- Academic Editor
Background: Chickpea is one of the most important leguminous crops and its productivity is significantly affected by salinity stress. The use of ecofriendly, salt-tolerant, plant growth-promoting rhizobacteria (PGPR) as a bioinoculant can be very effective in mitigating salinity stress in crop plants. In the present study, we explored, characterized, and evaluated a potential PGPR isolate for improving chickpea growth under salt stress. Methods: A potential PGPR was isolated from rhizospheric soils of chickpea plants grown in the salt-affected area of eastern Uttar Pradesh, India. The isolate was screened for salt tolerance and characterized for its metabolic potential and different plant growth-promoting attributes. Further, the potential of the isolate to promote chickpea growth under different salt concentrations was determined by a greenhouse experiment. Results: A rhizobacteria isolate, CM94, which could tolerate a NaCl concentration of up to 8% was selected for this study. Based on the BIOLOG carbon source utilization, isolate CM94 was metabolically versatile and able to produce multiple plant growth-promoting attributes, such as indole acetic acid, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, siderophore, hydrogen cyanide (HCN), and ammonia as well as solubilized phosphate. A polyphasic approach involving the analysis of fatty acid methyl ester (FAME) and 16S rRNA gene sequencing confirmed the identity of the isolate as Enterobacter sp. The results of greenhouse experiments revealed that isolate CM94 inoculation significantly enhanced the shoot length, root length, and fresh and dry weight of chickpea plants, under variable salinity stress. In addition, inoculation improved the chlorophyll, proline, sugar, and protein content in the tissues of the plant, while lowering lipid peroxidation. Furthermore, isolate CM94 reduced oxidative stress by enhancing the enzymatic activities of superoxide dismutase, catalase, and peroxidase compared to in the respective uninoculated plants. Conclusions: Overall, the results suggested that using Enterobacter sp. CM94 could significantly mitigate salinity stress and enhance chickpea growth under saline conditions. Such studies will be helpful in identifying efficient microorganisms to alleviate salinity stress, which in turn will help, to devise ecofriendly microbial technologies.
Historically, chickpea plant (Cicer arietinum L.) has been one of the
most significant component of the global cropping system. The total annual world
production of chickpeas is 15.0 million tons, with India being the major
chickpea-producing country, creating 73% of the global annual production [1].
Considering its high nutritional content, such as protein, energy, fiber,
vitamins, and minerals, it is considered a cheaper substitute for meat [2]. Thus,
it fills up the protein gap in the daily food intake of a growing Indian and
global population. Moreover, legumes, such as chickpea, fix a considerable amount
of nitrogen through biological nitrogen fixation, which has a significant
contribution towards sustainable agriculture production [3]. However, due to
their sessile nature, chickpea crop is considered susceptible to salt stress, and
during the past few years, has faced serious global yield losses of 8.0–10.0%
[4]. Globally, salt stress affects more than 800.0 million hectares (mha) of land
and 20% of irrigated agricultural land [5]. Nearly 6.74 mha of land in India is
under severe salinity stress [6]. Twelve states and one union territory in India
covering 44% of the land are affected by soil salinity. Kaur et al. [7]
reported that due to salinity, there has been a reduction of 36.1%–65.0% in
chickpea yield at EC
To mitigate salt stress throughout the world, conventional breeding or genetic engineering approaches are developing salinity-tolerant plants and reclamation of saline soil by applying chemicals or organic amendments. However, the development of tolerant varieties requires a long time as well as a significant number of resources. Further, engineered plants should also comply with the regulations relating to genetically modified organisms. Therefore, to mitigate the negative effects of salt stress, researchers are now focusing on beneficial salt-tolerant plant growth-promoting rhizobacteria (PGPR) as a bioinoculant [11, 12]. The PGPR performs a key role in plant growth through nitrogen fixation, augmenting nutrient (P, K, Zn, etc.) availability, the production of plant hormones (indole acetic acid and gibberellin) and 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase enzyme, secretion of exopolysaccharides (EPS), antibiotics, and hydrolytic enzymes, to aid in coping with various stresses [8, 13, 14, 15]. To date, many rhizobacterial genera, such as Azotobacter, Pseudomonas, Bacillus, Agrobacterium, Enterobacter, Streptomyces, Rhizobium, Exiguobacterium, Klebsiella, Serratia, and Ochrobactrum, are well known for their roles in plant growth and production under salinity stress [16, 17]. Among the different bacterial genera Enterobacter, which belong to the Enterobacteriaceae family, are adaptable and well-known root colonizers with multiple plant growth-promoting activities that have been found to help plants survive under different abiotic as well as biotic stress conditions [18, 19]. Different Enterobacter spp., such as E. cloacae, E. radicincitans, E. ludwigii, E. gergoviae, and E. asburiae have been reported in different cropping systems, to promote the growth of economically important crops by rendering direct or indirect benefits [14, 20, 21, 22]. In addition, some PGPR can induce the accumulation of osmolytes and modulate antioxidant enzymes to scavenge salinity-induced ROS and enhance plant growth [8]. Earlier studies have revealed that under salinity stress, salt-tolerant Enterobacter sp. P23 and Enterobacter sp. PR14 mitigated salinity stress in rice and millet seedlings by modulating defense-related antioxidant enzymes [21, 23]. Recently, in another study, Ali et al. [24] reported a defensive role of rhizospheric bacteria Enterobacter cloacae PM23 in improving salt-stress tolerance and growth of maize. Thus, using such beneficial PGPR strains as bioinoculants, instead of synthetic chemicals, will not only increase plant growth and development but will also help to manage soil health under salt stress conditions.
Therefore, under this hypothesis, the present work was planned to explore and characterize salt-tolerant rhizobacteria from the chickpea rhizosphere and evaluate it for mitigation of salt stress in chickpea plants under greenhouse conditions.
Chickpea (Cicer arietinum L.) plants grown in salt-affected fields of
Mau district (25.9417
Morphological and biochemical characteristics of the CM94 isolate were determined by
following Bergey’s Manual of Determinative Bacteriology. Additionally, the
culture was examined for colonial characteristics and subjected to microscopic
analysis. Biochemical characterization was carried out using a Hi25
The intrinsic tolerance of the CM94 isolate against different concentrations of
NaCl was evaluated by inoculating the bacterial culture (10
The sensitivity to different antibiotics viz. ampicillin (10 mcg),
chloramphenicol (30 mcg), azithromycin (15 mcg), gentamicin (30 mcg), kanamycin
(30 mg), erythromycin (15 mcg), penicillin (10 mcg), polymyxin-b (50 u),
rifampicin (5 mcg), tylosin (15 mcg), streptomycin (10 mcg), vancomycin (30 mcg),
nadifloxacin (10 mcg), and tetracycline (10 mcg) was determined by placing
different antibiotic discs on the bacterial lawn growing on NA. The plates were
incubated at 28
Plant growth-promoting traits were evaluated under normal (no NaCl) and saline (2 to 8% NaCl) conditions. NaCl treatment was supplemented in the respective growth medium.
IAA production was estimated following the protocol of Bric et al.
[26]. In brief, rhizobacterial cultures grown overnight were centrifuged (10,000
g for 5 min) and 2 mL bacterial supernatant, 4 mL Salkowski reagent (50 mL of
35% perchloric acid + 1 mL of a 0.5 M FeCl
ACC deaminase activity was determined as described by Penrose and Glick [27]. The pure rhizobacterial culture was spot inoculated on Dworkin and Foster (DF) salt minimal medium [28], amended with 0.10 M of ammonium sulfate or 3.0 mM of ACC for 48 h, and bacterial isolate growth was assumed as positive for ACC deaminase production. Quantitative activity of ACC deaminase at different NaCl concentrations was assayed by harvesting the bacterial cells through centrifugation (10,000 rpm for 10 min), followed by washing three times with Tris–HCl (0.1 M, pH 7.5), resuspended in 1 mL of Tris–HCl (0.1 M, pH 8.5). Toluene (30 µL) was mixed into the cell suspension and homogenized by vortexing. An aliquot of toluene labialized cells was used for the ACC deaminase protein and activity assays [27].
