- Academic Editor
†These authors contributed equally.
Background: Adinandra nitida, commonly known as Shiya tea, is
a healthcare drink enriched in several phenolic acids and flavonoids, with a
purple-red leaf variety possessing a unique flavor and a higher economic value.
However, the mechanisms underlying leaf coloration and senescence discoloration
remain unknown. Methods: Here, we compared both varieties of A.
nitida (purple-red leaf, RL, and green leaf, GL) at two stages of development.
To make sure the difference in leaf color in these four groups, several indexes,
leaf colorimetric differences, H
Shiya tea (Adinandra nitida) was originally cultivated in southwest China. In the Guangxi Province, it is a widely consumed traditional tea, the cultivation of which has become increasingly common worldwide [1]. The tea is prepared from young leaves harvested from the top of the shoots, with an annual global harvest yield of nearly 5 million tons [2]. The demand for beneficial compounds in young leaves has been steadily increasing with the increasing popularity of tea drinks [3]. Ceylon tea from Sri Lanka, acclaimed as the best tea in the world, has its inherent unique characteristics and reputation for over a century [4]. Compared with Sri Lankan tea, which only contains 12–24% flavonoids by weight, Shiya tea contains more than 20% [5, 6]. Additionally, Shiya tea possesses a higher concentration (20.9%) of polyphenols than tea plants, which contain 18–36% polyphenols in young shoots [7]. Moreover, the caffeine content in Shiya tea is two to five times lower compared to that in the tea plant, making it an appropriate drink for people with caffeine sensitivity and caffeine-induced insomnia. Moreover, it is an economic crop as a class of drink that produces numerous secondary metabolites, such as flavonoids and alkaloids, that boost immunity [3].
Leaf color is a critical trait for plants with edible leaves with respect to their nutrition, taste, and physiological resistance. Tea obtained from tea plants with purple leaves, called “special tea”, receives increased global attention owing to the pleasant taste and rich anthocyanin content; it contains increased nutrient compounds, thus fetching premium prices to farmers [8]. The synthesis and accumulation of plant secondary metabolites such as carotenoids, flavonoids, and betalains contribute to the leaf and flower color. Anthocyanins, a type of flavonoid, are the best-characterized plant secondary metabolites in terms of coloration, biochemistry, and molecular biology and were found to be responsible for red, blue, and purple pigmentation in petals, leaves, seeds, and fruits [9]. They are bioactive compounds that scavenge free radicals and reactive oxygen species (ROS), provide endurance against high light intensity, cold temperatures, pest attack, and pathogen infection, and contribute to pollen and seed dispersal by attracting birds and bees [10]. There are six main anthocyanidins in vascular plants, including pelargonidin, peonidin, petunidin, cyanidin, malvidin, and delphinidin, with red, purple, and magenta colors. The up-growth in the omics field established more accurately the link among genomic, transcriptomic, proteomic, and metabolomics data throughout the last decade. Integrating metabolome and transcriptome analysis, structure genes (UGT78D2, MYB12) were clarified to facilitate anthocyanins accumulation in purple leaflets of Camellia tachangensis [11]. Similar research found ANS critically related to anthocyanin content while several MYB TFs positively correlated to this gene; these genes co-regulated anthocyanin accumulation in the sepal of Heptacodium miconioides [12]. Therefore, transcriptome and metabolome integrative researches are a reliable approach for metabolic network analysis [13, 14, 15].
