- Academic Editor
The mammalian intestinal epithelium is a rapidly self-renewing tissue in the body and its homeostasis is tightly controlled by numerous factors at multiple levels. The RNA-binding protein HuR (human antigen R) is intimately involved in many aspects of gut mucosal pathobiology and plays an important role in maintaining integrity of the intestinal epithelium by regulating stability and translation of target mRNAs. Nonetheless, deregulation of HuR expression and altered binding affinity of HuR for target transcripts occur commonly in various gut mucosal disorders. In this review, we highlight the essential role of HuR in the intestinal epithelium homeostasis and discuss recent results that interactions between HuR and noncoding RNAs (ncRNAs), including circular RNAs, long ncRNAs, small vault RNAs, and microRNAs, influence gut mucosal regeneration and regulate barrier function in various pathophysiological conditions. These exciting discoveries advance our knowledge of HuR biological function in the gut mucosa and also create a fundamental basis for developing novel therapies to protect intestinal epithelial integrity in critically ill patients.
The mammalian intestinal epithelium is a single layer of columnar cells that function as a dynamic biophysical barrier to separate mucosal tissues from luminal bacteria and noxious substances, in addition to its crucial role in food digestion and absorption [1, 2, 3]. The intestinal epithelium undergoes a rapid and continual self-renewal process throughout the entire life, and the epithelium homeostasis depends on intestinal epithelial cells (IECs) to quickly alter gene expression to regulate proliferation, apoptosis, migration, differentiation, and cell-to-cell interaction in response to pathophysiological stresses. Intestinal stem cells and amplified progenitor cells divide continuously in the crypts and drive renewal process, while the newly divided cells differentiate into various mature cell types when they migrate up along the crypt-villus axis. Apoptosis takes place in both the crypt area, where it maintains the balance in cell number between survival cells and newly divided cells, and the luminal surface of the intestine, where apoptosis causes a loss of differentiated cells [4, 5]. The intestinal mucosa also displays a spectrum of responses after acute injury and can repair itself quickly to restore epithelial integrity. Nonetheless, disrupted homeostasis of the intestinal epithelium and severe mucosal wounds/erosions occur commonly in various pathologies, resulting in abnormalities of the epithelial structure and the translocation of luminal harmful substances and bacteria to the bloodstream [6, 7, 8].
The gene expression programs that govern homeostasis of the intestinal
epithelium are tightly regulated at the posttranscriptional level, although
transcriptional events also play an important role in this process. In
particular, regulation of mRNA stability and translation is essential for control
of intestinal mucosal defense, continual renewal, repair after acute injury, and
gut barrier function [9, 10]. The stability and/or translational control of labile
mRNAs require the association of specific mRNA sequences (cis-elements)
with trans-acting factors such as RNA-binding proteins (RBPs) and
noncoding RNAs (ncRNAs), specifically microRNAs (miRNAs). RBPs and miRNAs
directly bind to cis-elements that are frequently located at the
3
HuR (human antigen R) is a ubiquitous RBP and contains two RNA recognition
motifs (RRMs) at its N-terminus with high affinity for uridine-rich and adenine
uridine-rich elements located in the 3
The intestinal epithelium renews itself every 5–7 days in humans and 4–5 days in mice and this rapid process is tightly controlled by numerous factors at multiple levels. As with other tissues in the body, the intestinal epithelial renewal is uniquely regulated by growth hormones, cortisol, insulin, and thyroxin that modulate metabolism in all tissues in the body. However, the intestinal mucosa responds to a host of events triggered by the presence of luminal food that is one of the strongest stimulants of mucosal growth. Fasting in mice or food starvation in patients with critical disorders inhibits growth of the intestinal mucosa, leading to mucosal atrophy [9]. In cultured IECs, HuR silencing by transfection with specific small interference RNA (siRNA) targeting HuR (siHuR) inhibits cell proliferation and causes G1 phase growth arrest by controlling the expression of several genes associated with cell division and differentiation [25, 26, 27, 28, 29].