The phosphate solubilizing ability of CM94 was determined by spot inoculation of
the isolate on the National Botanical Research Institute’s Phosphate Medium agar
plate (NBRIP) [29] containing bromophenol blue and incubated for 48–72 h at 28
PSI = (colony diameter + halo zone diameter) / colony diameter
The ability to produce siderophores by the CM94 isolate was tested, as per a previously described method by Schwyn and Neilands [30], using a chrome azurol S (CAS) agar plate. The appearance of a yellow to orange colored zone surrounding the bacterial colony was considered a positive result for siderophore production. Further the quantitative estimation of siderophore production was determined through the CAS-shuttle assay in MM9 medium.
An actively grown CM94 culture was inoculated into sterile peptone water (10
mL), containing different NaCl concentrations in separate tubes, and incubated
for 48 h at 28
Production of hydrogen cyanide by the CM94 isolate was tested by adopting the
standard method by Lorck [32]. Briefly, the isolate was streaked onto a NA plate
supplemented with different salt concentrations and 4.4 g glycine/L. A strip of
Whatman filter paper presoaked in Na
EPS production was estimated using the method of Qureshi and Sabri [33]. Here, 100 mL rhizobacterial culture (48 h), grown in NB medium with different NaCl concentrations, was centrifuged at 10,000 g for 15 min at 4 °C. EPS was precipitated from the supernatant with the help of 3 volumes of prechilled acetone. After 2 days, EPS (precipitated) was filtered using Whatman filter paper. The wet EPS on the filter paper was dried overnight at 58 °C and weighed again. The increase in the weight of the filter paper with EPS biomass indicated the positive result of EPS production.
Based on the carbon (95 C-substrates) utilization pattern, the metabolic characterization of the selected rhizobacterial CM94 isolate was carried out using BIOLOG GNII (MicroPlate™ Biolog, Inc., Hayward, CA, USA), according to the manufacturer’s instructions.
To analyze the fatty acid profile, fatty acids were extracted from the isolate, following the methods described by Sasser [34], and analyzed by gas chromatography (Agilent GC 7820A) using MIDI Sherlock software (Sherlock TSBA Library version 3.9, MIDI Inc., Newark, DE, USA) for fatty acid methyl ester analysis (FAME).
Genomic DNA from the overnight grown pure culture of CM94 was extracted
following the method described by Pospiech and Neumann [35]. Amplification of the
16S rRNA gene was conducted using the universal forward primer PA (5
To evaluate the growth promotion and salt-stress mitigation efficiency of the
CM94 isolate on chickpeas, an experiment was conducted using 0, 75, and 150 mM
NaCl concentrations in the greenhouse at ICAR-NBAIM, Mau Nath Bhanjan (U.P.).
Briefly, the pure bacterial CM94 culture was grown overnight and harvested,
centrifuged, washed, and resuspended in phosphate buffer (0.1 M, pH 7.0), to
obtain a ~10
Fifteen days after germination, chickpea seedlings from each treatment, were carefully uprooted and washed, and biometric parameters, such as root/shoot length, and fresh and dry weight were recorded. The dry weights of the shoot and root were recorded after drying at 70 °C for 24 h.
Determination of Chlorophyll, Proline, Sugar, and Protein Content
Chlorophyll content was spectrophotometrically assayed by adopting the process
of Arnon [39]. In brief, 100 mg of leaves were completely homogenized in 80%
acetone and centrifuged at 5000 g for 5 min. Then, the supernatant was measured
at 645 and 663 nm, and total chlorophyll content was calculated and expressed in
mg g
Proline content was determined according to the protocol in Bates et al. [40]. Proline contents were spectrophotometrically measured by reading the
absorbance of the ninhydrin reaction at 520 nm and the quantity of proline was
examined using the standard curve of L-proline and expressed in µmol
g
The total soluble sugars (TSS) of the rhizobacterial-treated and non-treated chickpea
plants exposed to NaCl stress were estimated using the phenol sulfuric acid
method [41]. The optical density of the cooled reaction mixture was measured at
620 nm, and the amount of total sugar (µg g
The protein concentration was determined using the Bradford assay [42]. Spectrophotometrically the protein content was assayed at 595 nm and the protein concentration was determined using the BSA standard curve (bovine serum albumin).
The malondialdehyde content was estimated by adopting the procedure in
Kumari et al. [43]. A total of 500 mg of leaves were ground in 5% (w/v)
trichloroacetic acid and centrifuged at 12,000 g for 10 min. Then, 0.5% (w/v) of
thiobarbituric acid (TBA) was mixed in 2.0 mL of diluted supernatant extract.
Next, the reaction mixture was incubated at boiling temperature for 30 min and
immediately terminated by transferring the reaction mixture to ice. Then, the
reaction mixture was centrifuged at 12,000 rpm for 10 min, and measured
spectrophotometrically at 532 nm. The MDA content in the samples was calculated
using the MDA standard curve and expressed as nmol g
The crude extract for antioxidant enzymes present in the plant tissue was prepared by following the procedure in Kumari et al. [43]. Briefly, a 1.0 g leaf sample was ground with potassium phosphate buffer (0.1 M, pH 7.4) containing 1 mM EDTA, 1 mM PMSF (phenylmethylsulfonyl fluoride), and 2% polyvinylpyrrolidone (PVP). The homogenate was centrifuged (10,000 g for 15 min at 4 °C) to collect the enzyme extract, which was further used in the antioxidant enzyme analysis.
SOD activity was assayed spectrophotometrically, following the method of Beyer
and Fridovich [44]. The reduction in nitro blue tetrazolium (NBT) was determined
by recording the change in OD at 560 nm. Dark blanks were used to determine the
enzyme units (U). For the photoreduction of 50% NBT, one unit of SOD was
required. The activity of SOD was represented in U mg
The CAT activity was determined spectrophotometrically by monitoring the
disappearance rate of H
POD activity was assayed spectrophotometrically using guaiacol as the substrate
and recording the increase in OD at 436 nm at 30 °C [46]. The reaction
was started by mixing 50 µL leaf extract with 3.0 mL of a solution
consisting of guaiacol (1%), H
Plants from every treatment were uprooted carefully and washed 3–4 times with
sterilized distilled water and surface-sterilized, as described earlier.
Afterward, the root samples were homogenized in sterile phosphate buffer,
diluted, and spread (100 µL) on NA medium plates supplemented with a
combination of rifampicin, penicillin, kanamycin, and tetracycline (selected
based on initial intrinsic antibiotic resistance assay), using a concentration of
10 µg/mL for each. The plates were incubated for 24 h at 28
All data were produced in triplicate and analyzed by analysis of variance (ANOVA) using Duncan’s multiple range tests (DMRT) at the p = 0.05 level with SPSS (ver. 16.0, IBM SPSS Statistics, Chicago, IL, USA). The graphs were prepared using Microsoft Excel version 2019.
Based on the luxuriant and dominant growth on the NA medium amended with 4%
NaCl (as a selective additive for isolation of salt-tolerant rhizobacteria),
a CM94 rhizobacterial isolate was obtained from the rhizospheric soil of chickpea
plants. Primary characterization of the isolate was performed through
morphological and biochemical characteristics (Table 1). The results showed that
the selected rhizobacterial isolate CM94 had circular, smooth, convex, cream-colored
colonies with entire margins on the NA plates. Microscopic observation followed
by Gram staining revealed that it was a Gram-negative rod-shaped bacterium.