However, the mechanisms underlying leaf coloration and senescence discoloration remain unknown. In this study, we aimed to identify the difference between the two kinds of leaf colors at the transcriptional and metabolite levels to elucidate the different regulatory modules involved in the anthocyanidin biosynthesis pathway in the two leaf colors at two different leaf ages. To this end, we performed an integrated metabolomic and transcriptomic analysis of the green and purple Shiya tea leaves, of two different ages, from young shoots. The results of our study provide new insights into leaf color changes in A. nitida, which can aid in tea flavor improvement during Shiya tea breeding.
The Shiya tea (Adinandra nitida) leaves of purple and green color were
derived from Guangxi, Guangdong, Guizhou province from cliffs with short-day,
high-humidity, and low-temperature conditions. The Shiya tea leaves in this study
were harvested from Xinyi Forestry Research Institute, Guangdong province, China
(N22°19
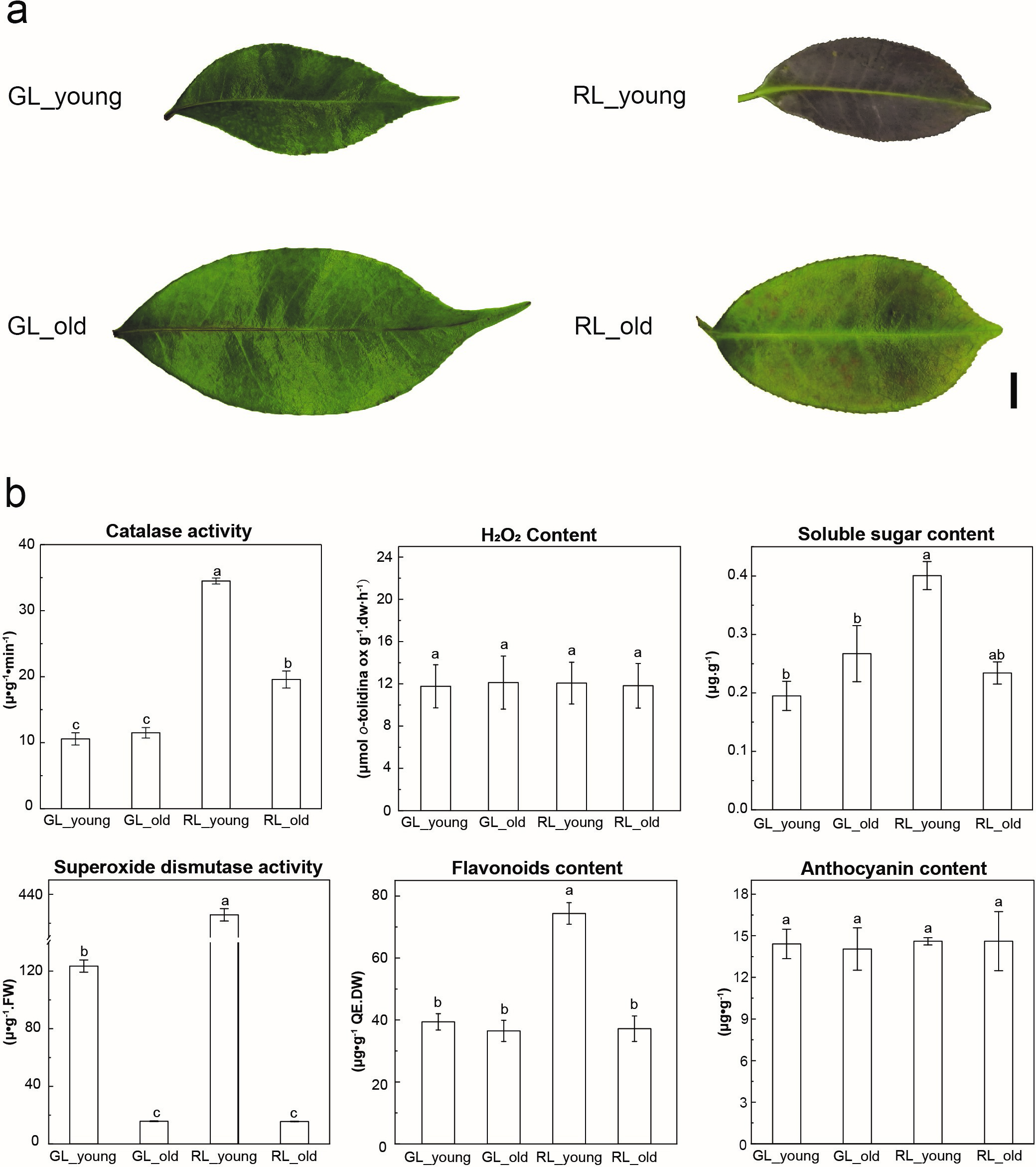 Fig. 1.
Fig. 1.Characteristics of phenotype and physiological traits at
different growth stages of red and green colored Adinandra nitida
leaves. (a) Shiya tea leaves at two different developmental stages in different
colors (RL, red leaves; GL, green leaves; young, young stage, 15 d; old, old
stage, 45 d). Scale bar = 1 cm. (b) Changes in the physiological traits of Shiya
tea leaves in the four groups. Different lower-case letters indicate a
significant difference (p
Approximately 0.2 g fresh leaf segments collected from each sample were used for
the determination of several physiological indices, including colorimetric
difference, superoxide dismutase (SOD), catalase (CAT), hydrogen peroxide
(H
H
Total flavonoid content was determined using Aluminum Chloride (AlCl
Anthocyanins were extracted according to the method of Zhang et al.
[20]. About 2.0 g of sample was immersed in 5 mL of 1% (v/v) methanol-HCl
overnight in the dark at 24 ℃, then the absorbance of each sample was measured at
530, 620, and 650 nm by a spectrophotometer (UV-1600, Shimadzu, Kyoto, Japan).
The anthocyanin content was quantified using the following formula: OD = (A530 –
A620) – 0.1
Proanthocyanin content determination was imitated by the method from
Hrnčič et al. [21]. About 2.0 g of the sample was grounded to
powder using liquid nitrogen. Then the powder was transferred into the reagent
prepared by 77 mg of Fe(SO
The activity of laccase (LAC) was estimated to imitate Fioretto et al. [22].
First, 0.5 g leaf sample was crushed to a fine powder using the liquid nitrogen
grinding method and transferred into a centrifuge tube, suspended in 10 mL of
buffer (50 mM acetate pH 5.0) and kept at 4 ℃. After incubation, the mixture was
homogenized for 1 min, centrifuged at 10,000
Peroxidase (POD) activity was determined using 4-methylcatechol as substrate.
