†These authors contributed equally.
Academic Editor: Graham Pawelec
Background: Long noncoding RNAs (lncRNAs) are closely associated with the initiation, progression, metastasis, and recurrence of hepatocellular carcinoma (HCC). They could therefore serve as markers for the early diagnosis and for the prognosis of HCC patients. Methods: This was an observational prospective cohort study. A total of 101 participants were included, comprising patients with HCC (n = 61), liver cirrhosis (LC) (n = 20), or healthy controls (HC) (n = 20). The baseline characteristics of participants in each group were compared. Serum levels of the lncRNAs HOTAIR, BRM and ICR were determined in each group by reverse transcription and quantitative real-time polymerase chain reaction (qRT-PCR). Correlations between the serum levels of the three lncRNAs and multiple clinical parameters were analysed. The receiver operating characteristic (ROC) curve was used to assess the diagnostic potential for HCC of each lncRNA individually, or in combination with AFP. Multivariate Cox regression analysis was used to evaluate the accuracy of these lncRNAs for predicting the outcome and survival of HCC patients. Results: The serum levels of HOTAIR, BRM and ICR were significantly higher in HCC patients compared to LC patients and healthy subjects. The HOTAIR level was positively correlated to tumour-node metastasis (TNM), Barcelona Clinic Liver Cancer (BCLC) stage, extrahepatic metastasis, vascular invasion, portal vein tumour thrombus (PVTT), and tumour size. The BRM level was positively associated with TNM stage, BCLC stage, vascular invasion, PVTT, and tumour size, while the ICR level was positively correlated with PVTT. A combination of the three lncRNAs and AFP showed the highest diagnostic accuracy for HCC, with an AUC of 0.998, sensitivity of 98.4%, and specificity of 100.0%. This combination showed a better diagnostic accuracy than the individual lncRNAs or AFP alone. Serum levels of the HOTAIR and ICR lncRNAs decreased significantly following surgery. Conclusions: Serum levels of the HOTAIR, BRM and ICR lncRNAs are potential prognostic markers for HCC. Upregulation of HOTAIR, BRM and ICR may facilitate early diagnosis and indicate poor prognosis for HCC. These lncRNAs could potentially serve as therapeutic targets for HCC. Combination of the three lncRNAs with AFP may increase the diagnostic accuracy for HCC. Further studies in larger cohorts of patients are needed to validate these findings.
Globally, hepatocellular carcinoma (HCC) is the third leading cause of
cancer-related deaths [1]. HCC often exhibits an insidious onset and rapid
progression making early detection and intervention difficult [2]. At present,
the most commonly used diagnostic markers for HCC include Alpha-fetoprotein
(AFP), Des-
Long noncoding RNAs (lncRNAs) regulate multiple biological processes and play important roles in the development and progression of various cancer types [8]. Aberrant levels of lncRNAs have been found in the tumour tissues and body fluids of patients with various cancers, and lncRNAs have therefore been investigated as candidate early diagnostic markers for cancers including HCC [9]. However, most of the lncRNAs studied so far in liver cancer have shown limited sensitivity and specificity as diagnostic markers. For example, the lncRNA UCA1 has low sensitivity (81.4%) and specificity (75.3%) for HCC diagnosis [10]. However, when combined with other markers such as Linc00152 and AFP, the diagnostic accuracy of this panel for HCC was significantly enhanced [11].
HOTAIR (homeobox transcript antisense intergenic RNA) is a lncRNA located in the Homeobox C gene cluster. It regulates epigenetic modification of multiple genes [12] and is involved in tumorigenesis, metastasis, and drug resistance in various cancer types [13]. The lncRNAs BRM (lncRNA for association with Brahma) and ICR (ICAM-1-related long noncoding RNA) are highly expressed in HCC tumours and in liver CSCs [14, 15]. Since lncBRM is necessary for the maintenance and self-renewal of liver CSCs and tumour initiation [14], it has been suggested as a potential marker for early diagnosis and progression of HCC. Increased expression of lncRNA ICR has been closely linked to the development of portal vein tumour thrombus (PVTT), HCC metastasis, and poor clinical outcomes in HCC patients [15]. Thus, lncRNA ICR may be a predictive marker for HCC metastasis.
Based on these earlier findings, we hypothesized that serum levels for the lncRNAs HOTAIR, BRM and ICR could comprise a useful panel for early diagnosis, monitoring of disease progression, and for predicting the outcome of HCC patients.
