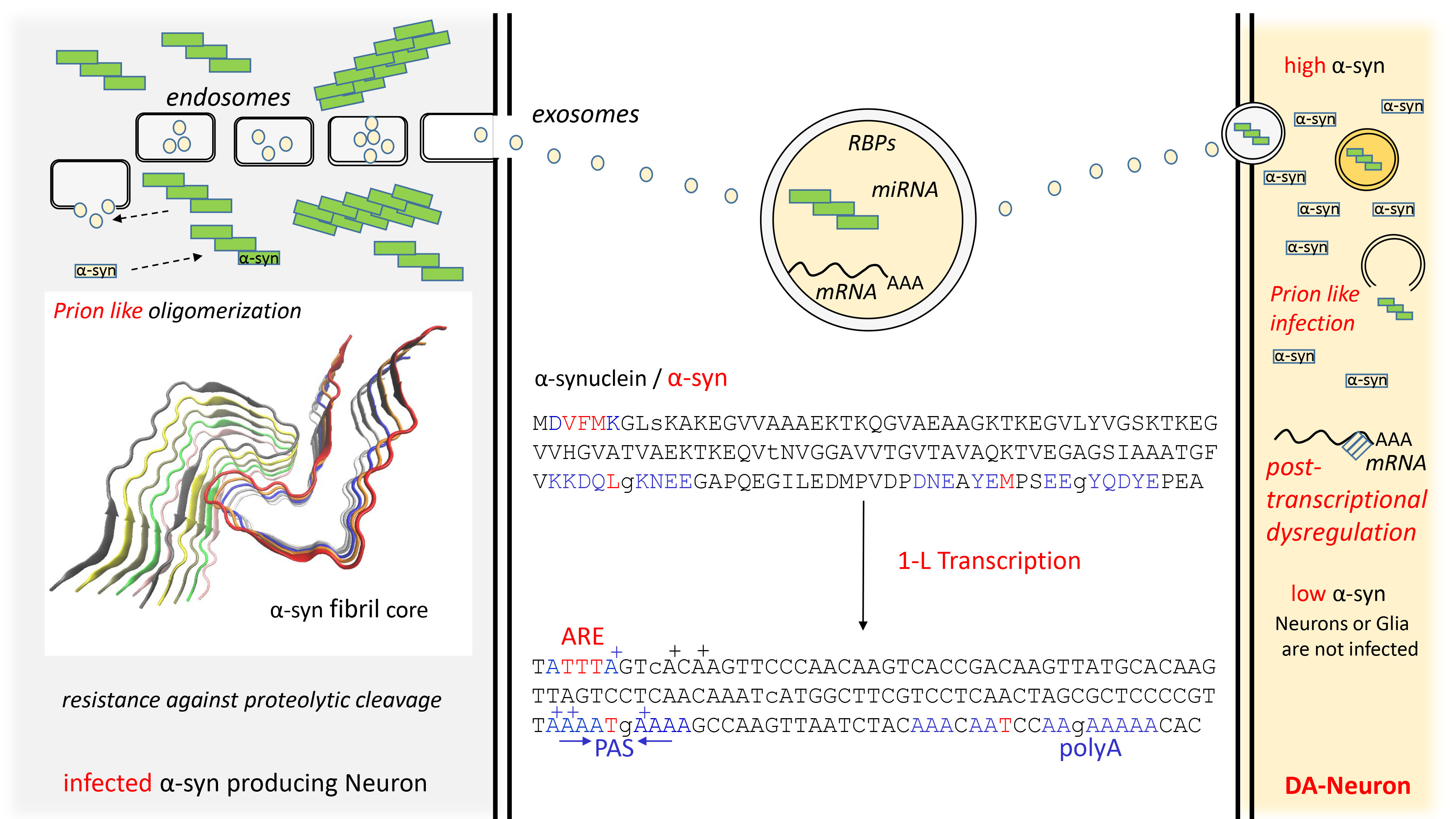 Fig. 3.
Fig. 3.Spread of PD by exosomes and 1-L transcription of
In summary, PD belongs to the
According to the one-letter key, one amino acid per nucleotide (1-L), RBPs use at least one amino acid sequence that is exactly compatible with the recognized RNA nucleotide sequence. A nucleotide is defined by the type of nucleotide at the second position in the amino acid codon (Fig. 2A). This fact can be used to identify RNAs that are potentially controlled/regulated by the tested RBPs (Fig. 4).