Further, CM94 was found to be positive for oxidase, catalase, citrate
utilization, indole production, nitrate reduction methyl red, and ornithine
utilization, while it was negative in the ONPG test, lysine
utilization, Voges–Proskauer test, arginine, H
| Characteristic | Enterobacter sp.CM94 |
| Shape | Rod |
| Motility | + |
| Color | Cream |
| Gram reaction | – |
| NaCl tolerance (%) | up to 8% |
| pH tolerance (optimum 7.0) | 6.0–9.0 |
| Temperature tolerance | 45 °C |
| Catalase activity | + |
| Oxidase activity | + |
| Citrate utilization | + |
| ONPG | – |
| Indole production | + |
| Methyl red test | + |
| VP test | – |
| Arginine | – |
| Nitrate reduction | + |
| Ornithine utilization | + |
| H |
– |
| Phenylalanine deamination | – |
| Lysine utilization | – |
| Industrial enzymes | |
| Protease | + |
| Lipase | – |
| Amylase | – |
| Gelatin hydrolysis | + |
| Cellulase production | + |
| Urease production | + |
(+) positive for test; (–) negative for test.
ONPG, O-Nitrophenyl-
Results of intrinsic NaCl tolerance revealed that CM94 could grow in various salinities, ranging from 0 to 8% NaCl (w/v), with optimum growth in terms of cellular protein concentration was recorded at 2% NaCl (Fig. 1). At higher NaCl concentrations, bacterial growth was sluggish over the first 24 h. However, after a few hours and up to 72 h of growth, it grew well in a saline medium before entering the stationary phase. In addition to the salinity, the CM94 strain could also grow in the liquid medium at a temperature range of 15–45 °C and a pH range of 4.5–9.0.
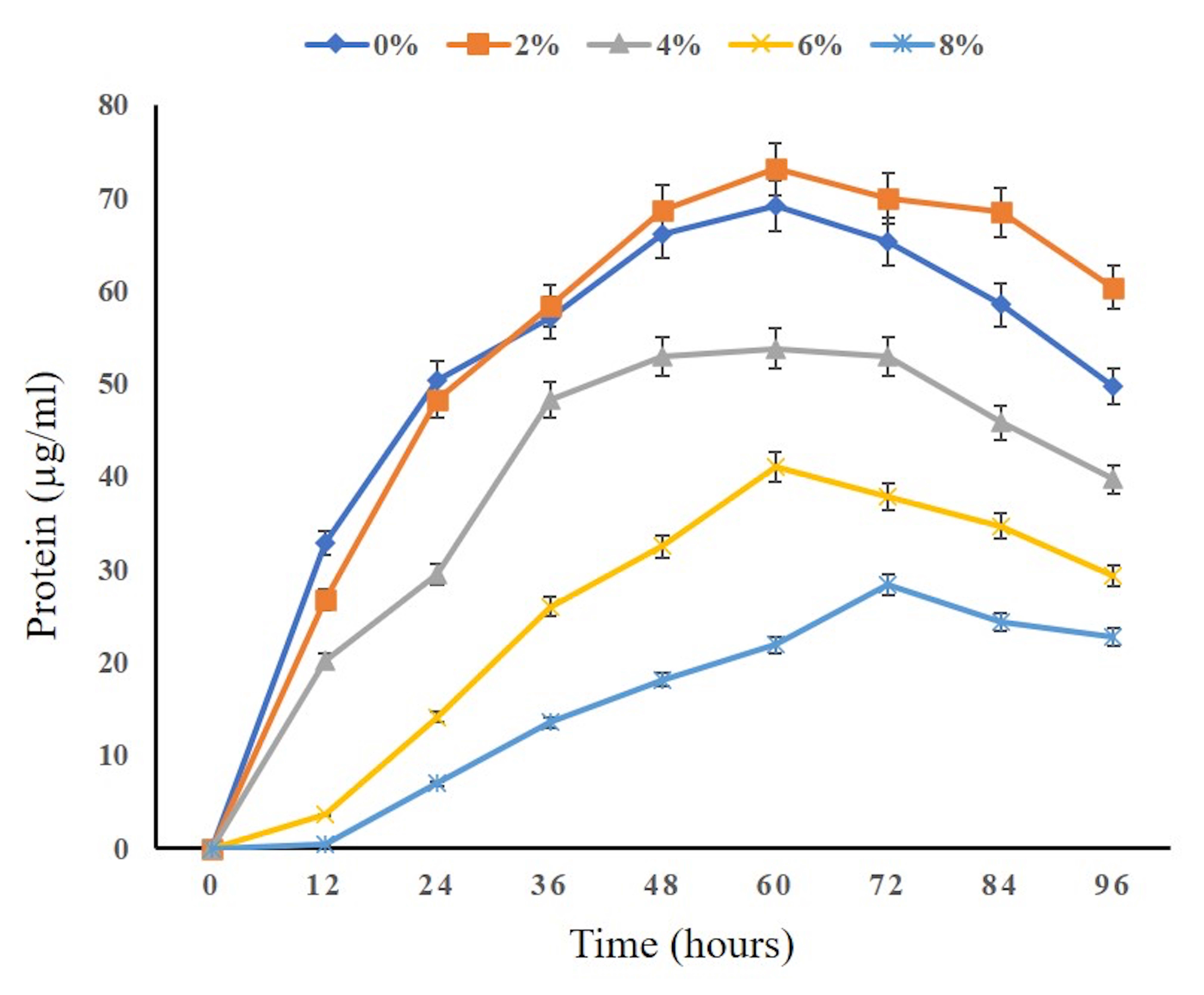 Fig. 1.
Fig. 1.Salt tolerance ability of the rhizobacterial isolate CM94
indicating cellular protein concentration under different salt concentrations at various
durations. The highest protein concentration was recorded after 60 hours at a
salt concentration of 2%. Each value in the graph indicates the mean of three
replicates. Error bars represent mean
Different plant growth-promoting traits, such as IAA production, ACC deaminase,
siderophore, ammonia, HCN, and P-solubilization by the CM94 isolate were
evaluated in vitro at various salt (NaCl) concentrations (Table 2). The
results showed that CM94 could produce 144.0 µg/mL IAA when grown
without NaCl, whereas as the salt concentrations increased to 2, 4, and 6% NaCl,
the IAA production decreased by 18.1, 44.6, and 56.3%, respectively. However,
CM94 was unable to produce IAA at a NaCl concentration of 8% (Table 2). The ACC
deaminase activity by CM94 was recorded as being 89.7
| NaCl concentration | IAA (µg/mL) | ACC deaminase ( |
P-solubilization index (PSI) | Siderophore (% unit) | Ammonia production | HCN production | EPS production (g/100 mL) |
| 0% | 146.04 |
89.71 |
4.1 |
69.16 |
+++ | +++ | 1.97 |
| 2% | 119.51 |
78.35 |
3.37 |
65.72 |
+++ | +++ | 2.53 |
| 4% | 80.89 |
66.42 |
2.36 |
58.57 |
+++ | ++ | 2.82 |
| 6% | 63.7 |
41.5 |
1.80 |
45.52 |
++ | ++ | 1.63 |
| 8% | -ND- | 13.56 |
0.53 |
27.14 |
+ | - | 0.76 |
‘+++’: high production; ‘++’: medium production; ‘+’: low production; ‘-ND-’: not detected.
IAA, indole acetic acid; ACC, 1-aminocyclopropane-1-carboxylic acid; HCN, h ydrogen cyanide; EPS, exopolysaccharides.
The CM94 EPS production was also determined. The CM94 isolate produced 1.97 g EPS/100 mL with no salt stress (0% NaCl). Interestingly, it was observed that CM94 could produce EPS at all NaCl concentrations, with the highest (2.82 g/100 mL) at 4% NaCl (Table 2). Overall, the results of the PGP attributes and EPS production showed that the isolate could retain its PGP activities even at higher salt stress conditions.
The metabolic potential of the CM94 isolate, based on the utilization of different C-sources, was determined using the BIOLOG (R) system. The results showed that CM94 was metabolically active and among the ninety-five different C-sources, isolate CM94 utilized 64 C-sources, including 2 amines, 15 different amino acids, 22 carbohydrates, 18 carboxylic acids, 5 polymers, and 2 miscellaneous carbon sources (Supplementary Table 2).