To examine the functional role of HuR in the intestinal epithelium in
vivo, we used a Cre-LoxP system to generate a mouse bearing with intestinal
epithelium tissue-specific HuR deletion (IE-HuR
Although the exact mechanism underlying function of HuR in the regulation of
intestinal epithelial renewal remains largely unknown, it has been shown that HuR
promotes growth of the small intestinal mucosa at least partially by activating
the Wnt signaling pathway [4, 29]. Targeted deletion of HuR in the intestinal
epithelium decreases levels of the mRNA encoding low-density lipoprotein
(LDL)-receptor-related protein 6 (LRP6) that functions as a Wnt-coreceptor and is
critical for constant gut mucosal growth. In addition, HuR deletion increases
expression levels of p65 and Smad7 in the intestinal mucosa without effect on the
levels of Frizzled-7 protein. Further experiments show that HuR physically
interacts with the Lrp6 mRNA via its 3
HuR functions as an anti-apoptotic factor in the intestinal mucosa and protects
IECs against apoptosis by altering expression levels of several
apoptosis-associated genes at the posttranscription level [31]. The products of
these genes regulated by HuR in IECs include X chromosome-linked inhibitor of
apoptosis protein (XIAP), transcription factor JunD, mitogen-activated protein
kinase kinase-1 (MEK-1) and activating transcription factor-2 (ATF-2). HuR
directly binds to the mRNAs encoding XIAP, ATF-2, JunD, and MEK-1 proteins and
enhances their expression, thus protecting cells against apoptosis. HuR
stabilizes the Xiap mRNA by interacting with both its 3
The integrity of the intestinal mucosa is quickly restored through a series of
coordinated processes after acute injury [36]. One primary way by which the
mucosa repairs itself rapidly and prevents further tissue damage is early
epithelial restitution, characterized by stages of cell spreading and migration
to quickly cover the wound area and restore the epithelial integrity [37]. HuR
plays an essential role in stimulating early epithelial restitution after acute
injury [17, 38, 39, 40]. In an in vitro model of epithelial
restitution, HuR silencing by transfection with siHuR inhibits early rapid
epithelial restitution after wounding. In an injury model in mice, exposure to
mesenteric ischemia/reperfusion (I/R) leads to small mucosal injury and erosions,
but IE-HuR
HuR stimulates rapid healing of the intestinal mucosa after acute injury by
enhancing expression of multiple signaling proteins such as stromal interaction
molecule 1 (Stim1) [28], cell division control protein 42 (Cdc42) [17],
14-3-3
The intestinal epithelium lines the luminal surface of the mucosa and acts as a physical and biochemical barrier that directly interfaces with a wide array of luminal toxic substances, pathogens, and microbiota. The structure of this specialized barrier in the intestinal epithelium is comprised of various intercellular junction proteins, including tight junctions (TJs) and adherens junctions (AJs). TJs are the most apical element of the junctional complex and form a selective permeable barrier that blocks even small molecules from leaking between epithelial cells [1]. Immediately below TJs are cadherin-rich AJs, which provide strong cell-to-cell interaction and integrate different signals to modulate epithelial paracellular permeability. Since TJ and AJ complexes are highly dynamic, maintenance of the levels of cellular TJ and AJ proteins is crucial for the integrity of the barrier function in stressful environments.