The increase in the absorption caused by the oxidation of 4-methylcatechol by
H
Twelve leaf samples from four groups with three independent biological
replicates were used for metabolome analysis. The freeze-dried samples were
crushed into powder using a mixer mill (MM 400, Retsch, Haan, Germany) with a
zirconia bead for 1.5 min at 30 Hz. Each 100 mg powder was dissolved in 1.2 mL 70
% methanol solution, vortexed 30 s every 30 min, repeating for a total of six
times, and finally placed in a refrigerator at 4 ℃ overnight. Following
centrifugation at 15,984
The analytical conditions were as follows, UPLC: column, Agilent SB-C18 (1.8
µm, 2.1 mm
Total RNA was extracted from the frozen leaf samples using TRIzol reagent
(Invitrogen, Carlsbad, CA, United States) following the manufacturer’s
instructions and then subjected to quantification and qualification by agarose
gel electrophoresis, an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo
Alto, CA, United States) and an RNA Nano 6000 Assay Kit of the Bioanalyzer 2100
system (Agilent Technologies, Palo Alto, CA, United States). Briefly, mRNA was
purified from total RNA using oligo dT attached magnetic beads, and then
first-strand cDNA was synthesized using hexamer and M-MuLV Reverse Transcriptase
(RNase H-), and the second-strand cDNA was synthesized using DNA Polymerase I and
RNase H. Finally, the amplified size-selected cDNA was purified (AMPure XP
system) and the cDNA libraries were sequenced using the Illumina sequencing
platform by Metware Biotechnology Co., Ltd. (Wuhan, China). Clean reads were
filtered, mainly to remove row reads and low-quality (Q
The DEGs and DAMs in the same group were mapped together to the KEGG pathway map
and screened using a p
Single-stranded cDNAs were synthesized from the RNAs using the PrimeScriptTM RT
reagent Kit, and quantitative real-time PCR was performed using ViiA7 (Thermo
Fisher Scientific, USA) and HieffTM qPCR SYBR Green Master Mix (Yisheng
Biotechnology, Shanghai, China). Primers were designed by Bioruqi (Guangzhou,
China) and synthesized by GENEWIZ (Suzhou, China). The AnActin gene
(LOC114296039) was used as an internal reference, and the relative expression was
calculated using the 2
The statistical significance of the difference between RL and GL in the physiological indices was tested by Duncan’s test using SPSS software (v17.0, IBM Corp., Chicago, IL, USA). The metabolite data and transcript expression were standardized to the Z score using TBtools software. We analyzed and compared the differences among the four sample groups (RL_young_vs._RL_old, GL_young_vs._GL_old, RL_young_vs._GL_young, RL_old_vs._GL_old). The correlation matrix using Pearson’s correlation with significance level was calculated and produced using the LinkET package (v0.0.5) in R Studio (v2022.07.1+554, RStudio, Inc., Boston, MA, USA).
The high-altitude growth region for A. nitida promotes the synthesis and accumulation of nitrogen-containing compounds and aromatic substances, with high protein and amino acid content in plants, rendering A. nitida favorable in taste. A purple-red A. nitida cultivar is popular among its peers for its unique red leaves and multi-stress tolerances. We investigated whether stress tolerance was associated with the red-leaf trait by analyzing a purple-red A. nitida cultivar ‘RL’ at two stages of development (young leaflets and mature leaves) and its green counterpart-‘GL’ as a control.
First, we plotted the absolute color coordinates and differences between the two varieties by applying CIE Lab color space consisting of three metrics, L*: lightness (+brighter, –darker), a*: redness–greenness (+redder, –greener), and b*: yellowness–blueness (+yellower, –bluer). As shown in Supplementary Table 3, we observed distinct a* values between the two varieties, while neither L* nor b* values presented significant differences, reflecting the “redder” hue in RL.
We further evaluated the intensity of red-causing components by extracting total flavonoids from plant leaves, followed by spectrophotometric determination. As expected, the TFC of RL_young was significantly higher than that of GL and RL_old (Fig. 1b). Moreover, the TFC of the “old” leaf was reduced compared with that of the younger leaf, indicating a mild fading trend with leaf development. This result indicated that a higher content of flavonoids was predominantly related to the red hue in RL.
Enhanced soluble sugar content and ROS scavenger occurred commonly at the leaf
behaving deeper color [28]. Moreover, we determined several physio-chemical
indices of stress resistance, including SOD, CAT, and soluble sugar content (Fig. 1b). The samples similarly suffered the oxygen partial pressure in these four
groups by quantifying the H
As a result, the soluble sugar content and ROS scavenger levels, including SOD and CAT, increased in RL and were positively correlated with flavonoid content. Visual observation and colorimetric determination suggested that anthocyanin (ACN) belonging to flavonoids may act crucially at the leaf pigmentation, and the ACN content may contribute to abiotic damage resistance, consistent with previous studies that reported on low temperature, drought, salinity, and heavy-metal stresses [29]. However, the ACN contents in these four groups were similar but not significantly different. Whether ACN content contributes to the Adinandra nitida leaf color needs to be further confirmed. Next step, we would notarize that using metabolomic data analysis.
The colorimetric and physio-chemical differences between RL and GL prompted us to investigate further the detailed metabolic data related to these traits. Thus, we performed Ultra performance liquid chromatography–tandem mass spectrometer (UPLC-MS/MS) based metabolome profile of RL and GL, focusing on differential pigments (see Materials and Methods).
First, principal component analysis (PCA) of the metabolite data (the normalized responses were used, accounting for 53.83% of the variation with a significance of 0.01) separated the two varieties in metabolite composition and content (Fig. 2a), indicating that leaf color differences existed within the RL and GL at the metabolite level.