In addition, isolate CM94 had a unique fatty acid composition, with 14:0, 14:0
3OH/16:1 iso I, 16:1
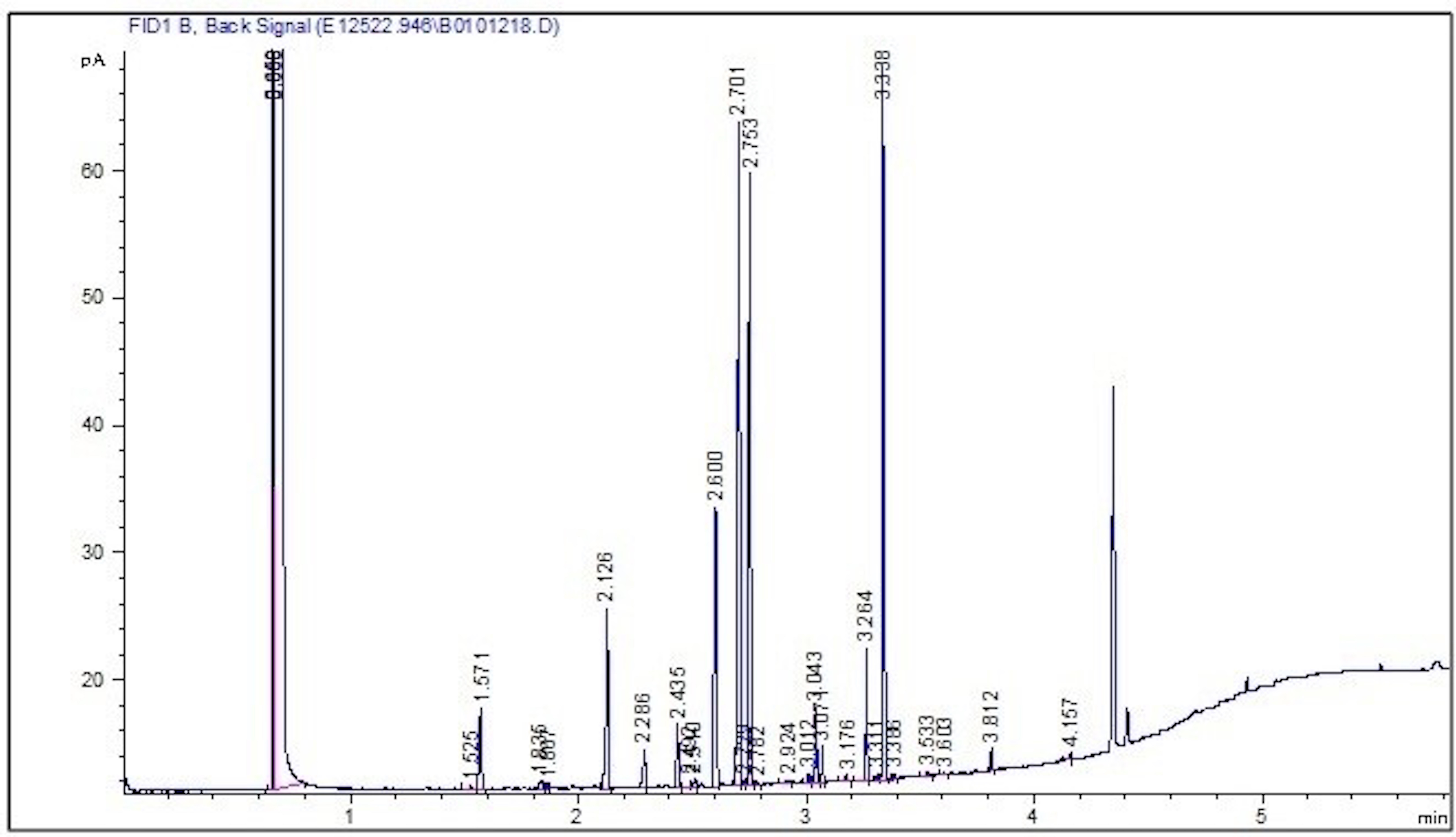 Fig. 2.
Fig. 2.Gas chromatography profile showing peaks of fatty acid methyl esters (FAME) for Enterobacter sp. CM94.
A ~1.5 kb sequence of the 16S rRNA gene was obtained by the sequencing analysis. The BLASTn search results showed that the CM94 isolate had a 99.0% similarity with the existing 16S rRNA gene sequences of the Enterobacter species in the NCBI GenBank database. Based on the neighbor joining method, a phylogenetic tree of the CM94 isolate containing similar sequences from the NCBI database was created by 1000 bootstrap sampling (Fig. 3). The obtained 16S rRNA gene sequence (1440 bp) of CM94 was deposited into the NCBI database with the accession number: KC504003.
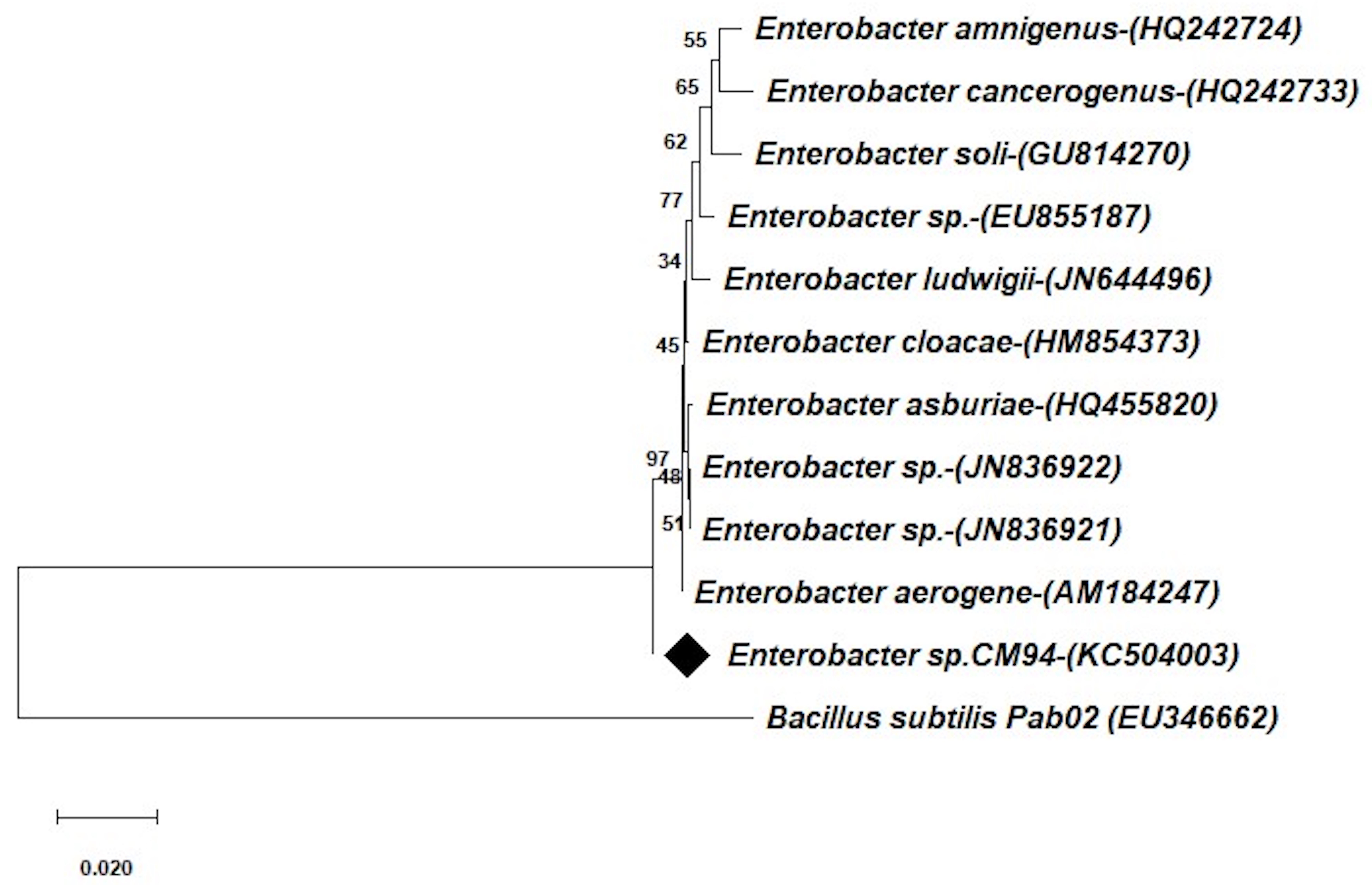 Fig. 3.
