1 Institute of Pharmaceutical Biology, Department of Biochemistry, Chemistry & Pharmacy, Goethe-University, 60438 Frankfurt am Main, Germany
Abstract
Since the end of 2019, the SARS-CoV-2 virus started to spread in different countries, leading to a world-wide pandemia, with today’s infection numbers of more than 690 million and with a case fatality rate of more than 6.9 million. In addition, about 65 million patients suffer from post/long-Covid syndromes after having infections with the SARS-CoV-2 virus or variants thereof. This review highlights the biology of the virus, summarizes our knowledge of some of the viral mechanisms that counteract our immune responses, and finally also discusses the different vaccines and their specific safety profiles. Also, the possibility to fight this virus with recently available drugs (Veklury, Lagevrio and Paxlovid) will be discussed. All these data clearly argue that SARS-CoV-2 variants still exhibit a dangerous potential—although with a lower case fatality rate—and that vaccination in combination with drug intake upon infection may help to lower the risk of developing chronic or temporary autoimmune diseases.
Keywords
- SARS-CoV-2
- interferon pathway
- immunology
- vaccines
- safety issues
- small molecules
The SARS-CoV-2 virus belongs to the a/ß-Corona family of viruses (n = 7) that are associated with typical respiratory infections. While 4 out of these 7 viruses cause harmless seasonal infections, the remaining 3 have a much higher lethal potential; these were SARS-CoV-1, MERS-CoV and the recently observed SARS-CoV-2 virus. SARS-CoV-1 appeared first in China in Nov 2002 and led to the infection of nearly 8100 human beings of which 774 died of their infection (lethality 9.6%). MERS-CoV appeared about 10 years later and was transmitted from camels to humans. Infections spread all over the world over a period of nearly 6 years, with 2143 infected patients of which 750 died (lethality 34.9%). Finally, SARS-CoV-2 appeared at a market in Wuhan (China) in 2019, spread rapidly around the world with more than 690.7 million infected humans today of which 6.89 million have died so far (lethality is ~1% world-wide) [1] (data retrieved 6.23.2023). This large number of infected people (nearly 8.5% of the world population) also led to ~10% of infected/recovered patients acquiring “Long-Covid” symptoms. Long-Covid is a synonym for a multitude of organ-specific autoimmune diseases which differ between different patients [2]. Long-Covid symptoms may encompass, e.g., cardiovascular, thromboembolic and cerebrovascular disease [3], type 2 diabetes [4] and myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) [5]. Symptoms may last for longer than a year [6], and cases with ME/CFS are expected to be lifelong [7].
Thus, we have to realize that this zoonotic virus—although described as a “respiratory virus”—has the potential to increase dramatically the prevalence to develop autoimmune diseases. Moreover, since the necessary angiotensin-converting enzyme 2 (ACE2) receptor is expressed on endothelial cells, the blood circulation system is a major target for viral infections, resulting in an increase of thromboembolic events. ACE2 is also expressed in the duodenum and small intestine (mucus producing cells), gall bladder, kidney, testis and heart muscle, explaining the various symptoms observed in infected and post-infected patients. Most symptoms are related to inflammatory conditions and tissue destructive processes in these organs. In addition, cells of our innate and adaptive immune system, such as mast cells, neutrophils and naive B-cells also express the ACE2 surface receptor (data retrieved from [8]) and may change their normal immunological activity after infection.
In 2020, when no vaccine was available, the situation was dramatic in several
countries. The mortality between different age groups differed significantly,
with a lethality of nearly zero in small children (0.0221%), 4.2% in adult
between 50–60 years and up to 40% in elderly patients (
This paper describes aspects of the biology of this virus, which in part explain the causal reasons for the onset and large heterogeneity of Long-Covid symptoms after real infections. Long-Covid describes the consequences of SARS-CoV-2 infection which are caused by inflammatory conditions and additional immunological mechanisms that all result in the onset of autoimmune diseases. These autoimmune reactions may affect all inner organs, blood vessels and nerve cells [2, 3]. This is caused by certain SARS-CoV-2 proteins that inhibit immunological defense mechanisms, and by the 2 proteases which generate cellular neoantigens. The latter process results in the production of auto-antibodies which are the major cause underlying the development of Long-Covid symptoms. Moreover, we propose Omicron-associated mechanisms explaining the immune suppression sometimes seen in patients following multiple infections. These result from observations made early in 2023 that some patients display either a prolonged infection time (bacterial or viral) or recurrent infections. In addition, our current knowledge about the mRNA and vector-based vaccines will be presented, along with their safety profiles. Finally, this article reflects on the available drugs that can be used to combat SARS-CoV-2 infections and which in part are able to avoid the development of Long-Covid symptoms. Alternatively, vaccination with the available bivalent mRNA vaccines against the Omicron-variant are also a protective measure against the development of Long-Covid, with a risk reduction of ~40% [11].
The original Wuhan strain of SARS-CoV-2 was early on described as a zoonotic virus which clusters together with the RaTG13 bat strain isolated from Rhinolophus, suggesting that RaTg13 could be the ancestral strain that circulated in bats long before the introduction of SARS-CoV-2 into the human population [12, 13]. However, some scientific opinions raise questions about origin of the SARS-CoV-2 virus, based on the presence of the Furin cleavage site in the Spike protein, which is missing in all natural ß-Corona virus variants [14], or the distribution of specific restriction recognition sites [15], a fact which has already been disproved by others [16, 17]. This is an ongoing debate, however, with more arguments on the side of a zoonotic disease spill-over into human beings and other mammals, rather than a leak of this virus from a research laboratory in Wuhan.