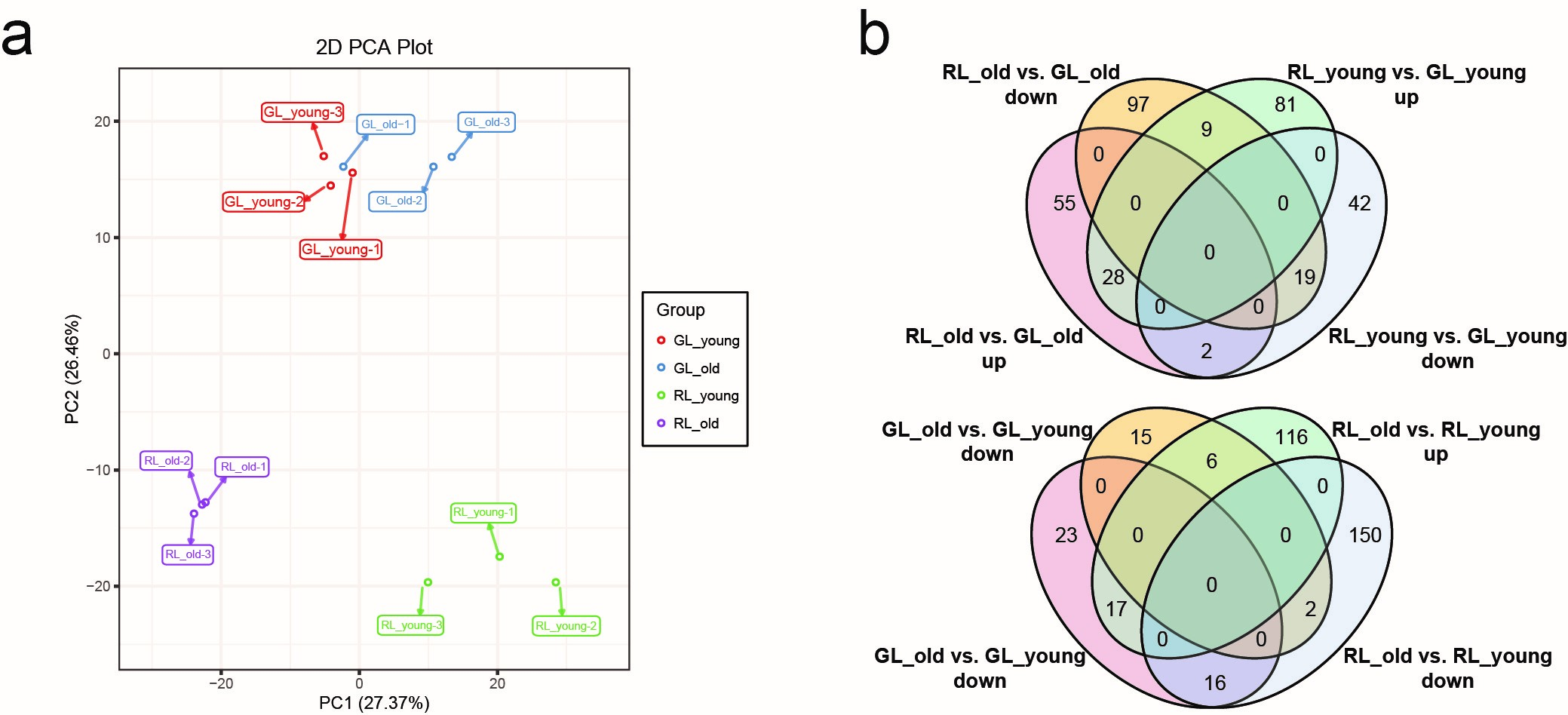 Fig. 2.
Fig. 2.Multivariate statistical analyses of the differences of metabolites in Adinandra nitida leaves. (a) Principal component analysis (PCA) of metabolite profiles. The x-axis represents the first principal component, and the y-axis represents the second principal component. (b) Venn diagrams of significantly upregulated and downregulated metabolites in four groups of Shiya tea leaves between different leaf colors (redness and greenness) and different developmental stages (young leaf and old leaf).
A total of 1079 distinct annotated metabolites were detected and classified into 11 main groups, including such detected anthocyanins and proanthocyanins (PAs) detected (cyanidin-3-O-glucoside, peonidin-3-O-glucoside, theaflavin, procyanidin A1, procyanidin A2, procyanidin A6, procyanidin B1, procyanidin B2, procyanidin B3, procyanidin B4, aesculitannin A, procyanidin C1, and procyanidin C2). However, the anthocyanins detected in this study (peonidin-3-O-glucoside and cyanidin-3-O-glucoside) expressed little difference. It is worth noting that the proanthocyanins found were all significantly different between RL and GL, likely contributing to ‘red’-hue due to their high enrichment in RL (Fig. 3 and Supplementary Tables 4,5). Phenolic acids and flavonoids were the two subclasses with the maximum number of relative upregulated metabolites in RL. Moreover, the upregulated organic acids were increased in old-stage RL since these compounds are one of the primary compositions in tissue formation [30].
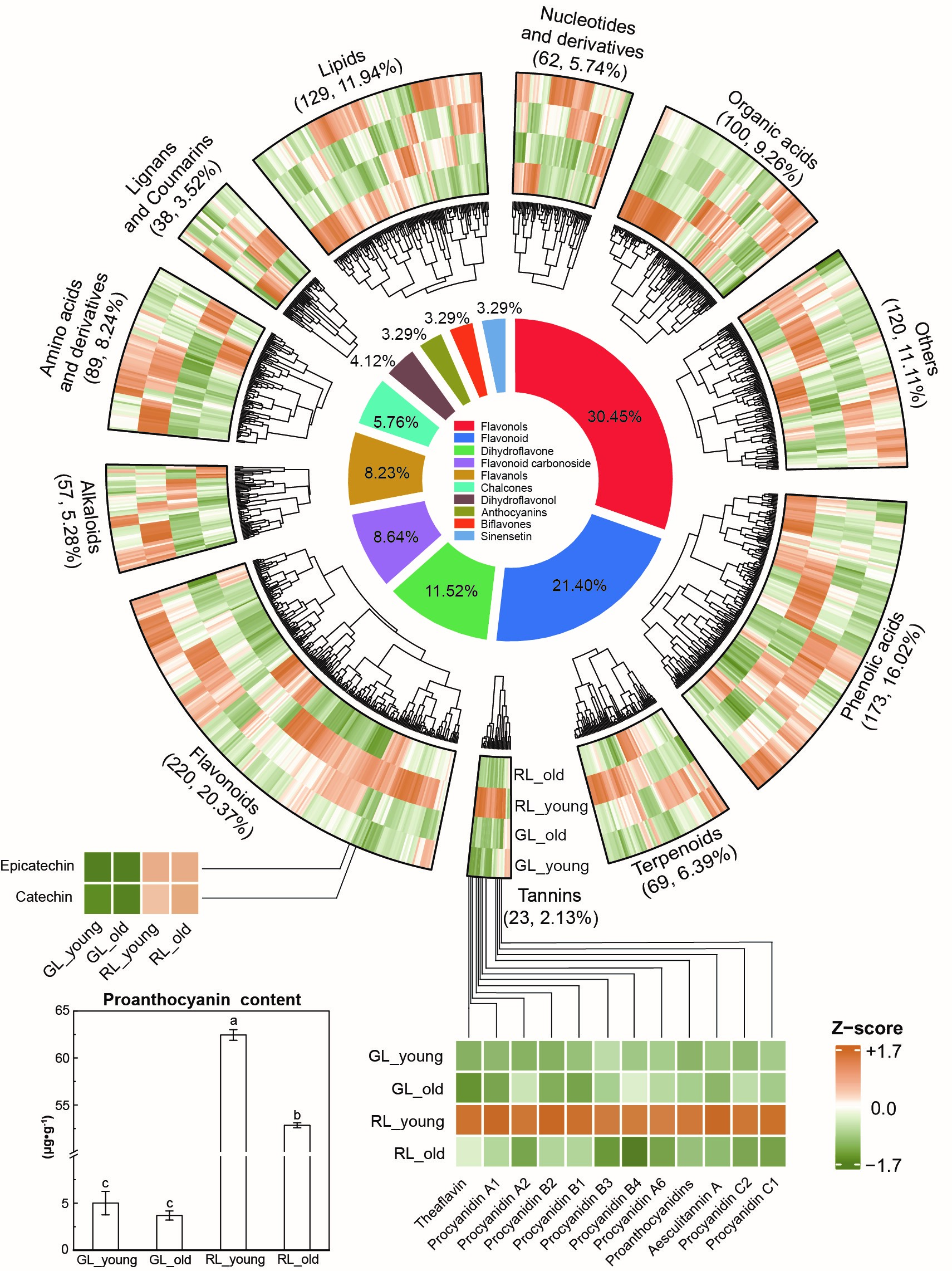 Fig. 3.
Fig. 3.Heatmap of metabolite expression profiles at different growth stages of red and green-colored Adinandra nitida leaves and the circle graph of percentage composition of the flavonoids category. The primary class, metabolite number, and percentage contained in the outer round correspond to the split sectors in the circular heatmap. The dendrograms in the third round denote the overall similarity of metabolite expression profiles by primary classes and the four groups. The proanthocyanins identified are shown in the bottom heatmap connected to the sectors by full lines. a, b, c, and d have achieved statistical significance at the 0.05 level, with p-values below 0.05.
We performed statistical tests to confirm key trait-related metabolites and
screened metabolites with a VIP