An increasing body of evidence indicate that HuR enhances expression of TJs and
AJs posttranscriptionally and its activation is essential for structure and
effectiveness of the epithelial barrier. HuR induces the stability and
translation of mRNAs encoding TJ proteins including occludin, claudin-1,
claudin-3, and JAM-1, and AJ protein E-cadherin, mostly via direct interaction
with their 3
Chk2-dependent HuR phosphorylation is necessary for HuR association with
occludin mRNA, and activation of Chk2 activity enhances formation of the
HuR/occludin mRNA complex and promotes occludin translation [42]. HuR
stabilizes claudin-1 transcript by directly interacting with its
3
Paneth cells are specialized IECs that are located at the bottom of the crypts
in the small intestinal mucosa. Paneth cells secrete various antimicrobial
peptides/proteins (AMPs) that are necessary for protecting the intestinal
epithelium from enteric pathogenic invasion [48, 49]. HuR enhances the production
and release of AMPs via control of Paneth cell function, thus promoting defense
of the intestinal epithelium [5]. Target deletion of HuR in IECs leads to defects
in Paneth cells, since intestinal tissues from IE-HuR
Autophagy is a process by which unwanted materials and pathogens in the cytoplasm are targeted to the lysosome for degradation. There are more than 30 members of autophagy-related genes (ATGs) identified in mammalian tissues. Autophagy activation is critical for the recognition and degradation of intracellular pathogens, which acts as an innate barrier of bacterial infection, whereas deregulation of autophagy compromises intestinal epithelial host defense and tissue homeostasis. Furthermore, the role of autophagy in preserving gut health is exemplified by the fact that certain bacteria can manipulate the autophagic process to promote their survival and colonization in the gut. Defects in autophagy-mediated clearance of intracellular bacteria may contribute to the chronic inflammation observed in IBD patients [52, 53], since genetic variations in the Atg16l1 gene induces the risk of developing Crohn’s disease [54].
HuR up-regulates autophagy activity by affecting ATG expression. HuR silencing
represses formation of autophagosome that relies on products of the Atg
genes and is critical for activation of autophagy. HuR directly interacts with
the mRNAs encoding ATG16, ATG12, and ATG5 primarily through their 3
Taken together, these exciting findings collected from cultured IECs, intestinal organoids, mice with ablated HuR, and human tissue samples strongly suggest a novel working model by which HuR is required for maintaining the intestinal epithelium homeostasis in various pathophysiological conditions (Fig. 1). According to this model, HuR activation enhances the intestinal epithelial integrity by stimulating mucosal growth, protecting IECs against apoptosis, promoting early epithelial restitution after wounding, increasing the barrier function, and elevating Paneth cell function and autophagy. In contrast, disrupted HuR activity impairs the processes of mucosal regeneration and adaptation, thus compromising the intestinal epithelial homeostasis and contributing to mucosal pathologies such as massive injury/erosions, delayed healing, gut barrier dysfunction, and inflammation.
 Fig. 1.
Fig. 1.Essential role of HuR in maintaining the intestinal epithelium homeostasis. HuR stimulates mucosal growth, IEC survival, epithelial repair after injury, gut barrier function, Paneth cell (PC) activity, and autophagy by altering expression levels of its target genes at the posttranscriptional level. HuR binds to and induces stability and translation of mRNAs that encode proteins involved in regulation of migration, proliferation, apoptosis, differentiation, and cell-to-cell interaction. HuR function is tightly controlled by multiple factors, whereas deregulation of HuR expression, its subcellular distribution, and binding affinity for target transcripts disrupts integrity of the intestinal epithelium, thus contributing to pathogenesis of different gut mucosal disorders. HuR, human antigen R; miRNAs, microRNAs; TC, tuft cells; EC, enterocytes; ISC, intestinal stem cells.