SARS-CoV-2 virus exhibits a ~30.000 nucleotide long RNA genome
[18] that encodes large polyproteins that are linked via a frameshift to produce
16 non-structural proteins (NSPs), including 2 proteases, a helicase (Hel) and an
RNA-dependent RNA Polymerase (RdRP). The 3
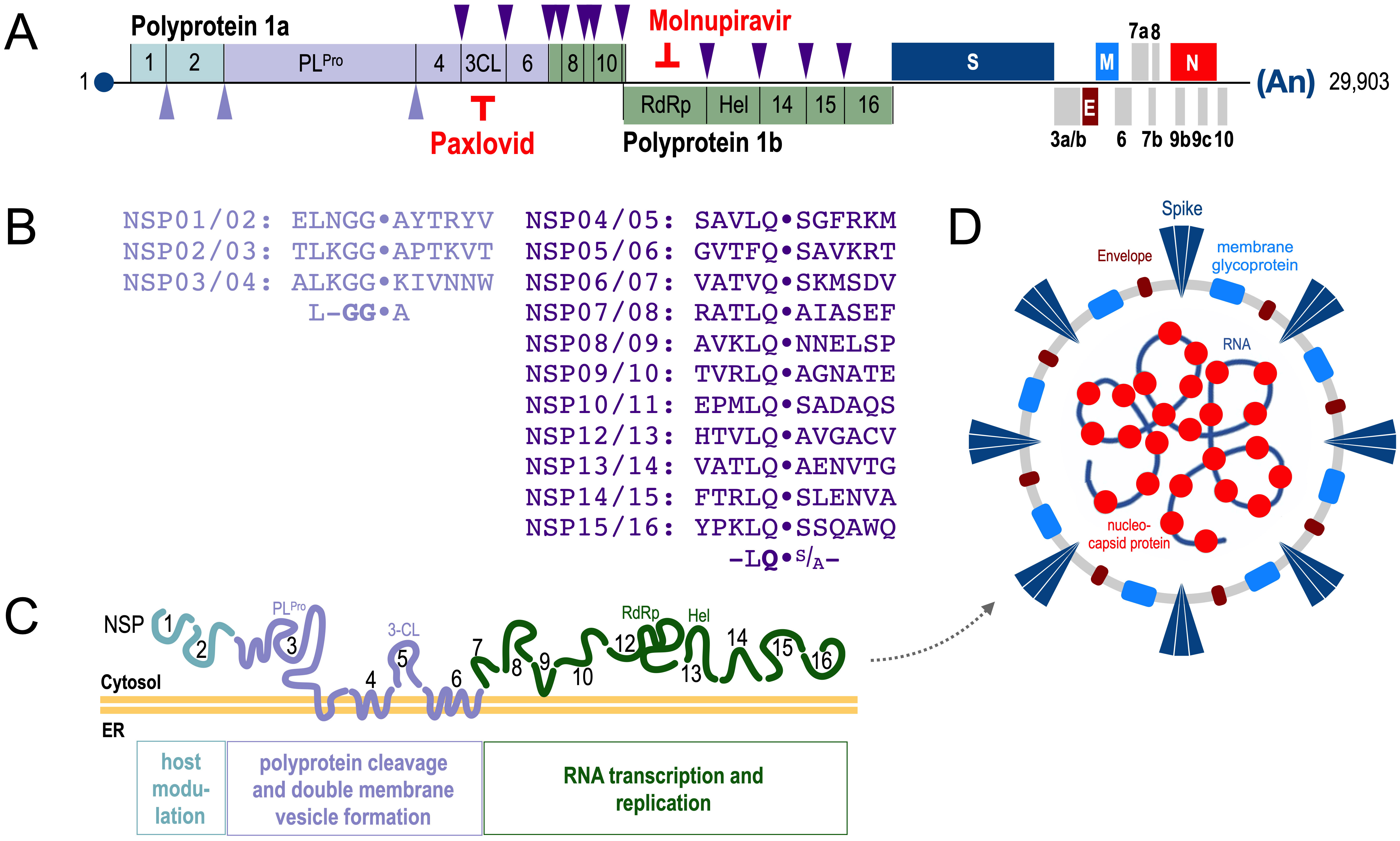 Fig. 1.
Fig. 1.Structure of the SARS-CoV-2 genome. (A,B) Cleavage
specificity of the two proteases PL
For some of these viral proteins a cellular function in infected cells is
already known. About 10 NSPs/orfs are involved in inhibiting the interferon
pathway [19], the two proteases (PL
One of the major dangers associated with SARS-CoV-2 infections is the fact that this virus encodes 2 viral proteases which appear to generate a massive pool of protein fragments (neoantigens) from host cellular proteins. This switches on a vicious pathway to steer autoimmune reactions [26], which is of course dependent on the affected host and their capacity to present such aberrant peptides to their immune cells. As a consequence, affected individuals experience autoimmune reactions against their own body organs. Such autoimmune reactions are then collectively termed “Long-” or “Post-Covid”, depending on how long these post-infection symptoms persist [11].
An important biological feature of SARS-CoV-2 is the final part of its replication cycle, namely how new virus particles are released from infected cells. As already mentioned above [20], the SARS-CoV-2 virus is assembled in the cytosol but leaves the infected cells by a rather unexpected route. Cell-internal lysosomes, which are usually rather important for the destruction of foreign or host proteins in acidic (pH 5) conditions, are hijacked for the egress of the virus. One of the reading frames of SARS-CoV-2, the orf3a protein, encodes a protein that inhibits the acidification of lysosomes. Therefore, the virus uses these de-acidified lysosomes as a shelter for newly produced viral particles, which are then released when these de-acidified lysosomes fuse with the outer membrane. This process is termed “lysosomal egress pathway” or “lysosomal exocytosis” (see Fig. 2, Ref. [20]). Interestingly, this pathway is dependent on RAB7 and ARL8B. Noteworthy, at least for RAB7 potential inhibitors are available [27].