Taken together, the ‘red’ color of RL leaves potentially results from the oxidation of 11 PAs, amongst which procyanidin C1 and procyanidin C2 were specifically accumulated in young leaves resulting in their redder coloration. This result may be caused by the different capacities of polymerization of polymerase or oxidase since the proanthocyanins were condensed by CC and ECC. The same results were discovered in red rice (Oryza sativa), which deepened redness in rice caused by the polymeric structures that range from 2-mers to 14-mers [31]. As a natural ROS scavenger, PAs represent a group of condensed tannins [32] and not only render RL highly tolerant against oxidation-induced stresses, which is consistent with variations in the physio-chemical indices of the two varieties (Fig. 1b) but played an extensive health-promoting effect in our body.
The quality of the total RNA extracted is shown in Supplementary Table
6. The transcriptome profiles of RL and GL were acquired using Illumina NovaSeq
6000 to decipher the transcriptional regulation of DAMs between the two
varieties relating to the ‘red’ trait. Three independent biological
replicates per group were analyzed (12 samples), outputting more than 5.93
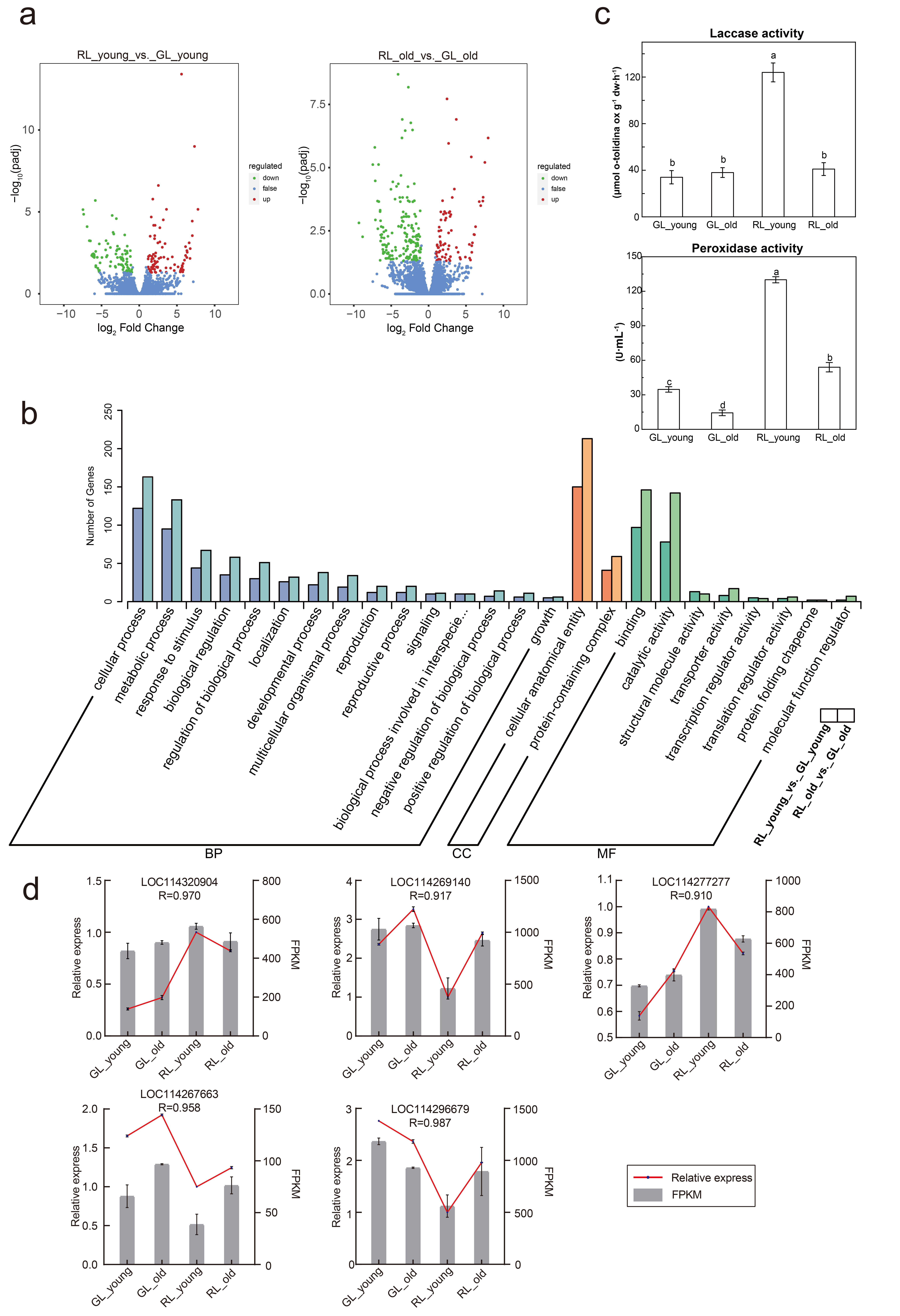 Fig. 4.
Fig. 4.Transcriptome data analysis in four groups of Adinandra
nitida leaves and quantitative real-time polymerase chain reaction (qRT-PCR)
verifying proanthocyanin biosynthesis-related genes. (a) Volcano plots of
transcripts between different leaf colors at the same developmental stage.
Transcripts with adjusted p-value
Gene Ontology (GO) functional enrichment analyses allotted 192 (in RL_young_vs._GL_young) and 258 (in RL_old_vs._GL_old) DEGs into biological processes (78.6% and 76.1%), molecular functions (86.7% and 84.0%), and cellular components (77.5% and 87.7%). In the biological processes term, genes associated with cellular processes were highly enriched. Additionally, most of the DEGs in molecular function terms of binding and catalytic activity were particularly highlighted, and the most abundant GO terms in the cellular component category were concentrated in the cellular anatomical entity (Fig. 4b and Supplementary Table 8).
Finally, the RNA-seq results were validated using qRT-PCR of five randomly chosen DEGs. The expression patterns of these genes obtained using qRT-PCR were consistent with those determined by RNA-seq (Fig. 4d).