A total of 61 HCC patients (50 men and 11 women) who attended HangZhou Xixi
Hospital from April 2019 to November 2019 were included in this study. Diagnosis
of HCC was based on clinical symptoms, serum AFP levels, imaging studies
(ultrasound, CT, and MRI) and histopathological examinations. Patients with a
history of other tumours, and those receiving radiotherapy or chemotherapy were
excluded from the study. Control groups included patients with liver cirrhosis
(diagnosed by ultrasound, CT and MRI) with or without complicating portal
hypertension and hypersplenism (LC group, n = 20), and healthy subjects
identified by physical examination at the Hangzhou Xixi Hospital (HC group, n =
20). Detailed clinical features of the study subjects are shown in Table 1. The
clinical staging of HCC patients was determined by TNM and BCLC classification
systems [16]. Serum samples were collected from 34 HCC patients before and one
week after surgery (27 men, 7 women; mean age: 54.88
| Categories | HC (n = 20) | LC (n = 20) | HCC (n = 61) | |
| Age (years) | 41.65 |
55.31 |
55.96 | |
| Gender | ||||
| Male/Female | 13/7 | 14/6 | 50/11 | |
| TNM stage | ||||
| I/II/III/IV | 31/5/19/5 | |||
| BCLC stage | ||||
| 0/A/B/C/D | 5/17/14/20/5 | |||
| ALT (IU/L) | 16.5 (12.0–22.0) | 24.5 (15.2–39.0) | 31.0 (21.0–54.5) | |
| AFP (ng/mL) | 2.30 (1.47–3.34) | 3.17 (1.94–9.94) |
54.39 (9.25–1291.50) | |
Five millilitres of venous blood were collected from each donor, centrifuged at 1000 g for 10 min, and the serum collected into a 1.5 mL centrifuge tube and stored at –80 °C until use.
All lncRNA assays were carried out in the HangZhou Adicon Clinical Laboratory
using qRT-PCR. Total RNA was extracted using the miRcute miRNA isolation Kit
(DP501; Beijing, China) according to the manufacturer’s protocol. Briefly,
complementary DNA (cDNA) was synthesized using the TIANGEN lnRcute lncRNA cDNA
kit (KR202; Beijing, China). One hundred ng of total extracted
serum RNA was converted into cDNA. The relative expression of various genes was
examined using the SYBR Green Realtime PCR Master Kit (QPK-201; Shanghai, China)
and a PCR ABI 7500 Sequence Detection System. Each reaction mixture consisted of
2
| Genes | Sequence (5’-3’) |
| HOTAIR | F: 5’-AAACAGAGTCCGTTCAGTGTC-A-3’ |
| R: 5’-TTCTTAAATTGGGCTGGGTC-3’ | |
| BRM | F: 5’-GAGGAGAGAAGTCACTGAAATGG-3’ |
| R: 5’-CTC-TTCAAAGCAGACCCTCTAC-3’ | |
| ICR | F: 5’-CCC-AGAAGGTCATAGAAAGTCCGA-3’ |
| R: 5’-TCTAAGCAGCCACAGCC-TGAT-3’ | |
| RNA 18S | F: 5’-CAGCCACCCGAGATT-GAGCA-3’ |
| R: 5’-TAGTAGCGACGGGCGGTGTG-3’ | |
| Abbreviations: F, forward; R, reverse. | |
The serum AFP levels were determined by chemiluminescence enzyme immunoassay (CLEIA).
A total of 47 patients were followed up till December 2021. Follow-up data were obtained through telephone calls and hospital records, and were updated every 3~6 months. Clinical information recorded in the follow up database included sex, age, AFP, Child-Pugh, Performance Status (PS), HBsAg, PVTT, tumour size, tumour number, type of treatment, tumour recurrence and metastasis. Patients who died from unexpected events or other diseases were excluded from the study.