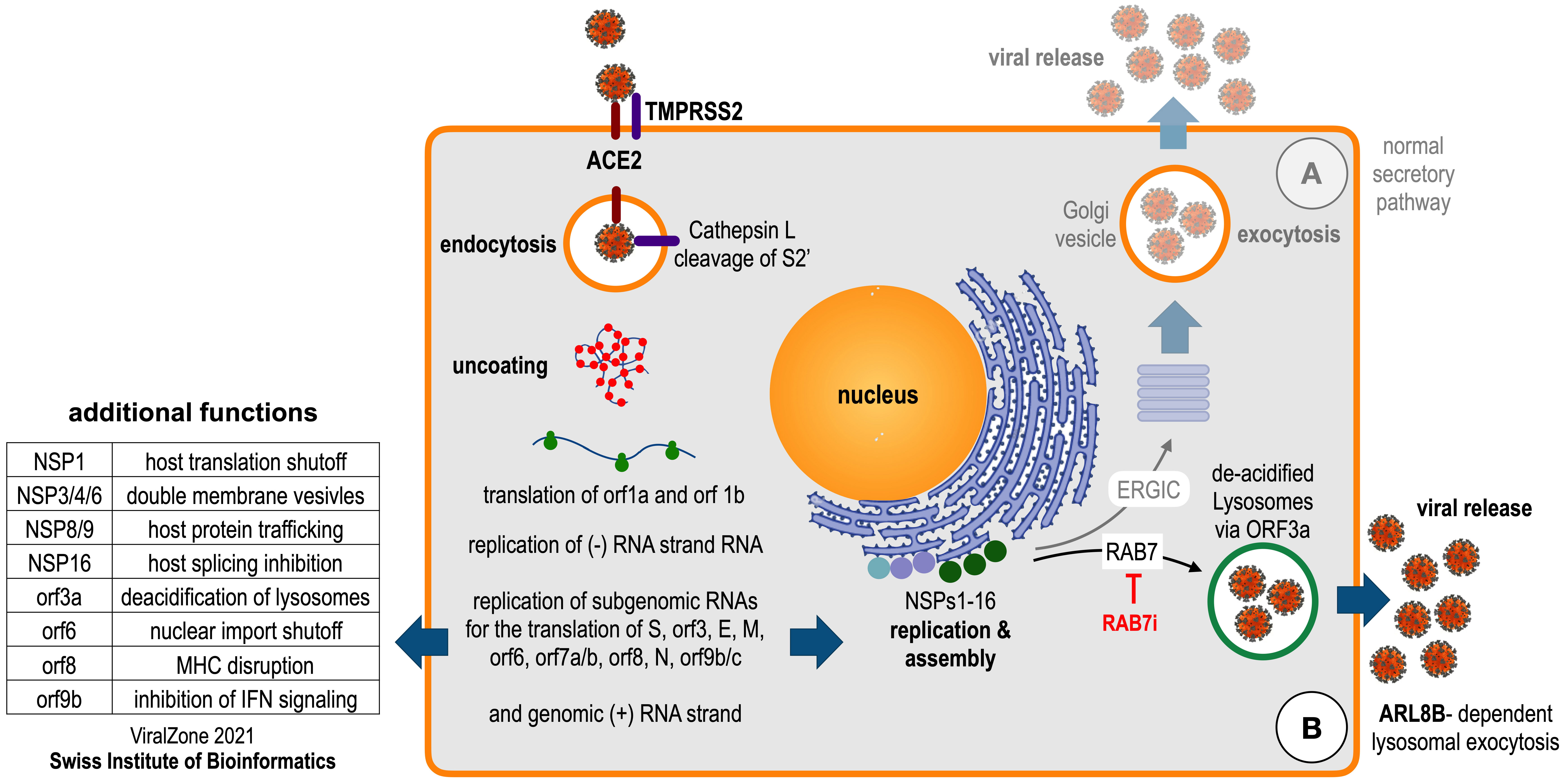 Fig. 2.
Fig. 2.The life cycle of SARS-CoV-2. Endocytosis of angiotensin-converting enzyme 2 (ACE2) receptor-attached SARS-CoV-2 virus and Furin cleavage of Spike protein by TMPRSS2; uncoating and translation leads to the assembly of replication machinery at the ER after translation of orf1a and orf1b, replication of subgenomic RNA leads to all necessary proteins. (A) Expected exocytosis for Virus release. (B) Observed Virus release via the lysosomal egress pathway. The latter pathway depends on the Ras-related protein Rab-7a (RAB7; can be inhibited by the drug CID1067700) and on the ADP Ribosylation Factor Like GTPase 8B (ARL8B). Additional functions known for SARS-CoV-2 proteins are indicated on the left (information retrived from the ViralZone 2021). The figure has been adapted from ref. [20].
This is important, because this trick bears a great danger for the infected
host. Lysosomes are urgently needed in Antigen-presenting cells (APCs) after
phagocytosing the virus or for B-cells after endocytosing surface antibody-bound
virus. The resulting membrane vesicles (phagosome, endosome)—containing the
infectious viruses—are then acidified to mature into lysosomes where all the
internal protein material becomes hydrolyzed into peptides. These peptides are
then used to load major histocompatibility complex (MHC) II complexes to present
the foreign peptides to T-helper (T
The augmented levels of TNF
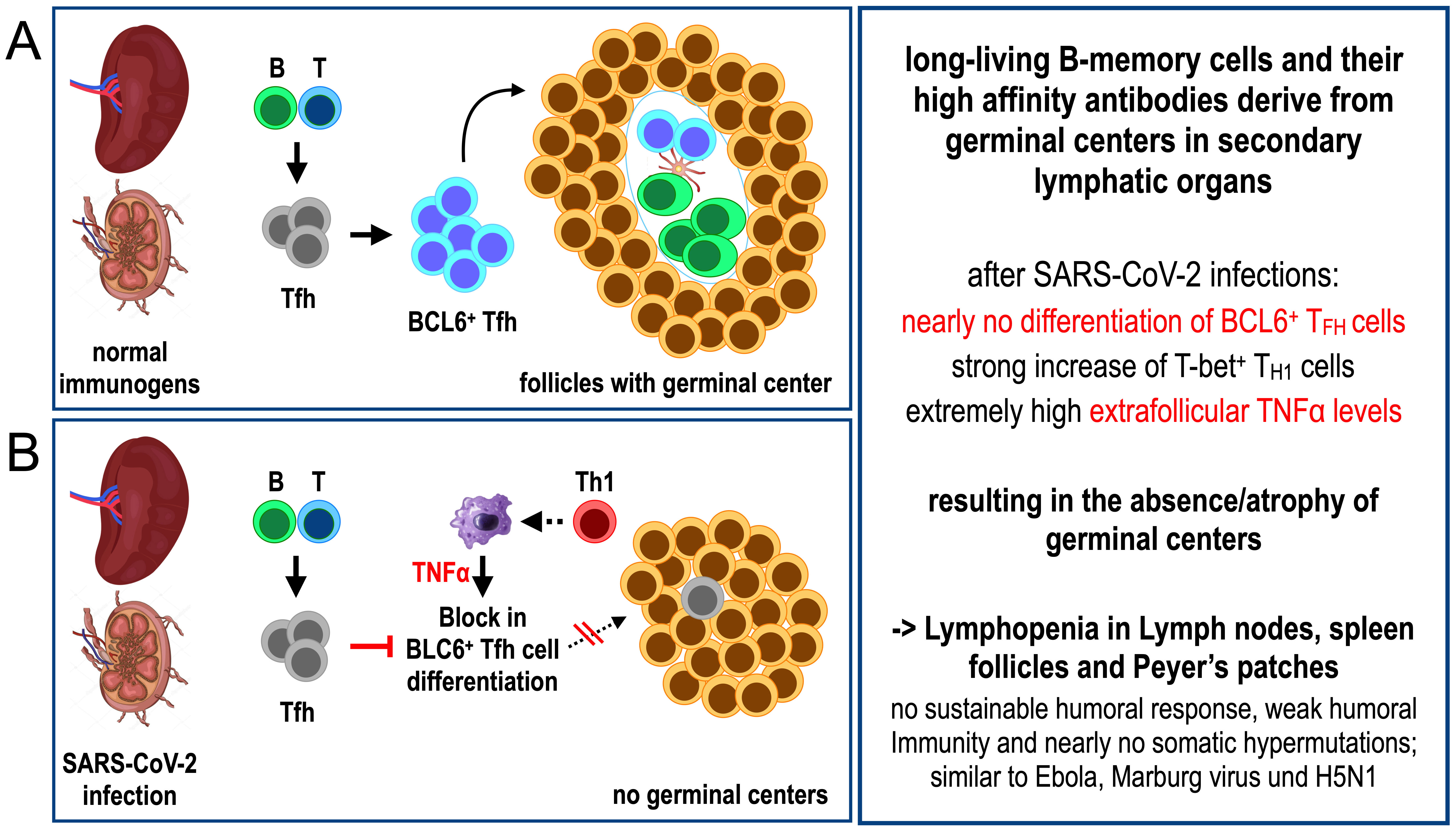 Fig. 3.