Fig. 3.Neighbor joining phylogenetic dendrogram based on 16S rRNA gene sequences showing relationships between CM94 and related taxa. Bacillus subtilis (EU346662) was used as an outgroup.
Effects of the rhizobacterium CM94 inoculation in the mitigation of any adverse
effects from salinity stress on chickpea growth was assessed under greenhouse
conditions (Fig. 4). Results showed that all the salt stress treatments
negatively affected the chickpea seedling growth in terms of a decrease in root
length (RL), shoot length (SL), root and shoot fresh, and dry weight. However,
isolate CM94 inoculation significantly (p
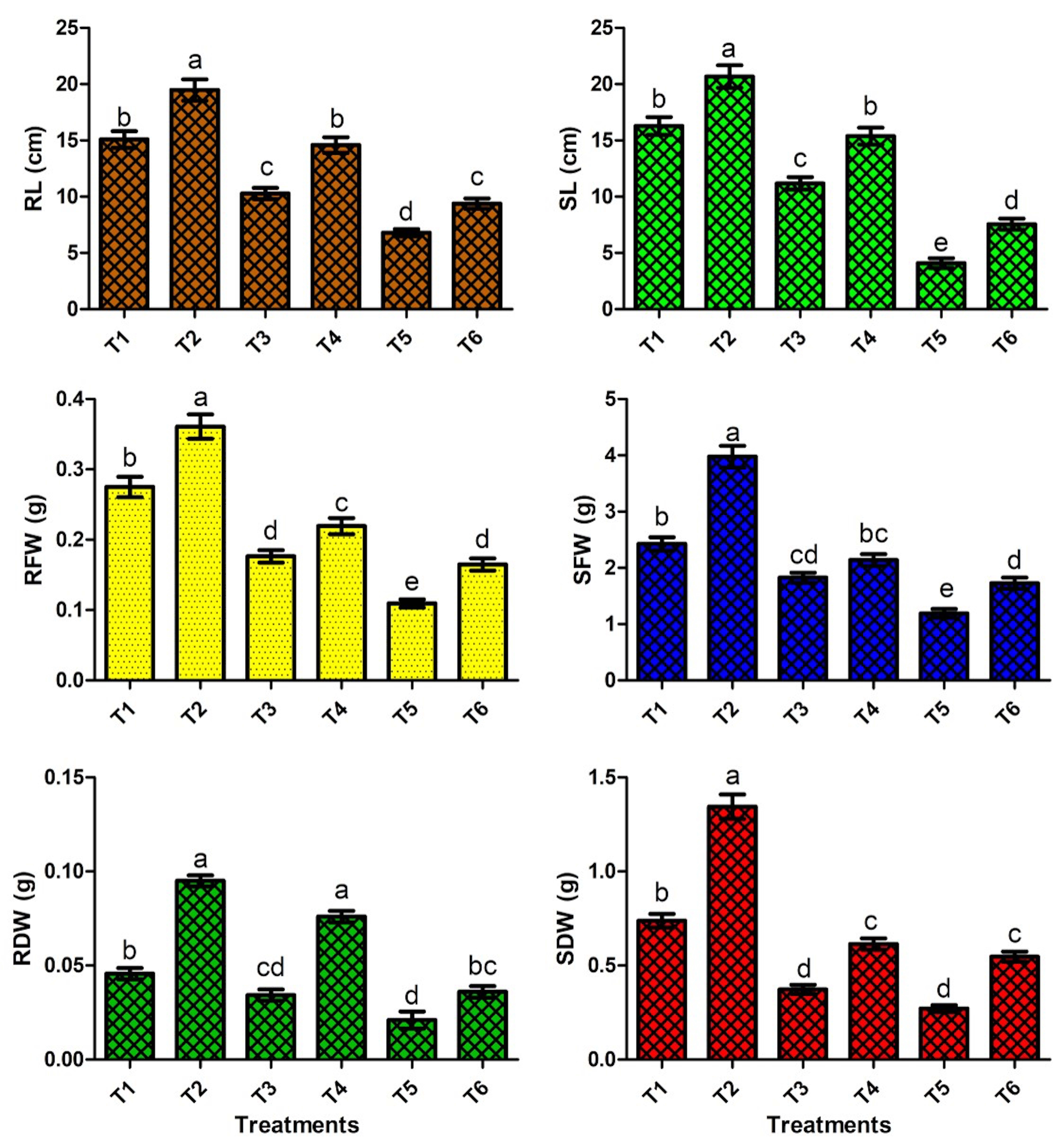 Fig. 4.
Fig. 4.Effect of Enterobacter sp. CM94 inoculation on chickpea
growth under different NaCl conditions. T1: uninoculated control (no NaCl); T2:
CM94 inoculation; T3: 75 mM NaCl; T4: 75 mM NaCl + CM94; T5: 150 mM NaCl; T6: 150
mM NaCl + CM94. RL, root length; SL, shoot length; RFW, root fresh weight; RDW,
root dry weight; SFW, shoot fresh weight; SDW, shoot dry weight. The mean
Chlorophyll content decreased by 21.1 and 49.6 % in the plants grown at 75 mM (T3) and 150 mM (T5) NaCl, respectively. Whereas increases of 23.7 and 31.6% were recorded in the chlorophyll content of the plants inoculated with the bacteria at 75 mM and 150 mM NaCl, respectively, compared to the respective uninoculated control plants (Fig. 5).