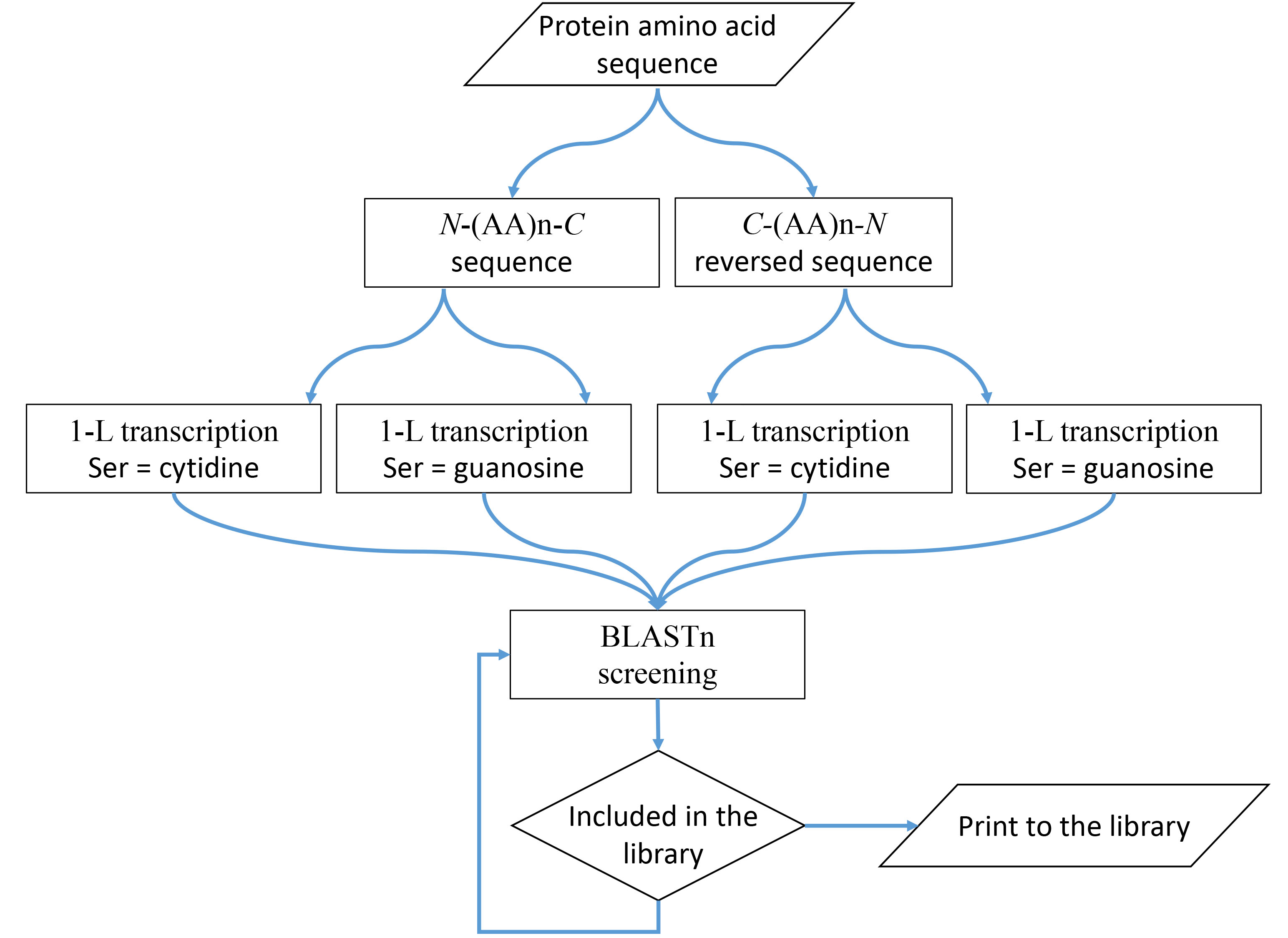 Fig. 4.
Fig. 4.1-L transcription method. A schematic diagram that visually represents the steps and concept.
The 1-L transcription method is very simple: the amino acid sequence of RBP is
transcribed into an RNA sequence, which is then used for classical BLASTn
screening in the human transcriptome. Reading of the 5
BLASTn screening of the human transcriptome was performed as a standard nucleotide blast at NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for the four nucleotide sequences separately. “Genomic + transcript databases” and “human genomic plus transcript”, “somewhat similar sequences” (blastn), word size 7, maximum number of target sequences 500, and expected threshold 500, were used for the search.
MicroRNAs are small endogenous RNAs that pair and bind to mRNA sites to induce post-transcriptional repression, such that reducing the level of miRNAs or