Fig. 3.Adaptive immune response under normal (A) and hyperinflammatory
condition (B) in secondary lymphatic organs (spleen or lymph nodes).
(A) Immune response under physiological
conditions. B-cells become activated in germinal centers by activated
follicular T-helper cells (BCL6
A similar immune suppressive mechanism has been recently discovered, namely a suppression of MHC I expression in SARS-CoV-2 infected cells [29]. Several SARS-CoV-2 VOCs (variants of concern) and Omicron variants have been tested using in vitro and in vivo infection models. They have identified the ability of orf8, in particular the mutant form of orf8 present in Omicron variants, to suppress MHC I expression in infected cells, thus impairing the cytotoxic T-cell response. This is probably due to an additional T91I mutation of the E protein which is common to all Omicron variants. The product of orf8 can be secreted into the cell culture medium, and its inhibitory capacity for type I interferon stimulated interferon-stimulated genes (ISG) expression or NFkB signaling has already been demonstrated [30], but conversely, also the induction of pro-inflammatory cytokines such as IL17 [31]. Thus, also the cytotoxic T-cell response could be impaired and inflammatory conditions augmented.
A very good overview on the diverse functions of SARS-CoV-2 proteins has been published already in 2020 [32]. Intracellular interactions with host proteins have been investigated using modern mass-spectrometry. These data have also provided potential drug targets for the treatment of SARS-CoV-2. As shown in Fig. 4 (Ref. [32]), most of the 28 SARS-CoV-2 encoded proteins are able to bind to multiple host factors, and thus, may have the potential to interfere with several cellular pathways. All this cellular interference helps the virus in its own replication and affects processes such as lipid and sugar metabolism, protein stability, vesicle trafficking, mitochondrial functions and control mechanisms for inflammatory and interferon responses.
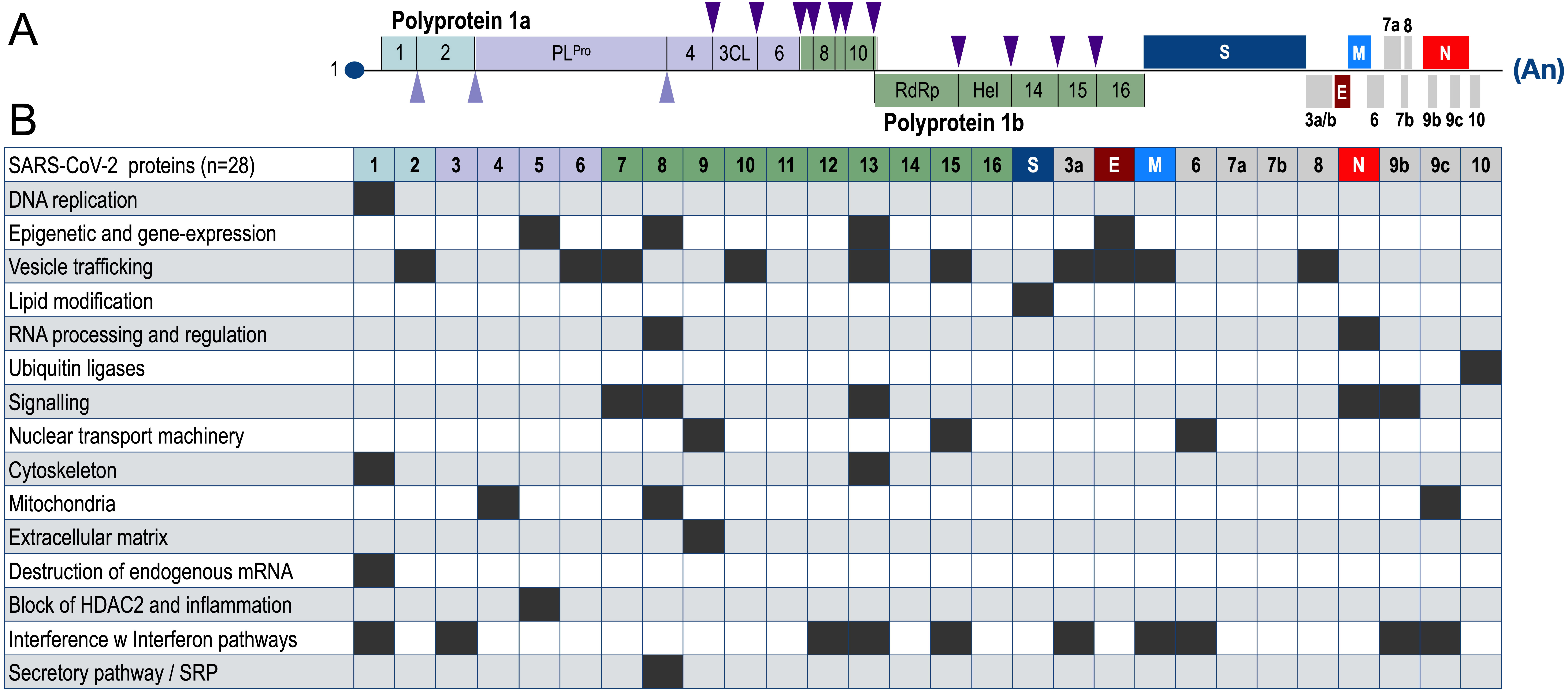 Fig. 4.
Fig. 4.Summary on protein-protein interactions of SARS-CoV-2-encoded proteins with host proteins and pathways. (A) structure of SARS-CoV-2 with a total of 28 proteins. (B) Summary of all the identified protein interactions between viral proteins and host proteins of major cellular pathways. Black boxes are indicating that a specific SARS-CoV-2 protein interferes with the highlighted pathway of the infected host cell. Noteworthy, about 10 viral proteins (NSP1, NSP3, NSP12, NSP13, NSP15, orf3a, M, orf9b and orf9c) are interfering with the interferon pathway. This figure has been adapted from ref. [32].