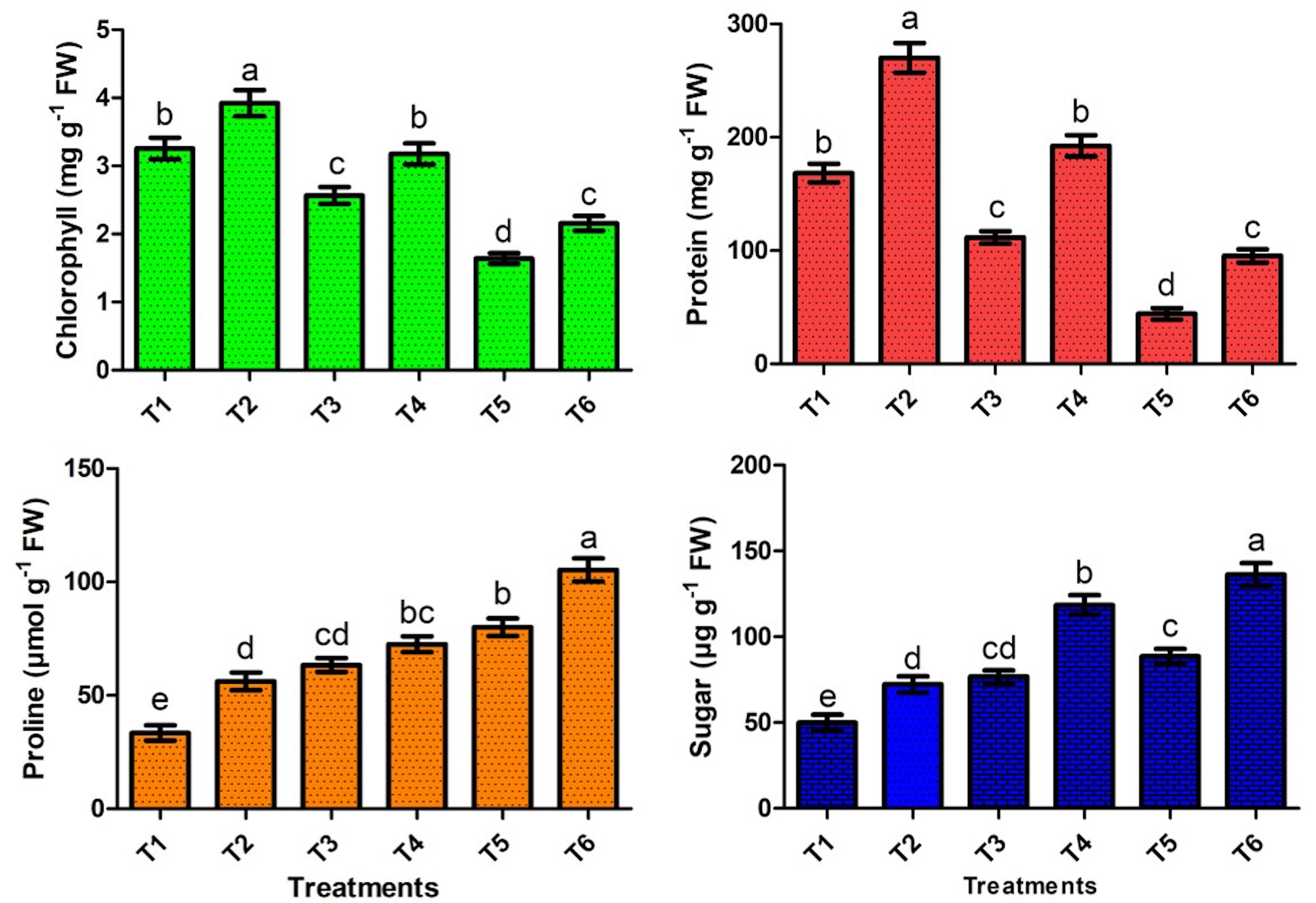 Fig. 5.
Fig. 5.Effect of Enterobacter sp. CM94 inoculation on
chlorophyll, proline, sugar, and protein content in chickpea plants grown under
different NaCl concentrations. T1: uninoculated control (no NaCl); T2: CM94
inoculation; T3: 75 mM NaCl; T4: 75 mM NaCl + CM94; T5: 150 mM NaCl; T6: 150 mM
NaCl + CM94. The mean
Also, significant increases (p
Similarly, the sugar content was also found to be remarkably (p
The effect of different salt stress with or without CM94 application on the
lipid peroxidation levels of the chickpea plants was studied by measuring the MDA
content. A significant (p
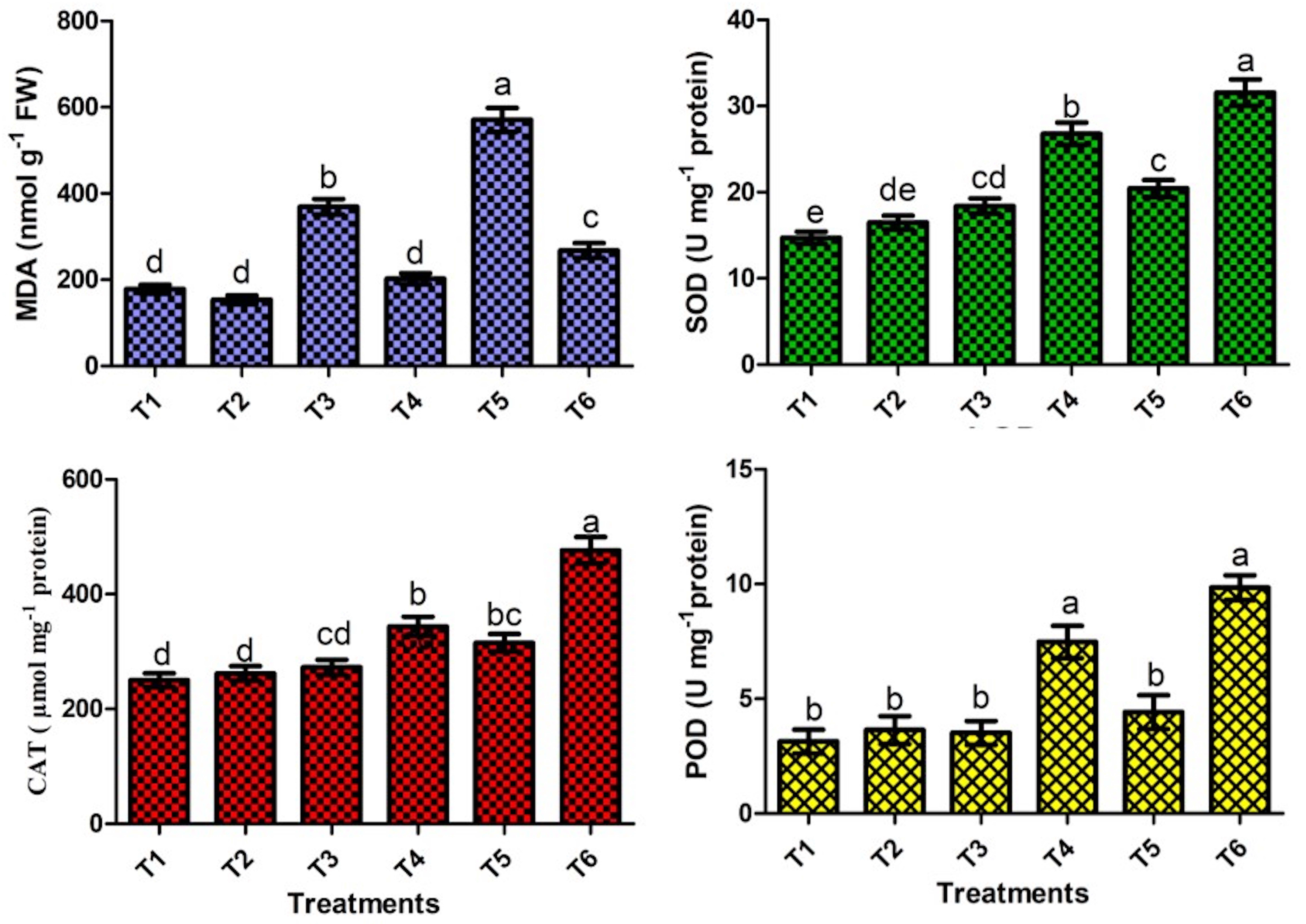 Fig. 6.
Fig. 6.Effect of Enterobacter sp. CM94 inoculation on MDA and
antioxidant enzyme activities in chickpea plants grown under different NaCl
concentrations. T1: uninoculated control (no NaCl); T2: CM94 inoculation; T3: 75
mM NaCl; T4: 75 mM NaCl + CM94; T5: 150 mM NaCl; T6: 150 mM NaCl + CM94. The mean
Modulation of antioxidant enzyme activities in response to CM94 inoculation was
determined at 0, 75, and 150 mM NaCl stress (Fig. 6). The results demonstrated
that as the salt stress increased from 75 mM (T-3) to 150 mM (T-5), all the
antioxidant enzyme activities increased in the NaCl-treated plants compared to
the uninoculated, non-stressed plants (T-1). Further, these antioxidant enzyme
activities were additively increased by the bacterial inoculation (Fig. 6). The
results showed that the activity of SOD increased by 25% and 38.9% in plants
treated with 75 mM and 150 mM NaCl, respectively, compared to the uninoculated,
non-stressed plants (T-1). However, compared to the uninoculated, stressed plants
(T3 and T5), the CM94 isolate significantly (p
From the roots of inoculated plants (both stressed and non-stressed), bacterial colonies were obtained on the antibiotic-supplemented NA plates. Conversely, no rhizobacterial cells were grown from the root samples of the uninoculated plants. Further, to confirm the identity of the obtained bacterial isolates, ERIC-PCR-based DNA fingerprinting was performed. It was observed that the obtained bacterial isolates from the inoculated root samples had the same ERIC-PCR banding patterns as the pure cultures of the wildtype CM94 isolate, thereby confirming the identity of the colonized bacteria (Fig. 7).