Several studies show that altered levels of tissue HuR in the intestinal mucosa are implicated in pathogenesis of Crohn’s disease (CD) and ulcerative colitis (UC), two types of IBD in [45, 55, 56]. We examined the levels of HuR and Paneth cells in human tissue samples of intestinal mucosa collected from ileal and colonic mucosal tissues in IBD patients [57]. HuR is distributed in both the cytoplasm and nucleus in normal small intestinal mucosa from control individuals. However, the intestinal mucosa with injury/erosions and inflammation obtained from CD patients exhibits a remarkable reduction in the levels of HuR protein, specifically in the cytoplasm, when compared with those from control individuals. Similarly, levels of HuR in the colonic mucosa also decrease dramatically in UC patients relative to control individuals. Interestingly, the decreased levels of small intestinal mucosal HuR are associated with defective Paneth cells in patients with CD, since lysozyme-positive cells are undetectable in the ileal mucosal tissue samples taken from CD patients. The reduced HuR levels and defective Paneth cells in patients with CD or UC are also associated with an inhibition of mucosal renewal and an inhibition of TJ expression. The levels of several proteins, including Bcl-2, nucleotide-binding oligomerization domain 2 (NOD2), and vitamin D receptor (VDR), also decrease in the intestinal mucosa of patients with IBD [40, 57, 58, 59, 60]; all mRNA encoding these proteins are targets of HuR. These findings suggest that reduced HuR and subsequent defects in Paneth cell function are involved in the pathogenesis of human IBD.
Altered HuR expression and deregulation of its binding affinity occur commonly
in gastrointestinal cancers, specifically colorectal cancer, but the exact roles
of HuR in carcinogenesis are cell type-dependent manner [19, 61, 62, 63]. For
instance, HuR promotes cell proliferation and inhibits apoptosis in gastric
cancer cells [19, 64], while HuR enhances angiogenesis and induces metastasis in
colorectal cancer cells [62], although the association of induced amounts of HuR
with cancer aggressive behavior is similar in both cancers. Moreover, HuR
up-regulates expression levels of cyclin D1, Bcl-2, c-Myc, cyclooxygenase 2
(COX-2) and increases the activity of Wnt/
Most (
LncRNAs can be transcribed from antisense, intergenic, or promoter-proximal regions but they have little potential to encode functional proteins. LncRNAs are poorly conserved in general and they modulate gene expression via a broad range of posttranscriptional and/or transcriptional mechanisms. LncRNAs can function as decoys, scaffolds, or signals and also act through cis- and trans-regulatory factors, genomic targeting, and antisense molecules. Enriched in the intestinal epithelium, several lncRNAs, including SPRY4-IT1, H19, TUG1, and GMDS-AS1, control mucosal renewal, adaptation, and repair after acute injury predominantly by interacting with HuR [23, 73, 74, 75].
H19 is a 2.3-kb lncRNA transcribed from the H19/Igf2 gene cluster and it is involved in regulating the gut epithelial barrier function [23]. The levels of tissue H19 increase dramatically in inflamed colonic mucosa and this induction in the level of tissue H19 damages small intestinal mucosa in patients with sepsis and IBD. Induced H19 affects intestinal epithelial renewal, wound healing, and gut barrier function. Ectopically expressed H19 suppresses expression of TJ ZO-1 (Zonula occludens-1) and AJ E-cadherin by reducing their mRNA stability and translation via miR-675, leading to gut barrier dysfunction. HuR directly binds to H19, prevents miR-675 processing from H19, thus promoting ZO-1 and E-cadherin expression and rescuing the barrier function. In contrast, target deletion of HuR in IECs increases miR-675 abundance in the intestinal mucosa and delays recovery of the gut barrier function in mice exposed to septic or mesenteric I/R stress [17, 23]. These results demonstrate the importance of HuR interaction with H19 in regulation of the gut epithelial barrier function.
LncRNA SPRY4-IT1 is derived from the SPRY4 gene, and it is shown to regulate proliferation and apoptosis in cancer cells. The expression of SPRY4-IT1 is upregulated in patients with gastric cancer or colorectal cancer, which is correlated with cancer metastasis. SPRY4-IT1 also plays a critical role in maintaining intestinal epithelium homeostasis by affecting gut barrier function through control of TJ expression. HuR directly binds to SPRY4-IT1 and enhances binding of SPRY4-IT1 to the mRNAs encoding TJs including occludin, claudin-1, claudin-3, and JAM-1 (junctional adhesion molecule 1), thereby promoting SPRY4-IT1-mediated stimulation of TJ translation [73]. HuR silencing decreases SPRY4-IT1 association with these TJ mRNAs and causes dysfunction of the epithelial barrier. These results indicate that SPRY4-IT1 and HuR stimulate TJ expression and promote the epithelial barrier function synergistically.