In particular, the potent inhibition of the interferon response pathway could
explain severe cases of COVID-19. SARS-CoV-2 infections turn on the Interferon
pathway via the RIGI/TLR3/IRF7 axis, leading to a profound type I/III interferon
response to block viral replication by specific defense proteins (different IFIT
proteins, ISG15 and Viperin). This usually defends against viral infections quite
effectively, because all these proteins recognize viral nucleic acids, covalently
modify viral proteins and target them for proteasomal destruction or synthesise
unusual RNA nucleotides (e.g., 3
Several studies have investigated the role of the interferon response in different COVID-19 patients. Based on single blood cell gene expression profiling, healthy controls can be clearly distinguished from mild, severe or critically infected patients by their gene expression profiles, the poorest interferon response being displayed in severe cases [33]. Moreover, genetic studies revealed the OAS1 gene as a restriction factor against SARS-CoV-2 infections [34, 35]. The p46 variant OAS1 gene (splice variant of the original OAS1 gene, OAS1 isotype) is encoded by a 75 kb piece of DNA located on chromosome 12q24.13. This OAS1 isotype gene variant originally derived from Neanderthals and protects against severe SARS-CoV-2 infections [36, 37, 38]. Patients that bear certain SNPs or display a genetic defect in OAS1 displayed a strongly impaired or absent interferon response [39], while children with such a genetic mutation develop the multi-inflammatory syndrome in children (MIS-C) [40]. Other patients with severe outcomes after infection displayed autoantibodies against the Interferon protein [41].
Most of these poor interferon-responding patients suffered from life-threatening
COVID-19 pneumonia [42] or multi-organ failure [43], which was responsible for
more than 20% of COVID deaths in elderly patients [44]. In general,
autoantibodies against different cytokines seem to correlate with severity of the
disease course [45], validating that these pathways are crucial for viral
defense. In summary, hospitalized patients with a fast
type I/type III interferon
response—accompanied with low levels of pro-inflammatory cytokines (IL1, IL6,
TNF
In order to prevent all the above-mentioned negative side effects of real SARS-CoV-2 infections on our immune system, only two possibilities exist. The first option is to vaccinate populations in order to develop immunological resistance to the virus and to prevent severe courses of disease. The second option is to take drugs that block viral replication which prevents fast spreading of the virus in infected hosts and promotes rapid clearance of the virus and virus-infected cells. The latter will be discussed in a separate chapter below.
Vaccination has been used as a strategy since the 20th century to fight and to prevent several diseases around the world. There are several vaccine developing strategies (attenuated living virus, antigen/adjuvant combinations, LPS-toxin conjugates) that have been successfully used in the past and are currently in use to combat major diseases. Similarly, the SARS-CoV-2 pandemia that arose in the beginning of 2020 required the development of a quick vaccine strategy world-wide. Luckily, two novel strategies were available, namely the adenovirus vector-based and mRNA vaccination technology. These novel strategies were used by different companies (e.g., Biontech/Pfizer: BNT162b2; Moderna: mRNA-1273; AstraZeneca: ChAdOx1 CoV-19; Johnsen/Johnsen: Ad26.COV2.S) and ended with successful clinical trials [47, 48, 49, 50]. These novel vaccine development strategies were in the end much faster than traditional ones. Therefore, I want to focus in this review on these two novel strategies, as both have a great potential for the future.
Adenovector-based vaccines use infectious adenoviral particles (Janssen/Johnson&Johnson: HAd26; Oxford Jenner Institute/AstraZeneca: ChAdOx1, Sputnik: HAd5 and HAd26) with the E1 gene replaced by a codon-optimized cDNA version of the Wuhan SARS-CoV-2 Spike gene. The ChAdOx1-encoded codon-optimized Spike cDNA was flanked by a cytomegalovirus (CMV) Enhancer, an intron-containing CMV Promotor and an BGH poly-A signal sequence. The whole cassette was located 174 bp upstream of the adenoviral pIX reading frame. The HAd26-encoded codon-optimized Spike gene is flanked by a CMV Enhancer, an intron-less CMV Promotor and an SV40 poly-A signal sequence, located 99 bp upstream of the adenoviral pIX reading frame. Both adenoviruses infect human cells and deliver the adenoviral vector DNA to the nucleus, with Spike mRNA transcripts then be produced and delivered to the cytosol of the cells. The mRNA is then translated at the endoplasmatic reticulum (ER) and the resulting Spike trimer is delivered via the ER-Golgi-axis as a membrane-bound, trimeric Spike protein to the outer surface of infected cells. The virus has a maintenance of 2–3 weeks and produces the recombinant Spike proteins over this time-period. One of the problems with this approach is the fact, that SARS-CoV-2 is an RNA virus, with a natural life cycle only in the cytosol of infected cells. Therefore, none of the genes encoded by SARS-CoV-2 has ever been selected or optimized by nature to be expressed in the nucleus of eukaryotic cells. As a consequence, the open reading frame of the Spike gene, coding for a 1274 amino acid long Spike protein, is full of potential splice donor and acceptor signal sequences. Some of them were point-mutated by Janssen, but none of them by AstraZeneca. Therefore, alternative splice events occur, leading to shorter Spike protein variants or even fusion by splicing to the downstream located pIX gene [51, 52]. None of these aberrant Spike protein variants can become a membrane-bound protein anymore, because all of these C-terminally shortened Spike protein variants lose the necessary membrane anchor, and thus, are secreted to the blood stream. Based on available RNA-Seq data (kindly provided by Prof. David A. Matthews, University of Bristol, UK), the amount of these aberrant Spike protein encoding mRNAs was estimated to be in the range of 1.5% of all Spike mRNA transcripts. Although it has not been formally proven, these shorter Spike variants —or the discovered Spike-pIX fusion protein—may be one reason for the observed thrombotic events (sinus vein thrombosis), which were observed shortly after vaccination with vector-based vaccines (see below).
Because of the associated adverse events, the Adenovirus-based vaccines were used only in the beginning of the vaccination campaign, but not used after summer 2021 for primary vaccinations in Germany. Therefore, the number of vaccinations with vector-based vaccines is quite low when compared to mRNA-based vaccines.