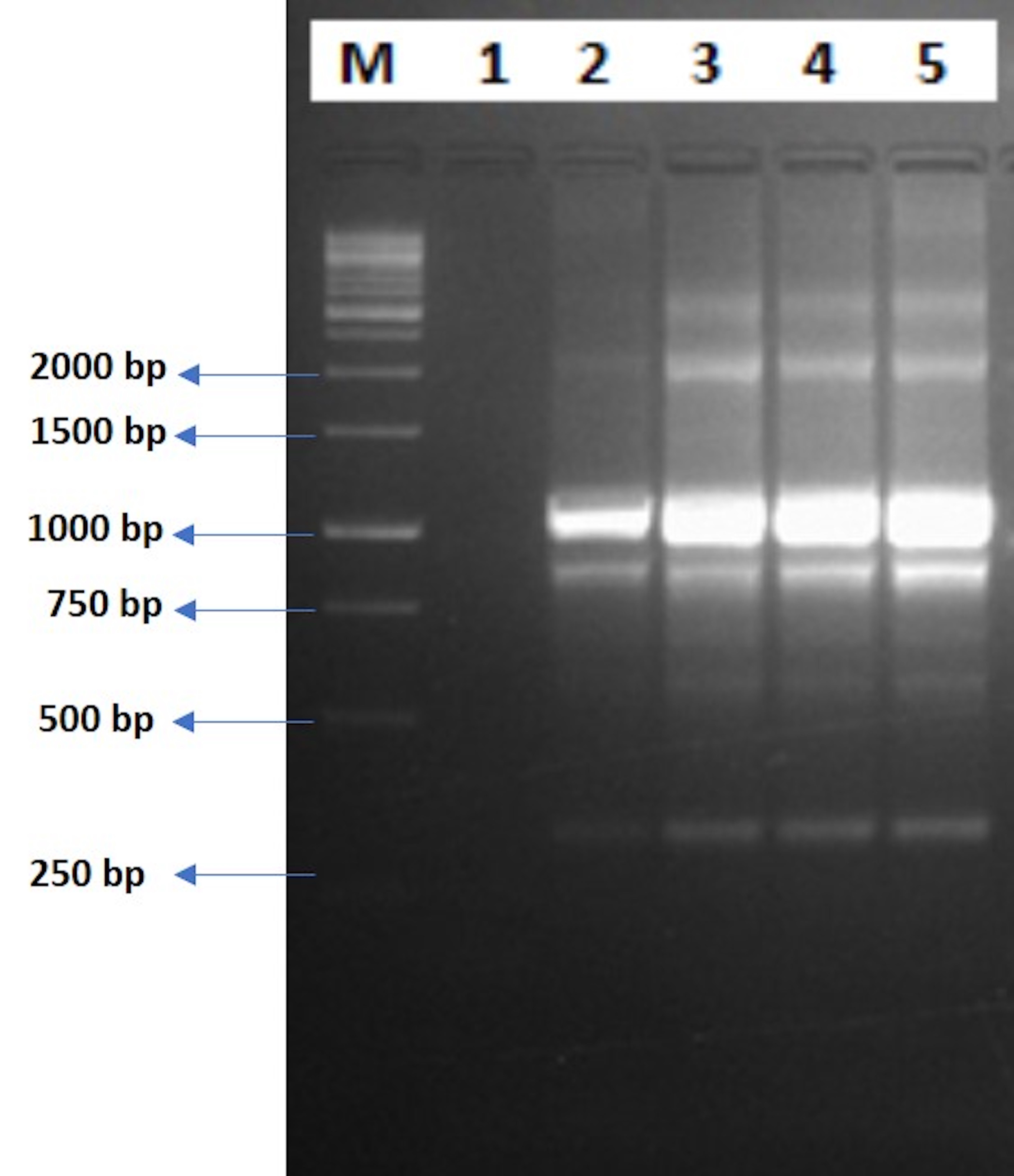 Fig. 7.
Fig. 7.Confirmation of ERIC-PCR-based root colonization. Lane 1: ‘M’: 1000 bp marker; lane 2: negative control; lane 3: wild CM94 isolate; lane 4: isolate obtained from 0 mM NaCl-treated CM94-inoculated plant (T2); lane 5: isolate obtained from 75 mM NaCl-treated CM94-inoculated plant (T4); lane 6: isolate obtained from 150 mM NaCl-treated CM94-inoculated plant (T6).
A significant part of India’s biological diversity is found in the fertile Indo-Gangetic Plain (IGP), along with the majority of its indigenous species. Additionally, the region has a complex geography with a large portion of infertile agricultural lands due to soil salinity. Improper management of the crops with indiscriminate and excessive application of synthetic chemical fertilizers further deteriorates crop production in such saline belts. In this context, PGPR has been considered as one of the possible alternatives for chemical fertilizers since they can mitigate the harmful effects of salinity in plants [47]. While screening novel isolates, it is crucial to consider their activity in a range of environmental conditions from where they are used. Therefore, it is of the utmost importance to study the indigenous rhizobacterial population in the same region where they will be employed as plant inoculants. Such climatic and temporal selection mechanisms will undoubtedly contribute to the evolution of a wide range of living beings, which are well adapted to shifting with climatic extremes. Thus, the search for salt-tolerant native rhizobacterial isolates in the present research work identified the salt-tolerant CM94 rhizobacterial isolate, which was isolated from the rhizosphere of chickpea plants grown in the salt-affected area of Uttar Pradesh. Isolated CM94 was able to tolerate and grow in conditions of up to 8% NaCl stress (Fig. 1). Hence, higher NaCl tolerances by CM94 in addition to PGP attributes can be used to enhance crop productivity in saline soils. The polyphasic characterization, based on the FAME (Fig. 2) and 16S rDNA sequence-based phylogenetic analyses, established the taxonomic position of the CM94 isolate as Enterobacter sp. (NCBI Accession Number: KC504003) (Fig. 3). Earlier studies showed that the rhizobacterial genus, such as Bacillus tended to be predominant in IGP saline soils [15]. However, less attention was provided to other genera, meaning they need to be further explored. Enterobacter species are an exceptionally diverse group of organisms that exhibit high levels of metabolic and genetic diversity. These bacterial groups possess multifarious plant growth-promoting traits, including the production of IAA, ACC deaminase, siderophore, ammonia, and HCN, as well as potentially improving the availability of important nutrients, such as phosphate, thereby making these bacteria very useful in the agriculture field [18, 21].
IAA is an important signaling molecule in microbe–plant interactions. IAA regulates cell division and lateral root elongation and has a major role in stress responses [48]. Soil salinity adversely affects the production of IAA in the roots and leaves of plants [49]. As a result, the reduced levels of IAA in the plant roots remarkably hampers the overall growth and development of plants. However, under salt stress conditions, some PGPR can synthesize IAA, which in turn, can enhance the root architecture and plant biomass [50]. Phytohormones, synthesized by microorganisms, induce the development of roots with higher surface area and biomass, which enables plants to extract more nutrients from saline soil. Masmoudi et al. [51] reported that the IAA producing Bacillus velezensis FMH2 significantly enhanced root length and lateral root production, and enhanced tomato plant growth under salt stress conditions. In congruence to this, the CM94 isolate also improved root length and biomass, which might be mainly attributed to IAA production.