TUG1 is a 7.1-kb lncRNA that is distributed across multiple tissues including the intestinal epithelium. TUG1 regulates expression of several target genes through distinct mechanisms such as chromatin remodeling, miRNA sequestration, and interactions with RBPs. Aberrant TUG1 expression is commonly observed in the intestinal mucosa of patients with IBD and colorectal cancers. HuR is able to interact with TUG1, and this association affects HuR function in various pathologies [74]. Ectopically expressed TUG1 attenuates mucosal inflammatory progression of DSS-induced colitis in mice partially by altering c-Myc expression via control of the balance between HuR and miR-29b-3p. The mucosal tissue levels of TUG1 are decreased by high glucose and high fat, which promotes apoptosis and release of pro-inflammatory cytokines in the intestinal epithelium. In contrast, TUG1 overexpression reverses these effects and protects IECs against the damage induced by high glucose and high fat.
LncRNA GMDS-AS1 is expressed in the intestinal mucosa and its overexpression plays an important role in the pathogenesis of colorectal carcinoma. Recently, GMDS-AS1 has been shown to physically interact HuR, and this interaction prevents HuR from being degraded by ubiquitination or the proteasome [75]. HuR association with GMDS-AS1 also stabilizes the Stat3 mRNA and results in the activation of GMDS-AS1-induced STAT3 signaling. Ectopically overexpressed HuR restores proliferation of colorectal cancer cells in the absence of cellular GMDS-AS1.
CircRNAs are a class of diverse endogenous ncRNAs in mammalian tissues and play critical roles in intestinal epithelium homeostasis and diseases [10, 76]. CircRNAs are produced via back-splicing reactions and have a covalently closed structure, preventing degradation and increasing their half-life. CircRNAs function as transcriptional regulators, miRNA sponges, protein decoys, scaffolds, and recruiters, and they also interact with RBPs such as HuR [77, 78].
CircPABPN1 is transcribed from the PABPN1 gene and initially identified to modulate PABPN1 expression by altering HuR interaction with the PABPN1 mRNA in HeLa cells [79]. CircPABPN1 also regulates intestinal epithelial autophagy by affecting ATG16L1 expression through interaction with HuR [78]. CircPABPN1 directly binds to HuR in IECs, and ectopically overexpressed circPABPN1 specifically decreases association of HuR with the Atg16l1 mRNA and represses the expression of ATG16L1 without effect on the expression levels of ATG5 or HuR. Furthermore, elevating the levels of cellular HuR rescues expression of ATG16L1 in cells overexpressing circPABPN1. Conversely, circPABPN1 overexpression and HuR silencing repress ATG16L1 expression synergistically. CircPABPN1 inhibits ATG16L1 translation primarily by preventing HuR binding to the Atg16l1 transcript, since the Atg16l1 mRNA does not have potential binding sites for circPABPN1. Importantly, human intestinal mucosal tissues from IBD patients display induced levels of circPABPN1 and reduced HuR abundances, along with a decrease in the level of ATG16L1 and autophagy inactivation. Since HuR targets multiple mRNAs in IECs, it is possible that circPABPN1 also regulates other transcripts via its association with HuR to modulate the intestinal epithelial autophagy and homeostasis.