By contrast, mRNA vaccines used a different strategy. In vitro
transcribed mRNA which was appropriately capped and poly-adenylated is packaged
into lipid-nanoparticles that contain specific lipid compositions of PEG-DMG,
DODMA, DSPC and Cholesterol. This particular composition guarantees the fusion of
these nanoparticles with target cells upon intramuscular injection. The mRNA was
synthesized with N
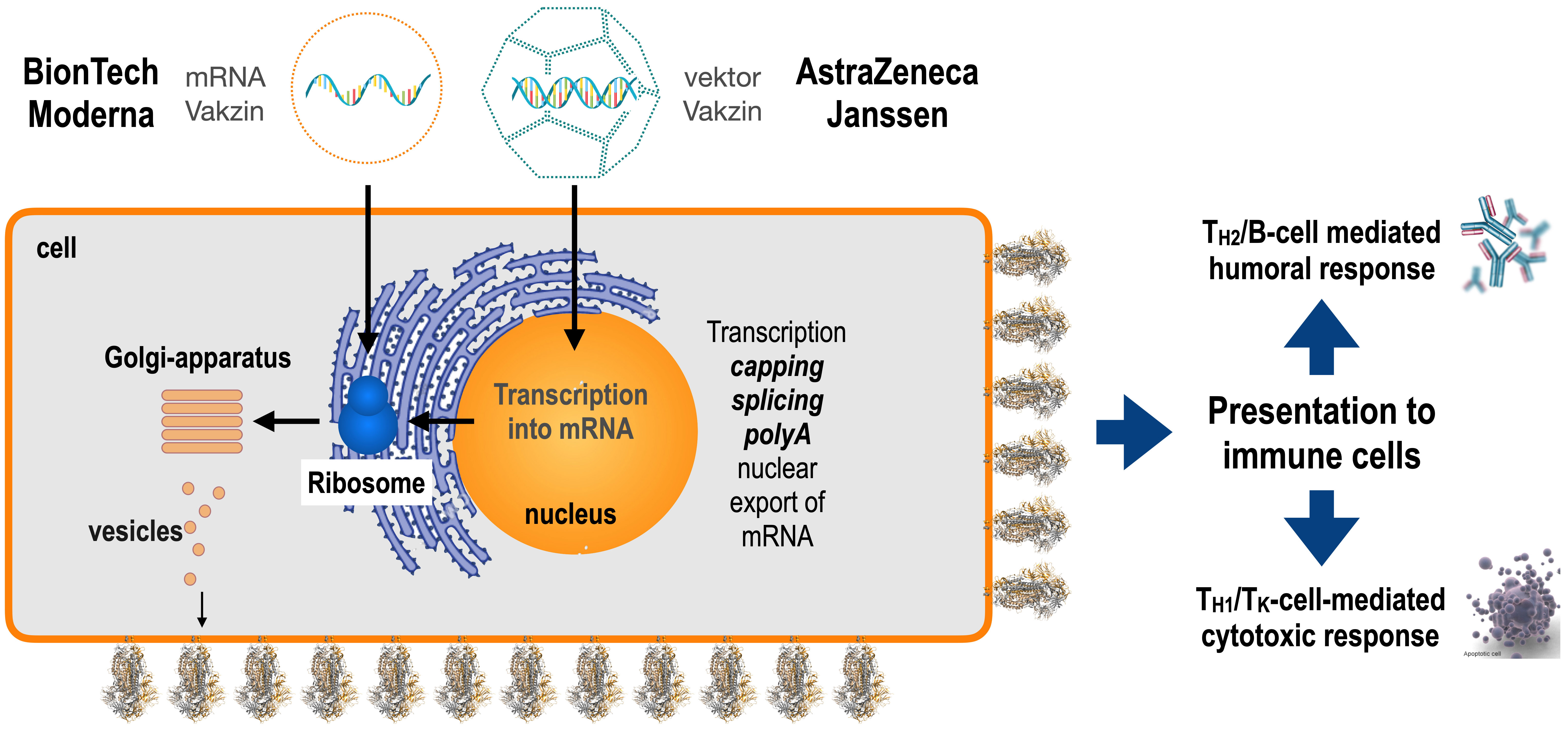 Fig. 5.
Fig. 5.Delivery of vaccines to target cells. mRNA-based vaccines deliver a perfect mRNA to the cytosol where it could be immediately translated into Spike protein at the ER, become post-translationally glycosylated and then exported as a trimeric, receptor-like protein via the Golgi system to the outer membrane of the producing cell (secretory pathway). Vector-based vaccines are delivered first to the nucleus where they are transcribed into Spike mRNA and then exported from the nucleus to the cytosol. As already mentioned above, the Spike protein is then translated at the ER, post-translationally glycosylated inside the ER and exported via the Golgi system to the outer membrane. This allows stimulation via antigen-presenting cells of corresponding T-helper and B-cells to initiate the production of cognate antibodies. Since peptides of the foreign Spike protein are also displayed via MHC I at the outer cell surface, appropriate T-killer cells are activated as well. Thus, these novel classes of vaccines allow a profound immune stimulation without the need of any adjuvant.
Both approaches were game changers for the field of vaccine development. In the
past, vaccine development was either based on attenuated living viruses for
life-long protection, or on the use of portions of viruses or bacteria
(recombinant) combined with adjuvant for short- to midterm immune protection.
Vector- or mRNA-based vaccines both deliver a nucleotide-encoded antigen into
human cells. By using the cell as a production unit for the protein antigen, a
profound stimulation of the humoral (T
The Federal Paul-Ehrlich Institute (PEI in Langen, Germany) has the important task to constantly evaluate the benefits or risks of vaccines on the market. By law, all physicians and companies are obliged to report all cases with negative side effects to this institution. Therefore, the PEI has a large data resource to monitor side-effects and efficacy of vaccines. Based on the last report in 2021 (23.12.21; about 1 year after the vaccination campaign started in Germany), all 4 vaccines were analyzed for their potential danger. This is summarized in Table 1 where total and adjusted numbers (cases per million injections in red) are summarized for the observed vaccine-associated complications. The 4 investigated vaccines differed quite dramatically in their usage. The Biontech mRNA vaccines was used for 96.6 million, the Moderna mRNA vaccine for 10.5 million. The Astrazeneca vector-based vaccine for 12.7 million, and finally, the Janssen vector-based vaccine for 3.4 million injections. When looking at the red marked lines where all side-effects are shown in numbers per million injections, it becomes quickly clear that the normal vaccine-related side effects (headache, pain at the injection site, fatigue, fever, etc.) were lowest for the Biontech (B) product, a little more for the Moderna (M) product, highest for the AstraZeneca (AZ) vaccine and again lower for the Janssen (J) product. The reason for the increased vaccine-related side effects with the AZ product could be explained by the identified impurities in several batches of the vaccine which may explain the increased inflammatory reactions [53]. Anaphylactic reactions were rare and between 3–8 cases per million injections. Noteworthy, myo- or pericarditis cases were observed only with mRNA vaccines, and were more than doubled in the M product when compared to the B product (50 µg vs. 30 µg mRNA). Instead, cases with Thrombosis with Thrombocytopenia Syndrome (TTS)—including the rare sinus vein thromboses—mainly occurred with the AZ product, were less frequent with the J product and nearly absent for both mRNA vaccines. Normal Thrombosis with immune thrombocytopenia (ITP) were again increased for the AZ product, while much less for the J, B and M products. Finally, cases with Guillain-Barré-Syndrom (GBS) were slightly elevated for the AZ and J products, while nearly absent for both mRNA vaccines. Finally, the total number of deaths was low for the B, M and J products, while strongly increased for the AZ product. This was the main reason for not using the AZ product later in the year 2021, and to switch the population-wide vaccination campaign completely to mRNA vaccines. In order to evaluate the data even further, the TTS incidence is ~3 for 1 million inhabitants in Europe per year, underlining again the security of mRNA vaccines with 0.4 and 0.5 cases after 1 year of mRNA vaccinations. The ITP incidence is ~20–40 for 1 million inhabitants in Europe per year, indicating that all 4 four vaccines had lower incidences. Also, the GBS incidence is between 8–19 for 1 million inhabitants in Europe per year, indicating that only the AZ vaccine was lying exactly in this range, while all other vaccines were below this number. Thus, based on this analysis, mRNA vaccines are very safe, apart from the observed risk for myo- or pericarditic events, mostly observed in young men between 25 and 30 years of age [54]. This also supports the notion that additional vaccinations with the now available bivalent vaccines from B and M should not only protect against the Omicron-variant but could also help to lower the number of Long-Covid patients.