We mapped the metabolic compounds and catalytic enzymes onto the pathway based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, focusing on the flavonoid biosynthetic pathway, to perform an in-depth analysis of the DAMs and DEGs involved in the red discoloration mechanism. According to the KEGG-enrichment analysis, 94 (in RL_young_vs._GL_young) and 122 (in RL_old_vs._GL_old) unigenes were clustered into 101 and 111 KEGG pathways. Among these pathways, phenylalanine, tyrosine, and tryptophan biosynthesis pathways were significantly enriched, indicating that 3-dehydroshikimate was activated prior to flavonoid biosynthesis to provide vital precursors [33].
Structural genes involved in the flavonoid biosynthetic pathway aiming at proanthocyanin biosynthesis were identified. The predicted proteins encoded by genes included two anthocyanin reductases (LOC114259095, LOC114297899), five multidrug and toxic compound extrusion proteins (LOC114273949, LOC114288619, LOC114298825, LOC114300598, LOC114302652), three peroxidases (LOC114255969, LOC114264567, LOC114309410) and two laccases (LOC114278581, LOC114290795). In the ‘red’ color of leaves, the structural genes of three PODs (LOC114255969, LOC114264567, LOC114309410) and two LACs were significantly upregulated at RL_young with a verifiable measurement of POD and LAC activities (Fig. 4c), while two ANRs, five MATEs were expressed with no significant difference.
Synthesis of PAs begins with the generation of flavan-3-ol units (e.g., catechin and epicatechin). The key enzymes, LAR and ANR, convert leucocyanidins and anthocyanidins to catechin and epicatechin, respectively. Additionally, the two ANR structure genes detected in this study were significantly different between green leavf groups and red leaf groups, though they showed ordinarily inner RL groups. That is to say; the downregulated ANRs caused little or no PAs to exist in GL. This result has been identified similarly in Arabidopsis [34], grapevine [35], tea [36], and soybean [37].
Based on the co-expression analysis of differentially accumulated flavonoids and DEGs, we identified several catalytic enzymes involved in biosynthetic pathways to eleven PAs. The upregulation of genes encoding POD64 (LOC114309410) and LAC17 (LOC114290795) may result in a higher accumulation of proanthocyanins in RL, especially in RL_young (Fig. 5b and Supplementary Table 9). In the ‘red’ leaflets, the high expression level of peroxidases (PODs) and laccases (LACs) against RL_old could be explained by the fact that oxidative polymerization of PAs dimers (procyanidin) was catalyzed by these enzymes that were homologous to LAC15, POD2, and POD25 in Arabidopsis [38].