CircRHOBTB3 plays a tumor suppressive role in the aggressiveness of colorectal cancer by regulating HuR. CircRHOBTB3 physically interacts with HuR and promotes degradation of HuR through ubiquitination [62]. CircRHOBTB3 also functions as a miRNA sponge in gastric cancer, although target genes by circRHOBTB3 remain largely unknown. It has been reported that circ0104103 inhibits cell growth of colorectal cancer and decreases metastasis at least partially by interacting with HuR [80]. In addition, expression level of circAGO2 is upregulated in patients with gastric and colon cancers, which is associated with poor clinical outcomes. CircAGO2 interacts with and induces HuR translocation from nucleus to the cytoplasm, thus altering expression of HuR target genes and enhancing aggressiveness of cancer [19]. On the other hand, decreasing the interaction between circAGO2 and HuR reduces tumorigenesis and cancer cell aggressiveness.
vtRNAs are small ncRNAs synthesized by RNA polymerase III and they are associated with giant cytoplasmic ribonucleoprotein (RNP) particles termed vaults. Humans express four vtRNA paralogs vtRNA1-1, vtRNA1-2, vtRNA1-3, and vtRNA2-1, while mice only produce one vtRNA [81]. vtRNAs have important roles outside of vault RNPs and have been linked to many cellular processes, including mRNA splicing, nuclear transport, drug resistance, synaptogenesis, lysosome function, apoptosis, and tumorigenesis. The free vtRNA1-1 regulates selective autophagy by directly interacting with the RBP p62 [82]. We have recently found that the expression patterns of vtRNAs in the intestinal mucosa change significantly in response to stress and that intestinal mucosa from patients with IBD exhibits increased levels of vtRNAs [43]. Further studies show that vtRNA2-1 inhibits expression of TJs occludin, claudin 1, and AJ E-cadherin posttranscriptionally and leads to gut barrier dysfunction. vtRNA2-1 represses the translation of TJs occludin and claudin 1 by disrupting interaction of the respective mRNAs with HuR. vtRNA2-1 associates with HuR and abolishes HuR binding to the mRNAs encoding occludin and claudin 1, thus decreasing their translation. These results indicate that induced vtRNA2-1 damages function of the intestinal epithelial barrier by inhibiting HuR-mediated translation of occludin and claudin 1.
MiRNAs are small ncRNA with ~22 nucleotides and induce gene
silencing by guiding argonaute proteins to bind to 3
MiR-195 is a key regulator of intestinal epithelial homeostasis in health and
pathologies by targeting different mRNAs encoding proteins involved in
proliferation and migration [28, 83]. Increased miR-195 also inhibits IGF
signaling by decreasing translation of the IGF2-receptor [84] but the levels of
cellular miR-195 are reduced by lncRNA uc.173 [85]. Ectopically
overexpressed miR-195 represses early rapid mucosal restitution after acute
injury by destabilizing the mRNA encoding Stim1, a protein necessary for
stored-operated Ca
MiR-29a/b is highly expressed in the intestinal epithelium and its tissue levels
change markedly in mucosal atrophy and gut barrier dysfunction. Ectopic
overexpression of miR-29b causes G1 phase growth arrest of IECs, whereas
decreasing the levels of tissue miR-29b by treatment with LNA-mediated silencer
stimulates renewal of the small intestinal mucosa in mice [89]. MiR-29a/b
interacts with the mRNAs encoding cyclin-dependent kinase 2, HMGB1, PTEN,
claudin-1, and ZO-1 via their 3
MiR-519 is downregulated in gastric and colorectal cancer, while its
overexpression inhibits migration, proliferation, and invasion of these cancer
cells by suppressing activity of the Wnt/
Together, these observations indicate that HuR and ncRNAs jointly regulate the stability and translation of target mRNAs in the intestinal epithelium. Interactions between HuR and ncRNAs, including lncRNAs, circRNAs, small vtRNAs, and miRNAs, alter HuR binding affinity for mRNAs and affect the turnover and translation of HuR target transcripts synergistically or antagonistically, which is essential for maintaining homeostasis of the intestinal epithelium in stressful environments (Fig. 2). In contrast, disruption of HuR association with ncRNAs is widely involved in many aspects of pathogenesis of various gut mucosal disorders. Although the precise mechanisms underlying the regulation of HuR function remain to be fully investigated, its subcellular localization is intimately linked to its effects upon target transcripts. Emerged evidence indicates that most HuR-regulated phenomena in the intestinal mucosa take place in the cytoplasm, while little is known about the roles of nuclear HuR in intestinal epithelium homeostasis and diseases. These findings strongly suggest that HuR and its binding to ncRNAs are novel therapeutic targets for intervention to protect integrity of the intestinal epithelium and gut barrier function in the clinical setting.