other small regulatory RNAs promotes translation; alignments with the reverse complement RNA sequences are considered promotive (yellow in the figures). In contrast, alignments with the RNA sequence of the gene are considered repressive (green in the figures) because sequestering and blocking of free mRNA by the test protein represses translation. The identified genes are shown in Figs. 5,6.
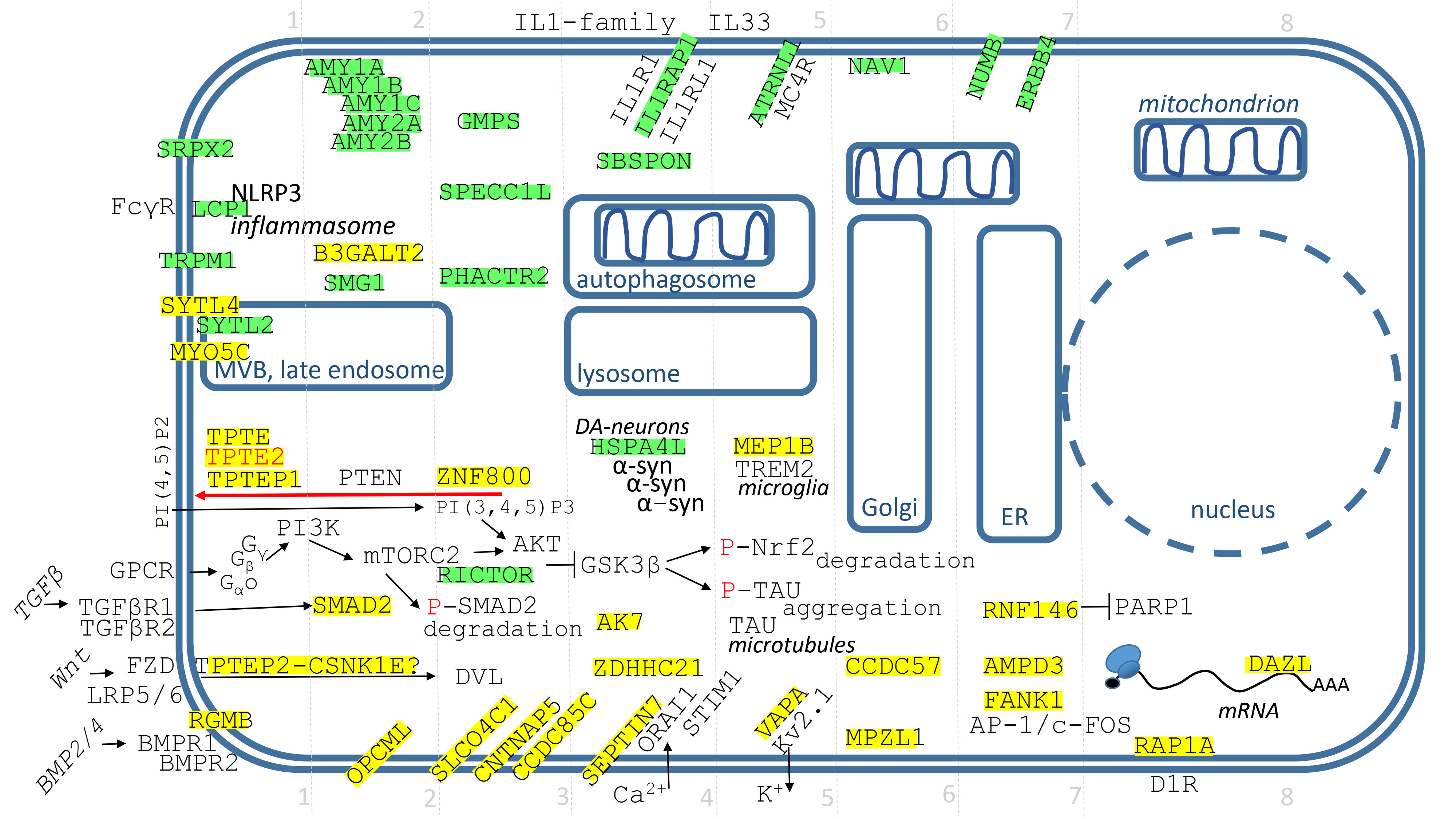 Fig. 5.
Fig. 5.Identified genes/proteins with function in the plasmatic membrane, cytosol or PM-cytosol interface. The description of genes/proteins is given in the text from the upper left corner down to the right. Green highlights show alignments with the gene transcript RNA sequence (post-transcriptionally repressed), yellow highlights show alignments with the reverse complement RNA sequences (post-transcriptionally promoted).
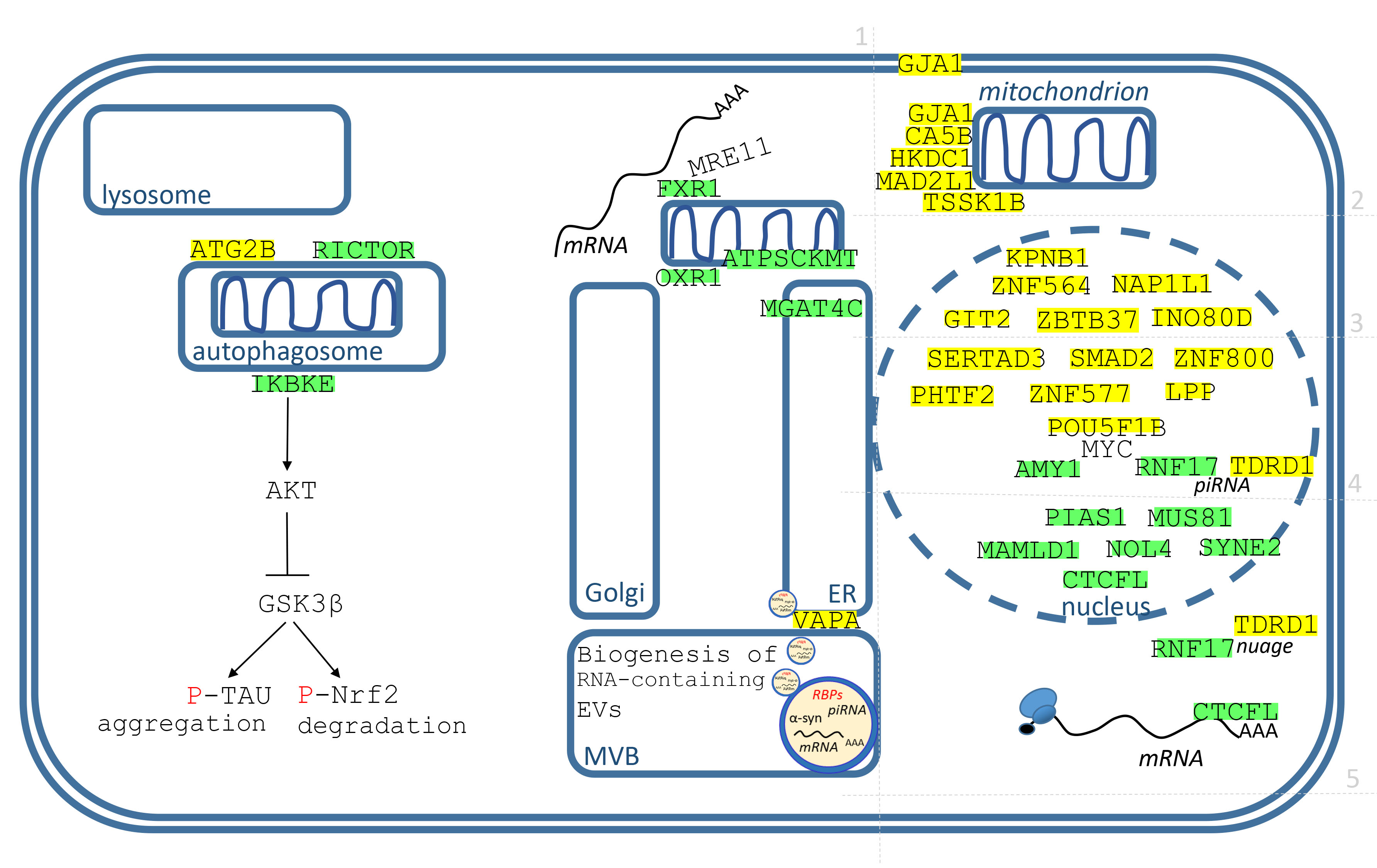 Fig. 6.