All statistical analyses were performed using SPSS software (version 22.0, IBM
Inc., Chicago, IL, USA). Nonparametric variables are presented as median and
interquartile ranges, and were analysed using the Mann-Whitney U test and
Kruskal-Wallis H test. Categorical variables were compared using chi square
tests. To determine the accuracy of the lncRNAs HOTAIR, BRM and ICR as biomarkers
for HCC, ROC (Receiver Operating Characteristic) curves were constructed and AUC
(Area Under the Curve) values were estimated. Multivariate analysis was performed
to identify independent prognostic factors. A p value of
Serum levels for the lncRNAs HOTAIR, BRM and ICR were all significantly higher
in HCC patients compared to LC patients and healthy controls (Fig. 1, p
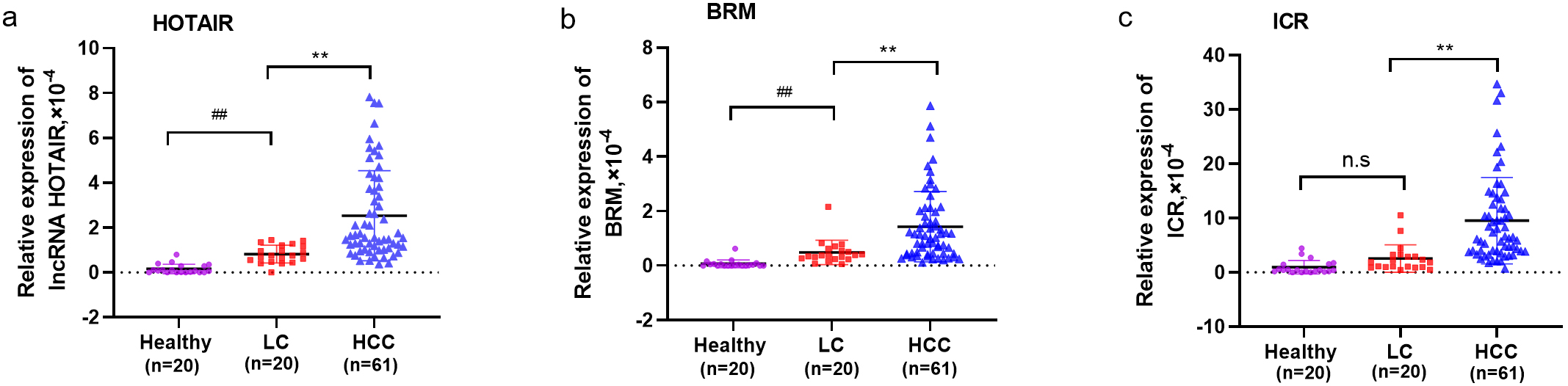 Fig. 1.
Fig. 1.Compariso of serum lncRNA levels in patients with HCC, LC, and
HC. Serum level of lncRNAs HOTAIR (a), BRM (b) and ICR (c) in HCC patients, LC
patients, and healthy controls. **: p
As shown in Table 3, the serum level of lncRNA HOTAIR was closely associated with TNM stage (p = 0.007), BCLC stage (p = 0.015), extrahepatic metastasis (p = 0.043), vascular invasion (p = 0.022), PVTT (p = 0.004), and tumour size (p = 0.039). The serum level of lncRNA BRM was significantly correlated with TNM stage, BCLC stage, vascular invasion, PVTT, and tumour size (p = 0.007, p = 0.009, p = 0.011, p = 0.005, p = 0.039, p = 0.036, respectively). The serum level of lncRNA ICR was closely correlated with portal venous invasion (p = 0.023). No significant associations were found between these lncRNAs and gender, age, PS, Child-Pugh score, serum HBsAg level, serum AFP level, and tumour number.