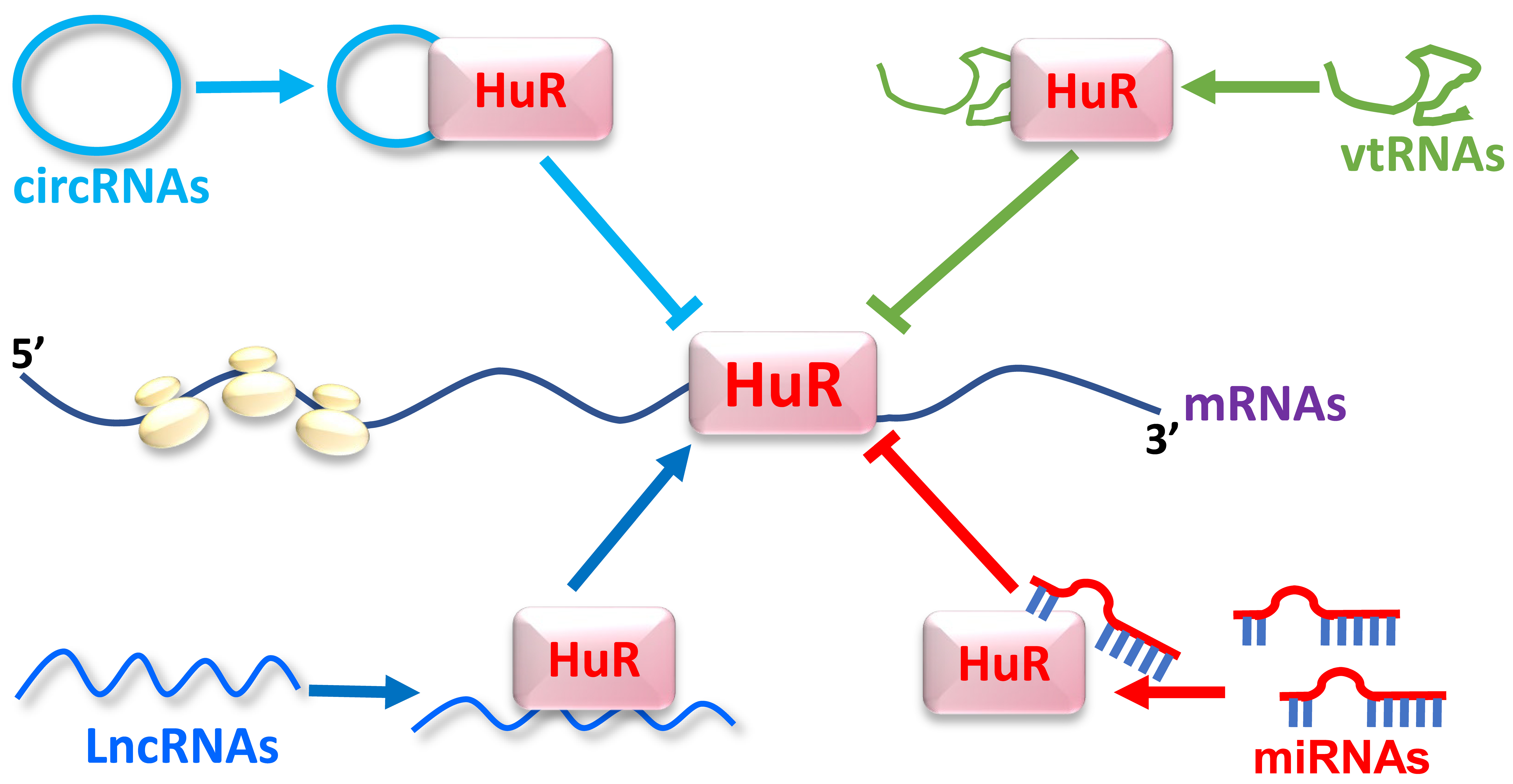 Fig. 2.