Fig. 6.Identified genes/proteins with function in membrane organelles, mitochondria and nucleus. The description of genes/proteins is made in the text, from endosomes to the nucleus, from left to right. Green highlights show alignments with the gene transcript RNA sequence (post-transcriptionally repressed), yellow highlights show alignments with the reverse complement RNA sequences (post-transcriptionally promoted).
The genes/proteins are displayed in Fig. 5 and their description is done from the upper left corner down to the right. For the better orientation, the grid divides genes to 8 groups.
SRPX2 (sushi repeat containing protein X-linked 2) is a neuronally expressed complement inhibitor that protects synapses from microglial engulfment and elimination [26, 27]. SRPX2 regulates complement-dependent microglial synaptic phagocytosis by blocking the classical C1q-mediated complement cascade [26, 27].
LCP1 (lymphocyte cytosolic protein 1)/L-plastin/LPL is an actin-bundling protein
and a critical regulator of immune cell function, for example, LCP1 can function
downstream of Fc
TRPM1 (transient receptor potential cation channel subfamily M member
1/transient receptor potential melastatin 1; initially named melastatin/MSLN) is
a founding member of the TRPM subfamily of ion channels. TRPM1 is believed to
positively regulate melanocyte pigmentation [31]. The main pathological features
of PD consist of the progressive loss of pigmented DA-neurons in the substantia
nigra pars compacta. Individuals with PD show reduced levels of neuromelanin in
both the ventral and dorsal tiers of the substantia nigra [32].
SYTL4/SLP4 and SYTL2/SLP2 (synaptotagmin-like 4 and 2) are Rab27 effectors responsible for docking secretory vesicles to the plasma membrane prior to exocytosis [34, 35]. SLP2 controls the localization of SLP4-mediated vesicle-tethering activity to a single PIP2-enriched initiation site; when SLP2 is disrupted, SLP4 targets vesicles ectopically to various plasma membrane locations [35]. Rab27a and SYTL4/SLP4 provide multivesicular body (MVB) docking to the plasma membrane [36].
MYO5C (myosin VC) is one of the three myosin V motors; myosin motors use energy derived from ATP hydrolysis to generate movement to control membrane transport, organelle trafficking, and actin dynamics. MYO5C differs from MYO5A or MYO5B, in that it colocalizes with secretory granule markers such as chromogranin A and RAB27b [37]. A unique function of MYO5C in melanosome biogenesis and secretion has been identified, depending on its interactions with RAB32 and RAB38 [38]. LRRK2 (mutations in this kinase represent the most common cause of PD) also interacts with RAB32 and RAB38 to regulate endo-lysosomal trafficking [8, 39].
TPTE, TPTE2 (transmembrane phosphoinositide-3-phosphatase with tensin homology)
are phosphatases that belong to the PTEN family (five members). PTENs oppose
PI3K-AKT activation; as such they activate GSK3
TPTEP1 (TPTE pseudogene 1) is a noncoding RNA silenced through DNA methylation in various cancers [43]. In addition, TPTEP1 suppresses high-glucose-induced dysfunction in retinal vascular endothelial cells [44]. TPTEP1 mRNA may function similarly to the PTENP1 pseudogene, which is targeted by PTEN-targeting miRNAs and acts as their decoy [45].
TPTEP2-CSNK1E (TPTEP2-CSNK1E readthrough) fusion may play a role in Wnt
signaling similar to CSNK1E, casein kinase 1
RGMB (repulsive guidance molecule b)/DRAGON is a coreceptor and regulator of
bone morphogenetic protein (BMP)-signaling [48]. In one epigenomic association
study (EWAS), RGMB was identified as a gene related to motor progression of PD
[49]. BMP2 has potential for use in the neurotrophic treatment of PD [50]. In
contrast to the RGMB-mediated enhancement of signaling observed for BMP2, RGMB
inhibits BMP14/GDF5 signaling [51] and BMP14/GDF5 exerts neuroprotection in the
AMY1A (amylase alpha 1A), AMY1B, AMY1C, AMY2A and AMY2B are
B3GALT2 (
SMG1 (SMG1 nonsense mediated mRNA decay associated PI3K related kinase) is a key
kinase involved in the nonsense-mediated mRNA decay (NMD)-pathway, which targets
mRNAs containing premature stop codons for rapid degradation and is essential for
mammalian embryonic development, brain development, and modulating the stress
response [60]. SMG1 was identified as a regulator of PD-associated
SMAD2 (SMAD family member 2) is one of the major components of the transforming
growth factor-
OPCML (opioid binding protein/cell adhesion molecule like) is a cell surface
glyco-phosphoinositol (GPI)-anchored protein that regulates spine maturation and
cognitive behaviors through the ephrin-EphB2-cofilin signaling pathway [66]. It
has been identified as one top genes most consistently underexpressed in the
aging brain [67] and as one GWAS candidate gene under selection for sporadic PD
[68]. OPCML interacts with S129- phosphorylated
GMPS (guanine monophosphate synthase) converts XMP (xanthosine 5
SPECC1L (sperm antigen with calponin homology and coiled-coil domains 1 like) associates with actin filaments and microtubules through its calponin homology domain and coiled coil domain 2 (CCD2), respectively. It has a role in regulating actomyosin organization during tissue morphogenesis. SPECC1L requires association with microtubules via CCD2 for transport in the cell. The absence of SPECC1L leads to a disorganized accumulation of actin at the cell periphery [73].