| Companies | Biontech/Pfizer | Moderna | Oxford/AZ | Janssen/J&J | ||
| Total vaccinations | 96,606,131 | 10,576,131 | 12,703,030 | 3,462,557 | ||
| complications | 113,792 | 28,289 | 46,325 | 7,758 | ||
| cases/Million injections | 1178 | 2675 | 3647 | 2241 | ||
| anaphyllaxia | 550 | 55 | 101 | 10 | ||
| cases/Million injections | 6 | 5 | 8 | 3 | ||
| myo/pericarditis | 1245 | 309 | 0 | 0 | ||
| cases/Million injections | 13 | 29 | 0 | 0 | ||
| TTS* | 36 | 5 | 200 | 24 | ||
| cases/Million injections | 0.4 | 0.5 | 15.7 | 6.9 | ||
| ITP** | 314 | 28 | 269 | 23 | ||
| cases/Million injections | 3 | 3 | 21 | 7 | ||
| GBS*** | 140 | 14 | 112 | 48 | ||
| cases/Million injections | 1 | 1 | 9 | 14 | ||
| death cases | 295 | 20 | 201 | 21 | ||
| cases/Million injections | 3 | 2 | 16 | 6 | ||
* TTS-Incidence in Germany: 3 diagnoses/ Million individuals per year.
** ITP-Incidence in Germany: 20–40 diagnoses/ Million individuals per year.
*** Guillain-Barré-Syndrome Incidence in Europa: 8–19 diagnoses/ Million individuals per year.
The displayed data were retrieved from the security report on the vaccination campaign in Germany, dated December 12 2021 of the Paul-Ehrlich Institute in Langen, Germany. It clearly shows that mRNA vaccines bear an elevated risk for heart muscle inflammation, while vector-based vaccines bear the increased risk for thrombotic events, Guillain-Barré-Syndrom (GBS) and vaccination-related death.
During the pandemia, many hopes have been set on drugs that could potentially be beneficial against SARS-CoV-2 infections. The use of bleach (HOCl), Hydroxychloroquine, Vitamin D or antihelminthic drugs developed for horses (Ivermectin) had no benefit in the treatment of COVID-19 patients. By contrast, the use of Dexamethasone was quite beneficial, as it rapidly counteracts hyperinflammatory conditions which was particularly helpful when hospitalized patients were at risk of severe tissue or organ damage during their initial inflammatory episodes. Also, the use of recombinant monoclonal antibodies (cocktails) such as Casirivimab/Imdevimab (Ronapreve®; Roche/Regeneron), Regdanvimab (Regkirona®; Celltrion), Tixagevimab/Cilgavimab (Evusheld®; AstraZeneca) or Sotrovimab (Xevudy®; GSK/Vir Biotechnology) prevented severe courses in 2020/21 and certainly saved lives of high-risk patients. However, many these recombinant antibody cocktails became ineffective when the Omicron-variant appeared in 2022, except Sotrovimab which still appeared to have neutralizing capacity against this novel virus variant.
I also want to place the focus on some oral drugs which were introduced for the treatment of COVID-19 patients: Remdesivir (Veklury), Molnupiravir (Lagevrio) and the combination of Normatrelvir and Ritonavir (Paxlovid). Remdesivir and Molnupiravir are both nucleotide analogues that should in principle hinder the RNA dependent RNA polymerase of SARS-CoV-2 (NSP12) to replicate efficiently the viral genome and to produce new viral particles. However, several studies have shown that the use of these drugs often increases the probability of generating new virus mutants [55, 56, 57, 58], and thus, their use should be carefully restricted for treatment. By contrast, Paxlovid, a mixture of Normatrelvir and Ritonavir, effectively blocks the activity of the main protease 3-CL of SARS-CoV-2 and has shown clear-cut benefits in clinical trials with COVID-19 patients [59, 60, 61, 62]. The 3-CL protease is absolutely necessary for virus replication, but the half-life of the specific protease inhibitor is too low. Therefore, it has been combined with Ritonavir, a well-known drug for the treatment of HIV-1 patients. Ritonavir is by itself also a protease inhibitor, but the main function is the inhibition of the liver enzymes CYP3A4 and CYP2D6 which are responsible for metabolizing Normatrelvir. Thus, the combination of both drugs enhances the pharmacodynamic behavior of Normatrelvir and allows the use of lower doses of both inhibitors. The only reason why it is not so easy to use Paxlovid is that inhibition of CYP2A4 also has dramatic effects for many other drugs (roughly one third of all available drugs which are today on the market). SARS-CoV-2 infected individuals need to take Paxlovid for 5 days in a perfect 12 h rhythm to effectively kill all SARS-CoV-2 viruses in their body, but if these patients are already taking several other drugs on a daily basis, then their treatment requires careful medical evaluation. The list of drugs that are dependent on CYP3A4 (either for drug destruction or activation) is rather long, and therefore, many physicians hesitate to prescribe Paxlovid, although it is as yet the only effective and available drug against SARS-CoV-2 virus [63]. However, the effectiveness of Paxlovid has been shown in a retrospective clinical study of the California Department of Public Health, as yet not peer reviewed, demonstrating that bivalent vaccines and Paxlovid could avert hospitalization (7.8% vs. 11.2%) and SARS-CoV-2 related death cases (16.2% vs. 25.2%) [64]. To this end, Paxlovid is a valuable drug in the treatment of SARS-CoV-2 infections.