ACC deaminase cleaves ACC (the immediate precursor of ethylene) into ammonia and
Nutritional imbalance is known to be a salinity-induced adverse effect on plant growth and production. Salt stress has adverse effects on P- uptake and transport in the plant. P fertilizers are generally suggested to be used for managing P deficiency in salt-affected soils. The use of salt-tolerant P-solubilizing-rhizobacteria can greatly improve P availability in saline soils. In the present study, isolated CM94 was found to be a highly efficient solubilizer of insoluble phosphates. The results are in accordance with earlier studies, where salt-tolerant PGPR strains representing various genera, such as Pseudomonas, Bacillus, Alcaligenes, Burkholderia, Pantoea, and Serratia, were able to solubilize phosphate and improve the growth of the plants [15, 53]. Furthermore, salt stress is a cause of iron deficiency and together they could impose a negative effect on the plant’s vegetative growth, stomatal conductance, photosynthesis, and transpiration rates [54]. Finding prospective siderophore-producing PGPR that are also salt-tolerant may be a beneficial non-transgenic method for reclaiming the salinity-affected soils for agriculture. In the present study, the CM94 isolate could produce a significant amount of siderophore, which was retained even at higher salinity levels. Thus, the siderophore-producing ability of microorganisms under stress conditions may be an auspicious alternative to chemical fertilizers and could aid in managing salt stress and iron limitations in salt-affected soils. Recently, a study by Sultana et al. [55] reported that salt-tolerant siderophore-producing PGPR supported rice growth.
The application of PGPR is one of the most prospective approaches to alleviate
salt stress in plants. When isolated CM94 was evaluated for growth promotion in
chickpea plants in the greenhouse, it displayed a significant increase in root
length, shoot length, and biomass. Particularly, increased root, shoot length,
and biomass are very important for improved nutrient uptake and are also
essential for coping with different environmental stresses, including salinity,
which is a serious problem in the IGP region of U.P. India. Li et al. [56] reported that salt-tolerant E. cloacae HSNJ4
significantly promoted vegetative growth, and improved photosynthetic pigment
content, and salinity tolerance by balancing the relative IAA and ethylene
content in plants. The findings in the present study are also in line with the
earlier reports where the use of salt-tolerant PGPR, such as
Sphingomonas, Bacillus, and Enterobacter enhanced the growth,
development, and yield of plants under saline conditions [8, 9, 14, 21]. It might
be due to improved nutrient uptake and water absorption, which helped plants cope
with the salinity stress. One of the possible mechanisms for mitigating the
salt-stress symptoms is the production of Na
In addition, an improvement in the physio–chemical parameters of plants was also observed in CM94-inoculated plants. The leaf chlorophyll content represents a primary indicator of photosynthesis, which correlates well with salt tolerance [58]. Under salt stress conditions, CM94-inoculation remarkably increased the chlorophyll content, compared to the respective uninoculated plants, which indicated an improvement in the photosynthetic activity. A similar increase in the plant’s photosynthetic activity and chlorophyll content under salt stress was also reported in earlier studies [59]. An improvement in photosynthetic activity is also one of the important contributors to enhancing shoot biomass.
Furthermore, in response to salinity stress, the accumulation of compatible solutes, such as proline, is a common stress-avoidance mechanism that guards the plant’s cell membrane, scavenges the free hydroxyl radicals, and stabilizes the structure of the proteins [60]. In addition, during NaCl stress, soluble sugar together with proline astonishingly maintained the osmotic equilibrium and formed around 50% of the total osmotic potential in plant cells [61]. Along with proline, soluble sugars, by making up 50% of their total osmotic potential, help plants maintain their osmotic homeostasis under NaCl stress. In the present study, inoculation of CM94 in chickpea plants enhanced the production of proline and sugar, which could significantly contribute to mitigating the salinity stress. Our results are in accordance with earlier studies, which also reported that the application of the Bacillus species improved the salt-stress tolerance and growth of wheat plants by modulating transpiration, photosynthesis, and proline accumulation [62, 63]. Furthermore, in PGPR-inoculated plants, the protein content was also increased under stressed and non-stressed conditions. The results are in congruence with the study by Sultana et al. [55], who reported that inoculation with the Pseudomonas strain provided differential protein concentrations in rice plants. The higher protein content is also related to improved photosynthesis, which in turn represents a function of higher chlorophyll content.
The malondialdehyde (MDA) concentration is also an indicator of oxidative injury under salinity stress and is directly related to the plant’s lipid peroxidation [64]. In the present work, it was observed that CM94 inoculation reduced the MDA level in stressed plants, compared to the uninoculated control plants. Reduced MDA levels by PGPR inoculation is an indication of a decrease in cell membrane damage, which enhances salt stress tolerance in plants. The results are consistent with the study by Singh and Jha [65], who reported that PGPR inoculation remarkably reduced the MDA content and alleviated lipid peroxidation in plants under stressed conditions.
Salt stress induces an unavoidable consequence of oxidative stress and generates
a higher amount of ROS, which poses a severe threat to the plant’s cells by
triggering lipid peroxidation, protein oxidation, nucleic acids, and enzyme
damage [66]. To detoxify excess ROS from cells, plants exhibit excellent
antioxidant enzymes, such as SOD, CAT, POD, APX, etc. These antioxidant enzymes
can scavenge the free radicals produced during oxidative stress [67]. Activation
of SOD leads to the conversion of O
Generally, the benefits of PGPR inoculation depend on its effective root colonization. Therefore, in the present study the colonization efficacy of the potential PGPR isolate CM94 was assayed following antibiotic-resistant and ERIC fingerprinting approaches. The initial and most important stage of a plant-microbe relationship is root colonization, during which bacteria travel toward the rhizosphere in response to root exudates. Antibiotic-resistance markers can be particularly helpful for colonization studies of PGPR isolates. In the present study, by using the combination of antibiotics amended in NA, the inoculated PGPR CM94 isolates were retrieved. Contrastingly, no bacterial colony was obtained from the uninoculated plants. These results showed that CM94 efficiently colonized chickpea roots. Further, the identity of the colonized bacterial isolates was confirmed by comparing the DNA fingerprints of wildtype PGPR CM94 isolate and the root colonized bacteria by ERIC-PCR. The results of the present study are in accordance with the study by Singh et al. [65], where they proved bacterial colonization through ERIC-PCR DNA fingerprinting analysis.
The results of the present study revealed that Enterobacter sp. CM94, with a salinity tolerance, possesses multiple plant growth attributes, which are even retained at higher salt concentrations. Inoculation with the CM94 isolate alleviated the salt stress and induced systemic tolerance in chickpea plants by significantly improving their growth and biochemical parameters as well as the expression of ROS-scavenging antioxidant enzymes. The results demonstrated that the utilization of such PGPR would enable a decrease in the use of chemical fertilizers, which will in turn help to achieve sustainable production of chickpea plants in the saline soils of eastern Uttar Pradesh, India. However, future studies are required to determine the nature of the isolate under different salt-affected areas and to harness their inherent positive capability as bioinoculants in farmer’s fields.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
AS, RK, and NK, conceptualized the idea and designed the experiments. AS, executed all the laboratory experiments, analyzed the data, and wrote the manuscript. AV, helped in software and data analysis. HC and AKS, helped in methodology and validation of results. YKB, helped in formal analysis, visualization, and investigation. NK and RK, helped in formal analysis, funding acquisition, writing, review, and editing. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
The chickpea plant seeds used in this study were procured from ICAR-Indian Institute of Seed Science (IISS), Mau Nath, Bhanjan (U.P.), India.
The authors are thankful to Director, ICAR-NBAIM, Mau Nath Bhanjan (U.P.) for providing necessary resources and facilities for this work.
This research received no external funding.
The authors declare that there is no conflict of interest. Given the role as Guest Editor and Editorial Board Member, Naeem Khan had no involvement in the peer-review of this article and has no access to information regarding its peer-review. Full responsibility for the editorial process for this article was delegated to Jorge M.L. Marques da Silva.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