PHACTR2 (phosphatase and actin regulator 2) belongs to the family of protein phosphatase 1 (PP1) and actin regulatory proteins [74]. PP1 is a multifunctional phosphatase with diverse roles in the nervous system, likely targeting neuronal substrates associated with the actin cytoskeleton. In the brain, the expression of PHACTR1 and PHACTR2 are remarkably complementary [74]. PHACTR2 has already been nominated as a risk factor for PD [75].
ZNF800 (zinc finger protein 800) is expressed as a long, non-coding RNA (lncRNA) that upregulates PTEN and inhibits the activity of AKT (Fig. 5) [76]. It can be also expressed as an oligodendrocyte-specific circular RNA [77].
RICTOR (RPTOR independent companion of MTOR complex 2/rapamycin-insensitive
companion of mTOR) is a scaffold protein that regulates the assembly of the
mTORC2 complex, which regulates cell survival and proliferation mainly through
PKC
SLCO4C1/OATP4C1 (solute carrier organic anion transporter family member 4C1/organic anion transporting polypeptide 4C1) is a renal organic anion transporter but can be detected in cultured neurons [80]. Asymmetric dimethylarginine, cAMP, CDCA, E1S, GCA, L-homoarginine, T3, and T4 are substrates for SLCO4C1/OATP4C1 [80].
CNTNAP5 (contactin-associated protein family member 5) is a member of the neurexin-like family of cell adhesion molecules and receptors expressed in vertebrate neurons [81]. A genome-wide scan in a cohort of 1130 patients with PD found an association signal at chr2q14.3. The only annotated gene within 1 Mb on either side of the DASH segment is CNTNAP5 [82].
CCDC85C (coiled-coil domain containing 85C) is a protein at the apical junctions
of radial glia that is disrupted in mice with hydrocephalus [83]. Hydrocephalus
is a disease characterized by the accumulation of cerebrospinal fluid in the
brain cavities. PD and
IL1RAP (interleukin 1 receptor accessory protein)/IL-1RAcP/IL1R3 forms a complex
with receptors for IL1-like cytokines and is essential for IL1 signaling, which
is involved in several neuroinflammatory diseases, including PD; therapeutic
strategies modulating IL1 activity could have neuroprotective effects in PD [85].
IL1
SBSPON (somatomedin B and thrombospondin type 1 domain containing): its Drosophila melanogaster ortholog (CG42339) was identified among the top-associated genes containing single nucleotide polymorphisms that render DA neurons vulnerable to PD [87].
HSPA4L (heat shock protein family A member 4 like) is a member of the Hsp110
chaperone family that alleviates
AK7 (adenylate kinase 7) is unusual in the adenylate kinase family because it is
a high molecular weight isoform. AK7 appears to be a marker of cilia; mutations
in the AK7 gene result in animals exhibiting characteristics of primary
ciliary dyskinesia and hydrocephalus [89]. Adenylate kinase and downstream
AMP-signaling function as an integrated metabolic monitoring system that senses
cellular energy states [89]. AK7 was among the three differentially expressed
genes in
ZDHHC21 (zinc finger DHHC-type palmitoyltransferase 21) is responsible for a
post-translational lipid modification of proteins called palmitoylation. For
example, ZDHHC21 palmitoylates PLC
SEPTIN7 (septin 7) is cytoskeletal GTPase, which can bind to other septins and
form a core component of most septin complexes. This septin is crucial to the
spine morphogenesis and dendrite growth in neurons [93]. SEPTIN7 regulates
extracellular Ca
ATRNL1 (attractin like 1) directly interacts with MC4R (melanocortin 4 receptor, energy homeostasis, erectile function) and has been proposed to act in MC4R signaling [95]. The link between dopamine and melanocortin systems of the brain has been documented at both structural and functional levels [96], and it appears that ATRNL1 signaling may be involved in the modulation of dopamine systems.
MEP1B (meprin A subunit beta) is a metalloprotease that cleaves TREM2 on
microglia and controls phagocytic activity [97]. MEP1B is also involved in APP
cleavage and plays an important role in the amyloidogenic pathway and A
VAPA [VAMP (vesicle-associated membrane protein) associated protein A] interacts with Kv2 channels. The Kv2.1 channel mediates cell death, which is associated with the loss of cytosolic potassium in neurons and, therefore, the targeted disruption of the Kv2.1-VAPA association provides neuroprotection [100].
NAV1 (neuron navigator 1) is an important member of the human neuronal navigator gene family and is involved in axon guidance. Growth-cone macropinocytosis of the neurotrophin receptor and neuritogenesis are regulated by NAV1 [101]. Growth-cone macropinocytosis is important for downstream signaling, neurite targeting, membrane recycling, NAV1 functions at the interface of microtubules, actin, and the plasma membrane organizing the cell periphery [101]. Expression profiling of long non-coding RNAs (lncRNAs) in PD revealed that lncRNA RPS14P3 interferes with miRNA-mediated NAV1 gene expression [102].
CCDC57 (coiled-coil domain containing 57) interacts with microtubules and microcephalic protein CEP63, and regulates centriole duplication and mitotic progression; centrosome dysfunction is associated with neurodevelopmental disorders [103].