Fig. 2.Regulation of HuR function by ncRNAs in the intestinal epithelium. NcRNAs, including lncRNAs, circRNAs, small Vault RNA (vtRNAs), and miRNAs, are able to directly interact with HuR and alter HuR binding affinity for mRNAs, thus jointly regulate stability and translation of target mRNAs synergistically and antagonistically. Some ncRNAs also regulate HuR function via control of HuR expression levels. On the other hand, interactions between HuR and ncRNAs affect ncRNA binding to given mRNAs and alter the regulatory effects of ncRNAs on target transcripts or genes.
HuR plays an essential role in maintaining the intestinal epithelium homeostasis
by regulating mucosal renewal, protection, repair after acute injury, barrier
function, Paneth cells, Tuft cells, and autophagy. HuR enhances stability and
translation of target mRNAs by directly interacting with their 3
Clearly, there is still a big gap in our knowledge regarding HuR function and
its regulation by ncRNAs in health and damaged intestinal epithelium. The exact
mechanisms underlying control of HuR expression, its subcellular trafficking, and
Chk2-dependent HuR phosphorylation remain to be fully investigated at the
molecular level. The applications of various state-of-the-art new techniques such
as tissue-specific genetic loss- or gain-function approach to examine HuR
functions in vivo and single cell RNA-sequence analysis to define novel
targets of HuR in individual cell type are badly needed and will point out new
directions. Consistent with this effort, targeted deletion of HuR in IECs is
recently shown to specifically alter gene expression profiles in Paneth cells and
impairs the integrity of the Paneth cell/intestinal stem cell niche [101]. More
studies are also needed to define the mechanisms by which HuR increases the
stability and translation of target mRNAs and determine if mutations in specific
domains of HuR alter its ability to interact with ncRNAs and affect HuR binding
affinity for 3
In addition, the influence of microbiota dysbiosis induced by target HuR deletion on regeneration and adaptation of the intestinal mucosa remains largely unknown and will provide useful information about the in vivo functions of HuR via interaction with microbiota. It is possible that the presence of certain microbial species can enhance or suppress HuR expression, thus indirectly shaping cellular responses and contributing to gut mucosal-related pathologies. Recently, it has been reported that C. difficile infection induces gut mucosal injury and increases gut permeability, whereas treatment with Lacticaseibacillus casei strain T21 reduces C. difficile toxin effect by altering HuR expression [102]. Finally, experiments using various human mucosal tissue samples from patients with critical illnesses such as IBD, sepsis, leaky gut, and cancers will increase the impact of HuR on pathogenesis of intestinal mucosal disorders and potential clinical application.
RBPs, RNA-binding proteins; IECs, intestinal epithelial cells; ncRNAs, noncoding RNAs; lncRNAs, long ncRNAs; IBD, inflammatory bowel diseases; RRM, RNA recognition motif; miRNA, microRNA; CRs, coding regions; UTRs, untranslated regions; circRNAs, circular RNAs.
SS: Conceptualization, data curation, and original draft of manuscript. LX: Conceptualization, data curation, and preparation of Figures. JYW: Conceptualization, data curation; formal analysis and supervision, original draft, review, and editing of manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity.
Not applicable.
Not applicable.
This work was supported by Merit-Review Award from US Department of Veterans Affairs (to JYW) and by NIH Grants DK-57819, DK-61972, and DK-68491 (to JYW).
The authors declare no conflict of interest. Jian-Ying Wang is a Senior Research Career Scientist, Biomedical Laboratory Research & Development Service, US Department of Veterans Affairs.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