Humanity has faced the largest world-wide pandemia of the 21th century. In 2020, we needed to learn how dangerous SARS-CoV-2 infections really were, how hospitalized SARS-CoV-2 patients had to be treated and how we could protect ourselves against airborne SARS-CoV-2 infections. But even with all these efforts, we still faced a case fatality rate of ~2.4% worldwide. Luckily, one year later we already had several vaccines available of which 4 were mainly used in the Western world. Therefore, the second year of the pandemia was accompanied by large vaccine campaigns in order to get the whole population convinced and vaccinated. Many hopes but also a much criticism was heard in 2021, because mankind has never produced, tested and legally approved new vaccines within such a short time-frame. However, all clinical studies demonstrated the efficacy of these vaccines, even after a first injection. The efficacy to combat the virus (an all subvariants thereof) increased with the second and third vaccination steps. However, at the end of 2021, a new game changer appeared in form of the Omicron-variant. Omicron appeared first in Africa, probably deriving from a single immuno-suppressed patient in which SARS-CoV-2 had evolved over months to perfectly adapt to the human host [65]. The Omicron-variant represented an immune-evasive derivative of the original Wuhan SARS-CoV-2 virus. The Omicron-variant displayed a much higher infectivity and out-competed all yet existing strains and variants within 2–3 months. Since Omicron had so much more mutations in the ACE2 binding domain (~18 mutated amino acids in the ACE-2 binding interface alone), all neutralizing antibodies failed to neutralize the virus, and thus, could not prevent novel infections. Thus, infections in 2022 occurred equally in vaccinated and not-vaccinated people, and at least 50% of the populations in different countries were re-infected at least once. Most countries could not handle this situation anymore and things became statistically uncontrollable. At the end of 2022, most people—if not all—were vaccinated several times, infected or both. Thus, the Omicron-variant did two things: due to the high infectivity, it infected most people, but its lethality was dramatically reduced. Therefore, the CFR is now in most countries between 0.3–0.4% which is slightly higher than that for flu-infections. Thus, the SARS-CoV-2 became endemic and the SARS-CoV-2 pandemia is ending in 2023.
Despite this hopeful conclusion, we have to acknowledge a few things. First, airborne infections can be handled well by simply wearing masks, and many studies have shown this clearly [66]. As a scientist, I am quite thankful that most journals made SARS-CoV-2 studies freely available to anyone in order to speed up scientific progress. We also have realized the value of pre-print servers (like e.g., https://www.medrxiv.org/, https://www.biorxiv.org/, https://www.researchsquare.com) to rapidly spread scientific information. We should also be thankful to companies that took the efforts to produce these novel vaccines which do not need adjuvants, are dead vaccines by definition but will give a long-lasting protection against harmful infectious agents. At least the mRNA vaccines are not only very rapid in terms of development but provide also a high safety and efficacy when compared to existing vaccines. This is important because with global warming, many new viruses will come to the Western world, for which we have yet no effective vaccines available (e.g., West-Nil-virus, Krim-Kongo virus, Dengue fever, Zika virus, Chikungunya virus, etc.).
The last thing to mention is that SARS-CoV-2—regarded as a respiratory
infection virus—should be classified as systemic disease causing virus. It
mainly affects our blood circulation system. Blood vessels are internally lined
with a thin monolayer of endothelial cells that control blood flow, blood
pressure (via the smooth muscle cells behind them), blood clotting reactions and
integrity of the complete circulation system. Endothelial cells are also involved
in the control of the early steps of inflammatory reactions, e.g., by controlling
the transmigration of white blood cells. Infection of these cells not only causes
pro-thromboembolic situations, it also destroys the important barrier functions
of the endothelial cell layer. Thus, the effects for the body are dramatic
(damage, inflammation, etc.) and cause the aging of affected organs. So
even if the pandemia is over, we will still face in the future the results of all
this organ damage across the whole population. Also the proteases encoded by this
virus, PL
The SARS-CoV-2 pandemia has taught humanity an important lesson for our lives in modern times: viruses can easily spread around the world and can cause an enormous physic, emotional and economical damage. This pandemia has displayed the fragility of our medical systems, has produced a large number of patient deaths, has shed light on our product delivery chains and the strong influence on economic activity in the case of lock-down decisions taken by governments. We should be aware that global warming, the way we destroy nature and the way we live today, harbour a big danger for ourselves. We need to learn this lesson before the next pandemia arrives. In addition, we should be aware that not every virus is amenable to rapid and effective vaccine development (see HIV-1).
On the other hand, we have learned how basic and medical Science—in combination with the pharmaceutical industry—were able to develop and produce novel vaccines, which were then rapidly evaluated for their efficacy in clinical trials. This gives hope for the future in which similar pandemic situations may emerge. Noteworthy, these vaccines are able to avoid all the above-mentioned adverse effects of real SARS-CoV-2 infections which ended with a high rate of case fatalities. The effects on our immune system are dramatic and ~10% of all infected patients have developed Long-Covid symptoms. This is in strong contrast to the very low numbers of severe vaccination-related side effects (post-vac syndrome) which is in the range of several hundreds to thousands in Germany (~0.0015% of at least 1-time vaccinated individuals).
In contrast, we have also seen negative aspects in this pandemia: social media has been misused to spread fake news or hate-speech. In addition, social media has influenced the recognition of vaccination as a key element to combat any kind of pandemia. Based on the available data, the CFR between unvaccinated and vaccinated individuals (a factor of 15 to 20-fold) tells a clear story for most european countries [68]. Therefore, effective education of young scientists has been also recommended to block the spreading of fake news [69]. This could help in the future to get more rapid responses of populations to viral threats and to be prepared for the next pandemia—as this is only a matter of time.
The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
Not applicable.
I would like to express my gratitude to Professor Dr. Owen Williams for meticulously reviewing the manuscript. I am also thankful to the Corona Task Force at Goethe University in Frankfurt for their financial support during the SARS-CoV-2 pandemic, which has been instrumental in advancing our research on SARS-CoV-2 and SARS-CoV-2 vaccines.
This manuscript is dedicated to Professor Dr. Theo Dingermann on the occasion of his 75th birthday, who has been one of my esteemed scientific mentors. As the editor-in-chief of the ”Pharmazeutische Zeitung” (PZ) at Avoxxa Group, he has authored numerous articles on the SARS-CoV-2 virus, the pandemic, methods of protection against airborne infections, and information about available vaccines. This contribution has been invaluable in raising awareness among the public, pharmacists, and physicians regarding the threat posed by SARS-CoV-2, as well as the efficacy and safety of mRNA vaccines, which are central to the global effort against this pandemic.
I extend my heartfelt thanks to Theo for his outstanding work in this regard.
RM is supported by grants from the DFG (Ma 1876/12-1) and Wilhelm Sander foundation (2018.070.2).
The author declares no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.