MPZL1 (myelin protein zero like 1) is an immunoglobulin that promotes tumor cell proliferation and migration [104, 105]. In bladder cancer, SNCA was highly correlated with MPZL1 protein in activated CD8+ T cells [106].
NUMB (NUMB endocytic adaptor protein) is an evolutionarily conserved protein originally identified as an inhibitor of Notch signaling [107]. It determines cell fate for neural progenitor cells, their maintenance and self-renewal. It interacts with the endocytic machinery to control Notch, with cadherin and integrins to control cell adhesion and migration, and with the E3 ubiquitin ligase Itch for ubiquitination and degradation of the Notch ICD [107]. Although NUMB interacts with Itch to promote intracellular trafficking and subsequent degradation of the NOTCH1 receptor ICD [107], NUMB appears to enhances Notch signaling by regulating the ubiquitinating activity of the BAP1–BRCA1 complex [108]. In Drosophila, loss-of-function NUMB mutations lead to loss or reduction of DA neurons [109].
ERBB4 (erb-b2 receptor tyrosine kinase 4) is a neuregulin-1 (nerve growth and
differentiation factor, NRG1) receptor that regulates extracellular dopamine
through p38/MAPK14 signaling [110]. The decreased expression of ERBB4 by RNA
interference leads to increased extracellular dopamine [110]. The total number of
nigral ERBB4-positive neurons is reduced in PD, but ERBB4 rose in the remaining
functional neuromelanin-containing DA-neurons [111]. In microglia, NRG1-ERBB4
signaling exerts anti-inflammatory effects and inhibits the release of
inflammatory factors, most likely via ERBB4-dependent inhibition of the
NF-
RNF146 (ring finger protein 146) is a poly (ADP-ribose)-directed E3 ligase that
regulates axin degradation and Wnt signaling. It directly interacts with poly
(ADP-ribose) and promotes the degradation of PARylated proteins [113]. In cell
pellets from the nasal lavage of PD patients, the transcript level of RNF146
(mRNA) is decreased, although the transcript level of
AMPD3 (adenosine monophosphate deaminase 3) is a cytosolic enzyme that catalyzes
the thermodynamically irreversible deamination of AMP to IMP and ammonia
(AMP
FANK1 (fibronectin type III and ankyrin repeat domains 1) is a co-regulator of AP-1 in the AP-1-signaling pathway. The transcription factor AP-1 is a dimeric complex composed of members that belong to the Jun, Fos, ATF, and MAF protein families. This TF complex is involved in the regulation of cell proliferation, differentiation, and apoptosis. FANK1-AP-1 can mediate pro-apoptotic and anti-apoptotic signaling [118, 119].
RAP1A (member of RAS oncogene family/repressor-activator protein 1a) is a small monomeric GTP-binding protein of the RAS family known to switch between an inactive GDP-bound form and an active GTP-bound form. Acting as a molecular switch linking extracellular signals to intracellular responses, RAP1A is associated with multiple signaling pathways including proliferation, integrin activation, cell adhesion, and migration (https://www.genome.jp/pathway/hsa04015+5906), but it also plays role in inflammation, oxidative stress, and apoptosis involved in the pathogenesis of PD. Actually, RAP1A activation is involved in L-DOPA signaling in medium spiny neurons in D1R (dopamine receptor)-PKA (cAMP-protein kinase A)-RAP1 GEF (Rasgrp2)/RAP1A-MAPK intracellular signaling [120].
DAZL (deleted in azoospermia like) is an RBP (Fig. 2) that mediates a broad
translational program regulating the expansion and differentiation of
spermatogonial progenitors [121]. DAZL binds the 3
The genes/proteins are displayed in Fig. 6 and they are described in order from the endosomes to the nucleus. For the better orientation, the grid divides genes to 5 groups.
ATG2B (autophagy related 2B) participates in the early formation of
autophagosomes, has membrane binding activity, and lipid transfer activity, which
is accelerated by negatively charged lipids and WIPI4 [122, 123]. Impaired
autophagy significantly contributes to
IKBKE/IKK
RICTOR (RPTOR independent companion of MTOR complex 2/rapamycin-insensitive companion of mTOR) is a scaffold protein regulating the assembly of mTORC2 complex; mTORC2-SGK1 signaling integrates external signals to regulate mitochondrial autophagic turnover through reactive oxygen species (ROS). Loss of RICTOR induces mitochondrial ROS and leads to mitophagy [79].
FXR1 (FMR1 autosomal homolog 1) is an RNA-binding protein that can be present in an amyloid form in the brains of various mammalian species, including humans [127]. This RBP regulates synaptic homeostasis [128] and is involved in oxidative stress responses; depletion of FXR1 by siRNA increases sensitivity to the mitochondrial ROS [129].
OXR1 (oxidative resistance 1) is present in mitochondria and cytoplasm and is essential for protection against oxidative stress-induced neurodegeneration [130]. Hypothetically, OXR1 functions as an oxidative stress sensor [131].
ATPSCKMT (ATP synthase c subunit lysine N-methyltransferase) is a mitochondrial
methyltransferase that trimethylates Lys-43 in the c-subunit of ATP synthase and
optimizes mitochondrial ATP synthase function [132]. The C-subunit is an
amyloidogenic protein that can spontaneously fold into
MGAT4C [mannosyl (
VAPA [VAMP (vesicle-associated membrane protein) associated protein A] and its binding partner CERT (ceramide transporter) control the biogenesis of vesicles containing extracellular RNA at endoplasmic reticulum (ER) membrane contact sites [136].