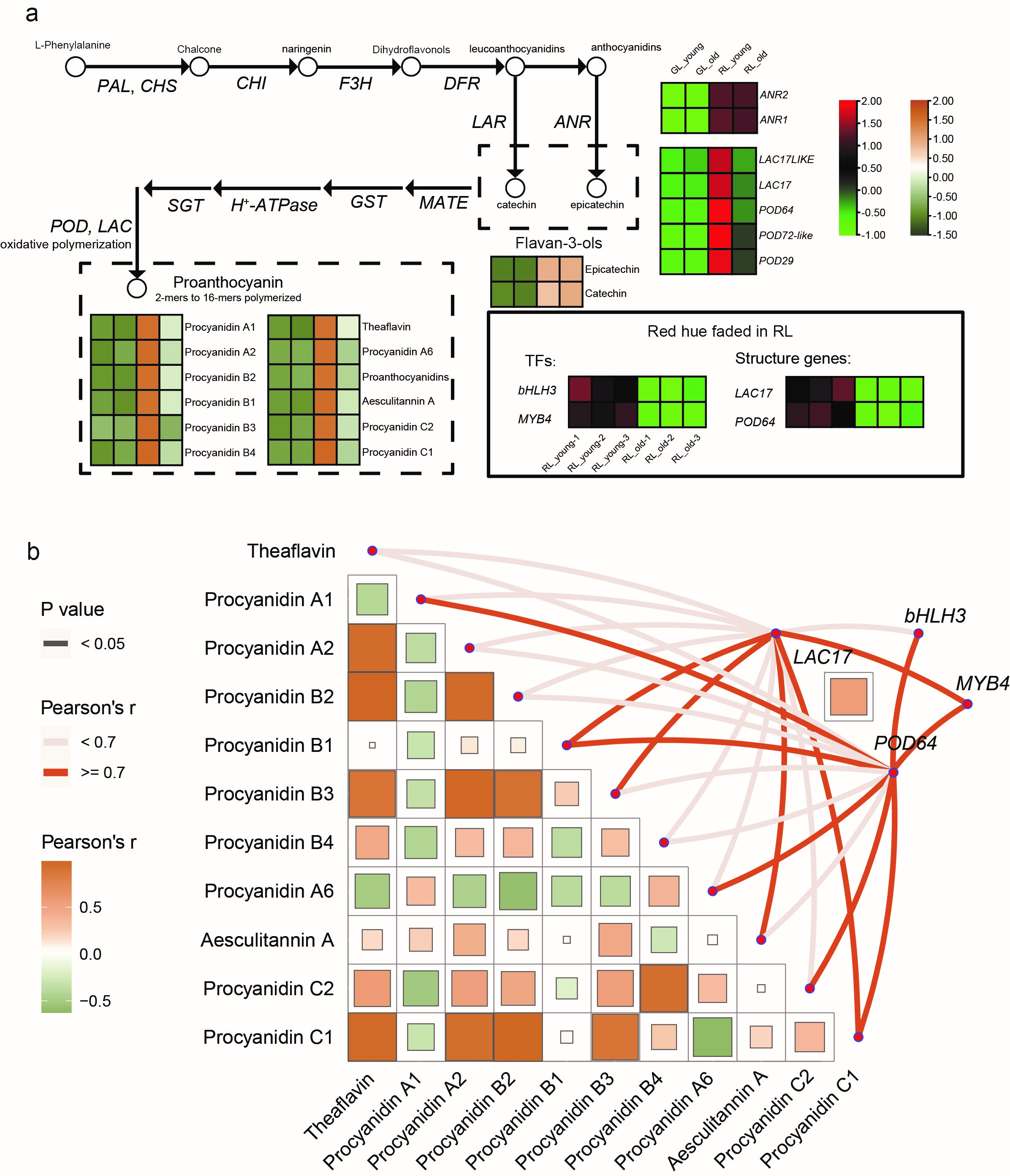 Fig. 5.
Fig. 5.Simplified model of proanthocyanin biosynthesis pathway in
Adinandra nitida and the correlation comparisons of metabolites, key
enzymes, and transcription factors in proanthocyanin biosynthesis pathway at red
leaves of Adinandra nitida for discoloration exploration. (a)
Comparison of the differential metabolites and differential genes in anthocyanin
biosynthesis pathway in four groups of leaves. The heatmaps show the expression
pattern; the metabolite expression patterns were based on the Z-score in “brown
- white - green”, and the gene expression patterns were based on the Z-score bar
in “red - black - green”. (b) Pairwise comparisons of metabolites are shown,
with a color gradient denoting Pearson’s correlation coefficients. The genes
corresponding to the enzymes in this pathway were related to each metabolite by
correlation and significance analysis. Edge width denotes the statistical
significance based on p
Proanthocyanin biosynthesis is usually regulated by various transcription
factors (TFs), particularly the MBW complex [39]. We further explored the TFs,
including MYBs (MYB4, MYB61, MYB308,
MYBCDC5, MYBCC, MYB3R-5, MYB-like 4, and
MYBCD5) and bHLHs (bHLH1, bHLH3,
bHLH130, bHLH104, and bHLH116), regulating flavonoids
metabolism in RL by performing another co-expression network analysis to direct
differentially expressed TFs toward flavonoids biosynthesis and polymerization
closely related enzymes (CHI, LDOX, ANR, MATE, H
As did POD64 and LAC17, the synthesis of procyanidin C1 and C2 significantly correlated to the expressions quantity of POD64 and LAC17 (Fig. 5b), which could condense the catechin or epicatechin to a high degree of procyanidin polymerization products [47]. Multiple varieties of proanthocyanins possess essential roles in the growth and development of leaves and fruits [3]. Colorless PAs are converted to red-brown PA derivatives due to the oxidative polymerization of monomers by PODs and LACs [41]. Trimer procyanidin C1 and C2 contain a higher intensity of UV-Vis absorption than the lower polymerization degree of procyanidins like dimer procyanidin B-type, which suggests that the degree of procyanidin polymerization significantly influences the color-deepening phenomenon [30]. Therefore, the downregulation of these two catalysts (POD64 and LAC17) in old leaves was considered the driving factor for the decreasing trend in PA content (Fig. 5a). This result is common in other plants, including bilberries, persimmons, and blueberries [48]. However, several studies have indicated that partial PODs and LACs are responsible for the degradation of anthocyanins in Brunfelsia calycina and Pyrus bretschneideri [49].