GJA1 (gap junction protein alpha 1) is a member of the family of connexins,
which are gap junction and hemichannel-forming proteins involved in cell death,
proliferation, and differentiation. GJA1 higher expression in motoneurons was
negatively correlated with genes associated with neuronal activation profiles
[137]. GJA1 was strongly associated with
CA5B (carbonic anhydrase 5B) is a mitochondrial isoform of carbonic anhydrase
(CA, conversion of CO
HKDC1 (hexokinase domain containing 1) is a novel fifth hexokinase (the first step in glucose metabolism) that, like HK1 and HK2, has been shown to interact with mitochondria [142]. Flunarizine depletes mitochondria and reduces dopamine concentrations in the striatum [143]. A genome-wide CRISPR KO library and loss-of-function genetic screen revealed that HKDC1 is important in a mitophagy-independent mitochondrial quality control and mitolysosome exocytosis in flunarizine-induced parkinsonism-like symptoms [143].
MAD2L1 (mitotic arrest deficient 2 like 1) is a component of the mitotic spindle assembly checkpoint that prevents the onset of anaphase until all chromosomes are properly aligned in metaphase. This protein also targets the MYC gene and is significantly downregulated in tumor dormancy [144], and may be associated with mitochondrial function [145]. Mitochondrial dysfunction is a well-established player in the pathogenesis of PD [146].
TSSK1B (testis-specific serine kinase 1B) belongs to the TSSK group, which is part of the AMPK protein kinase family, but currently there is not much information about TSSK substrates in vivo [147]. Targeted deletion of Tssk1 and 2 causes male infertility due to developmental dysregulation and collapse of the mitochondrial sheet in late spermatids [147].
KPNB1/IPO1 (karyopherin subunit beta 1/importin-1) is a component of
karyopherin/importin; karyopherin abnormalities have been identified in
synucleinopathies including PD and dementia with LBs [148]. KPNB1 is in the list
of genetic markers in PD [149]. In PD patients, the proportion of dopaminergic
neurons with immunoreactive NF-
ZNF564 (zinc finger protein 564) is a transcription factor of unknown function, but it was identified among potential core genes in PD by bioinformatics analysis [152].
NAP1L1 (nucleosome assembly protein 1 like 1) has been identified as a molecular marker of early PD based on gene expression in blood [153]. NAP1L1 has been seems to control the proliferation and differentiation of embryonic neural progenitor cells in the developing brain [154].
GIT2 (GIT ArfGAP 2) acts as a key protein in the aging process and is an ARF-GAP (ADP-ribosylation factor GTPase-activating protein) that controls both DNA repair and glucose metabolism [155, 156]. GIT1/2 proteins were originally identified as regulators of GPCR internalization through their influence on ARF-GTP-binding proteins, thereby inactivating all subtypes of ARF proteins. GIT2 potentially plays a multidimensional role in coupling neuronal and energy-regulatory functions in aging. GIT2-KO mice show anxiety-like behavior and advanced aging. GIT2 appears to protect against oxidative stress, metabolic dysregulation, and inflammation, and appears to promote DNA repair [155].
ZBTB37 (zinc finger and BTB domain containing 37) belongs to a family of 49 transcription factors that are sequence-specific repressors of gene expression that play an essential role in the development of the immune system [157]. ZBTB37 is not yet characterized.
INO80D (INO80 complex subunit D) is an undefined subunit of the INO80-nucleosome complex; an INO80 complex/chromatin remodeler is responsible for nucleosome sliding, histone exchange, and nucleosome spacing [158]. The long noncoding RNA CR933609 acts as a decoy to protect the INO80D gene from downregulation by miRNA-5096 [159].
SERTAD3 (SERTA domain containing 3) is an interferon-inducible type I antiviral protein that disrupts the formation of the RNA-dependent RNA polymerase complex and inhibits influenza A virus replication [160]. SERTAD3 also promotes the growth, invasion, and migration of prostate cancer cells through the p38-p53-p21 pathway [161].
PHTF2 (putative homeodomain transcription factor 2) may be an immune-related gene; PHTF2 is correlated with immune cell infiltration in cancer [162], but on the other hand, it significantly enriches the fatty acid metabolism pathway in gastric cancer [163].
ZNF577 (zinc finger protein 577) methylation levels in leukocytes from women with breast cancer is modulated by adiposity, menopausal state, and the Mediterranean diet [164].
LPP (LIM domain containing preferred translocation partner in lipoma) is a
proto-oncogene that belongs to the zyxin family of LIM domain proteins that
mediate actin cytoskeleton remodeling and migration. LPP interacts with
POU5F1B (POU class 5 homeobox 1B), also known as OCT4 pseudogene 1 or POU5F1P4, is located near MYC on human chromosome 8q24. POU5F1B, like the transcription factor POU5F1/OCT4, can control the pluripotency and self-renewal capacity of mammalian stem cells [167]. One Oct4 pseudogene transcript in mouse neural stem cells, mOct4pg9, promotes the loss of neural stem cell identity and mediates the inhibitory effects of ethanol on development [167].
AMY1A (amylase alpha 1A), AMY1B, AMY1C are
RNF17/TDRD4 (ring finger protein 17/Tudor domain containing 4) contains both a RING finger and a Tudor domain and is part of the mammalian germ cell nuage (characteristic cytoplasmic ribonucleoprotein bodies) essential for spermatogenesis [168]. PIWI-interacting RNAs (piRNAs) have a role in the regulation of transposable elements, especially in spermatogenesis [168]. In the piwi pathway, RNF17/TDRD4 represses the ping-pong cycle, an adaptive amplification loop that generates secondary piRNAs and blocks piRNA responses against protein-coding genes [168]. In addition, RNF17 interacts with all four Mad proteins, enhances c-MYC function, and co-regulates some target genes common to glucocorticoids [169]. The ‘survival motor neuron’ (SMN) protein has been implicated in the formation of membraneless organelles/bodies via its dimethylarginine (DMA)-binding Tudor domain [170]. RNF17/TDRD4 has two Tudor domains that bind DMA and two Tudor domains that do not bind DMA. The inherent multivalency of RNF17/TDRD4 implicates a germinal condensate formation mechanism [170].
TDRD1 (Tudor domain containing 1) belongs to Tudor proteins that contain multiple Tudor domain repeats and form an evolutionarily conserved class of germinal nuage, a characteristic cytoplasmic ribonucleoprotein complex/germinal granule [171]. TDRD1 is required for efficient piwi pathway activity and proper nuage formation [172]. It helps regulate the entry of transcripts into piRNA biogenesis [173]. TDRD1 protein is expressed in most human prostate tumors but not in normal prostate tissue [174].
PIAS1 (protein inhibitor of activated STAT 1) can block the DNA-binding activity
of STAT1 and is a critical regulator of innate immune responses mediated by
IFN-
MUS81 (MUS81 structure-specific endonuclease subunit) binds EME1 and generates a structure-specific endonuclease involved in the rescue of stalled replication forks. MUS81 expression appears to be important in maintaining genomic integrity. MUS81-KO mice show susceptibility to spontaneous chromosomal damage [177].
MAMLD1 (mastermind like domain containing 1) on the X chromosome is one of the causative genes for 46, XY sex differences/disorders; it acts as a transcriptional coactivator and enhances testosterone production in Leydig cells [178]. Using transcriptome data from PD patients, MAMLD1 was identified as gene responsive for Braak stages in PD [179].
NOL4 (nuclear protein 4) is a transcriptional cofactor that regulates LCOR [180]. LCOR is a major transcriptional activator of APM genes (e.g., MHC, major histocompatibility complex class I) that binds to IFN-stimulated response elements (ISREs) in an IFN signaling-independent manner, mediating interferon-independent immunogenicity [181]. LCOR is one of the most differentially expressed genes in PD [182].
SYNE2 (spectrin repeat containing nuclear envelope protein 2)/nesprin-2 is a KASH domain-containing protein critical for nuclear-envelope integrity [183]. Vertebrate skeletal-muscle fibers contain hundreds of nuclei, three to six of which are functionally specialized and stably anchored under the postsynaptic membrane at the neuromuscular junction. SYNE1 and SYNE2 play critical roles in the anchoring of both synaptic and non-synaptic myonuclei, which are important for proper motor neuron innervation and respiration [184].
CTCFL/BORIS (CCCTC-binding factor like/brother of regulator of imprinted sites) is a transcription factor associated with microglia-exclusive enhancer regions [185] and an RBP that is associated with polysomes and active translation in both neural stem cells and young neurons [186]. As a transcription factor, CTCFL regulates membrane receptors that have AKT as a downstream target, the PI3K-AKT pathway [187].
As mentioned in the introduction,
As discussed above, in PD individuals,
mTORC2-SGK1 signaling integrates external signals in order to regulate
autophagic turnover of mitochondria through ROS; loss of RICTOR induces
mitochondrial ROS and leads to mitophagy [79]. It is interesting that FXR1 and
OXR1, which protect against oxidative stress-induced neurodegeneration [129, 130],
are repressed by
In PD, impaired autophagy significantly increases
Intracellular accumulation of
Nrf2 (NFE2L2) is a key redox-regulated transcription factor. Its activation
leads to the upregulation of cytoprotective and antioxidant enzymes/proteins
[191]. GSK3
Inhibition of PARP1 energy-exhaustion-induced apoptosis is considered a relevant
approach for the treatment of PD [192]. LBs post-transcriptionally over-promote
GIT2 (Fig. 6, group 3), the positive regulator of poly (ADP-ribose)-polymerase-1
(PARP1). GIT2 appears to control multiple aspects of the complex ageing process
and PARP1-mediated DNA repair capability contributes to mammalian longevity
[155]. PARP1 catalyzes the covalent attachment of poly (ADP-ribose) (PAR) on
acceptor proteins (PARylation), which affects DNA repair, chromatin remodeling,
and gene transcription [192]. PARP1 protects neurons from cell death under mild
oxidative stress as a result of DNA repair, but an excessive activation of PARP1
leads to mitochondria-related energy-exhaustion-induced apoptosis [192].
Intracellular accumulation of
DAZL (deleted in azoospermia like) is post-transcriptionally over-promoted by
LBs (Fig. 5, group 8). Like
As mentioned above, the neuronal protector of the neuronal synapse against
microglial engulfment, SRPX2 (blocking the classical C1q-mediated complement
cascade), is post-transcriptionally over-repressed in “infected” DA-neurons.
Consequently, increased LB-phagocytosis may lead to the accumulation of these
degradation-resistant
In all meta-analyses of cerebrospinal fluid (CSF), the evidence suggests that
soluble and total
According to the PD model presented here,
The present bioinformatics study showed that
In
Although the presented concept will need to be verified in vivo, this bioinformatic study revealed potentially “groundbreaking” findings that will, I hope, initiate the necessary experimental studies.
The datasets generated and/or analysed during this study are listed in the Supplementary Material, further information is available from the corresponding author.
The single author had the sole role in designing, data collecting, analyzing, and writing the manuscript.
Not applicable.
I kindly acknowledge the Centre of Excellence for White-Green Biotechnology (ITMS26220120054), supported by the Research and Development Operational Programme funded by the ERDF.
This research was funded by Vedecká grantová agentúra MŠVVaŠ SR a SAV (VEGA), grant number 2/0064/22.
The author declares no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