In summary, PAs deemed as tannins causing red coloration in RL leaves were regulated by flavonoid biosynthesis pathways (ANR, LAC, MYB4, and bHLH3). The expression of the key gene ANR strongly influenced the flavan-3-ol accumulation in PA biosynthesis. That could be one of the key points resulting in the different leaf colors of Shiya tea varieties. An increased degree of catechin/epicatechin polymerization in the chromophore of PAs in RL could be induced by oxidative polymerase [50]. And higher polymerization degree of PAs may result in color deepening. The activities or the expressions of oxidative polymerases, POD64 and LAC17, were affected directly or indirectly by the polymerization of PAs.
To understand the altered metabolite compositions contributing to leaf coloration in red and green leaf tea, we associated significant changes in redness-related metabolites with the expression of structural genes involved in flavonoids biosynthesis pathway and associated TFs (bHLH3 and MYB4) at two stages of development of Adinandra nitida leaves. The accumulations of flavonoids and tannins, especially the increased accumulation of PAs in young stage of RL, were observed. Integrated with the selected enzyme genes and TFs, our results revealed that six structural genes found in proanthocyanin biosynthesis, two reductases (ANR), two oxidative polymerases (POD64, LAC17) and two TFs (bHLH3 and MYB4) related to biosynthesis and polymerization of proanthocyanins were associated with not only the difference of GL and RL but also the faded coloration in two RL groups (RL_young and RL_old). Among these genes, the enzymes (POD64 and LAC17) and TFs (bHLH3 and MYB4) were positively correlated to the accumulations of PAs in these two groups. Moreover, several oxidative polymerases were downregulated in the old stage of RL compared to its young stage, though the flavan-3-ol exhibits little difference in these two groups. Thus, these results provide explanations for the appearance of a red hue in RL and the faded red hue in the old stage of RL. This study provides a foundation for further research to investigate the PA biosynthesis regulatory networks via transcriptome and metabolome analysis in Adinandra nitida, offering insights into the regulatory mechanisms underlying beneficial metabolite accumulation in plants.
ACNs, Anthocyanins; ANR, Anthocyanin Reductase; bHLH, Basic Helix-Loop-Helix; CAT, Catalase; CE, Collision Energy; CHI, Chalcone Isomerase; CUR, Curtain Gas; DAMs, Differentially Accumulated Metabolites; DEGs, Differentially Expressed Genes; DP, Declustering Potential; FPKM, Fragments Per Kilobase of Transcript Per Million Fragments Mapped; GL, Green leaf; GO, Gene Ontology; GST, Glutathione S-Transferase; GT, Glycosylation; KEGG, Kyoto Encyclopedia of Genes and Genomes; LAC, Laccase; LAR, Leucoanthocyanidin Reductase; LDOX, Leucoanthocyanidin Dioxygenase; MATE, Toxic Compound Extrusion Transporter; MRM, Multiple Reaction Monitoring method; MT, thylation; PAL, Phenylalanine Ammonialyase; PAs, Proanthocyanins; PCA, Principal Component Analysis; POD, Peroxidase; QQQ, Triple Quadrupole scans; qRT-PCR, Quantitative Real Time Polymerase Chain Reaction; QTRAP, Quadrupole-Linear Ion Trap; RL, Purple-red leaf; ROS, Reactive Oxygen Species; SGT, UDP-Glycosyltransferase superfamily protein; SOD, Superoxide Dismutase; TFs, Transcription factors; UFGT, UDP Glucose-Flavonoid 3-O-Glucosyl-Transferase; UPLC-MS/MS, Ultra performance liquid chromatography - tandem mass spectrometer; VIP, Variable Importance in Projection.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, SAMN32601060 - SAMN32601083.
Conceptualization, YL, JAH, DZ and BZ; methodology, YL, JAH, DZ and BZ; validation, RH, YS and HS; formal analysis, RH, YS and HS; investigation, RH, YS, HS, WL, JXH and ZK; resources, WL, JXH and ZK; data curation, RH, YS and HS; writing—original draft preparation, RH, YS and HS; writing—review and editing, RH, YS, HS, WL, JXH, ZK, YL, JAH, DZ and BZ; visualization, YL, JAH. and DZ; supervision, BZ; project administration, DZ and BZ; funding acquisition, JAH, DZ and BZ. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Shiya tea (Adinandra nitida) plants were used in this study. The purple-red leaf (RL) and green leaf (GL) varieties selected from Xinyi Academy of Forestry (Maoming, Gungdong, China)
We thank Wuhan Metware Biotechnology for assistance with metabolic data analysis.
This research was funded by the Provincial Natural Science Foundation of Guangdong, grant number 2021A1515110296, the Commercial Research Funds, grant number D12120260, and the Guangzhou Municipal Science and Technology Project, grant number 202002020028.
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