| Factors | N | HOTAIR | p | BRM | p | ICR | p | |
| Age (y) | 0.575 | 0.859 | 0.707 | |||||
| 24 | 1.47 (0.96, 3.59) | 1.13 (0.44, 1.51) | 5.79 (3.30, 15.80) | |||||
| 37 | 1.63 (1.20, 4.04) | 1.03 (0.27, 2.07) | 6.69 (3.98, 12.20) | |||||
| Gender | 0.599 | 0.985 | 0.113 | |||||
| Male | 50 | 1.55 (1.14, 3.93) | 1.01 (0.44, 2.16) | 7.84 (3.99, 14.61) | ||||
| Female | 11 | 1.68 (0.73, 3.93) | 1.16 (0.44, 1.99) | 4.84 (3.34, 8.05) | ||||
| TNM stage | 0.007 | 0.007 | 0.188 | |||||
| I+II | 38 | 1.36 (0.91,2.44) | 0.71 (0.35, 1.42) | 5.81 (3.79, 10.90) | ||||
| III+IV | 23 | 2.96 (1.54, 2.44) | 1.44 (0.81, 2.83) | 9.62 (4.03, 16.4) | ||||
| BCLC stage | 0.015 | 0.009 | 0.822 | |||||
| 0–A | 22 | 1.27 (0.85, 2.18) | 0.56 (0.27, 1.25) | 6.59 (3.79, 12.43) | ||||
| B–D | 39 | 2.10 (1.30, 4.40) | 1.23 (0.67, 2.13) | 6.27 (3.92, 13.50) | ||||
| PS | 0.293 | 0.786 | 0.119 | |||||
| 0~1 | 50 | 1.47 (0.98, 4.23) | 1.01 (0.43, 2.14) | 5.81 (3.52, 12.20) | ||||
| 2~4 | 11 | 2.15 (1.54, 3.39) | 1.16 (0.46, 1.81) | 10.40 (7.35, 13.50) | ||||
| Child-Pugh | 0.810 | 0.781 | 0.215 | |||||
| A | 39 | 1.63 (1.04, 4.25) | 1.03 (0.36, 2.57) | 7.35 (4.06, –14.80) | ||||
| B + C | 22 | 1.55 (1.22, 3.46) | 1.15 (0.68, 1.48) | 5.50 (3.27, 11.08) | ||||
| AFP (g/L) | 0.658 | 0.848 | 0.315 | |||||
| 24 | 1.89 (1.02, 4.79) | 1.04 (1.02, 4.79) | 8.67 (4.13, 14.40) | |||||
| 37 | 1.55 (1.14, 3.42) | 1.14 (0.43, 2.13) | 6.14 (3.68, 11.75) | |||||
| Extrahepatic metastasis | 0.043 | 0.102 | 0.605 | |||||
| Yes | 12 | 3.60 (1.26, 5.88) | 1.48 (0.72, 3.08) | 10.01 (3.02, 19.40) | ||||
| No | 49 | 1.54 (1.03, 3.28) | 1.03 (0.41, 1.70) | 6.27 (3.92, 11.75) | ||||
| Vascular invasion | 0.022 | 0.011 | 0.714 | |||||
| Yes | 19 | 4.22 (1.30, 5.56) | 1.63 (0.81, 2.84) | 9.54 (3.29, 16.30) | ||||
| No | 42 | 1.46 (1.01, 2.62) | 0.79 (0.35, 1.47) | 6.25 (3.92, 12.23) | ||||
| PVTT | 0.004 | 0.005 | 0.023 | |||||
| Yes | 11 | 4.25 (1.55, 5.65) | 2.57 (1.16, 3.91) | 10.40 (9.29, 16.30) | ||||
| No | 50 | 1.46 (1.01, 2.71) | 0.81 (0.43, 1.47) | 5.81 (3.52, 11.73) | ||||
| HBV | 0.338 | 0.691 | 0.894 | |||||
| Yes | 57 | 1.63 (1.10, 4.02) | 1.03 (0.44, 2.07) | 6.27 (3.89, 13.65) | ||||
| No | 4 | 1.14 (0.91, 3.10) | 1.15 (0.62, 1.35) | 7.70 (4.34, 10.79) | ||||
| Tumor size (cm) | 0.039 | 0.036 | 0.239 | |||||
| 40 | 1.43 (1.00, 2.62) | 0.79 (0.37, 1.52) | 5.81 (3.88, 11.40) | |||||
| 21 | 2.96 (1.42, 4.97) | 1.39 (0.81, 2.70) | 9.62 (3.90, 14.95) | |||||
| Tumor number | 0.398 | 0.337 | 0.884 | |||||
| Single | 41 | 1.55 (1.03, 3.69) | 0.84 (0.38, 2.00) | 6.49 (3.97, 11.20) | ||||
| Multiple | 20 | 1.61 (1.26, 4.36) | 1.16 (0.67, 2.05) | 7.95 (3.22, 16.00) | ||||
| TNM, Tumor node metastasis; BCLC, Barcelona Clinic Liver Cancer; PS, Performance Status; AFP, Alpha fetoprotein; HBV, Hepatitis B Virus; PVTT, portal vein tumor thrombus. | ||||||||
The serum levels of the three lncRNAs in 34 HCC patients were measured on day one of their hospital visit (pre-operative) and one week after tumour resection (post-operative). A significant decrease in the serum level after surgery was observed for the lncRNAs HOTAIR and ICR, but not for BRM (Fig. 2).
 Fig. 2.
Fig. 2.Comparison of serum Levels of HOTAIR, BRM and ICR in Pre- and Postoperative HCC Patients. Serum levels of three lncRNAs HOTAIR (a), BRM (b) and ICR (c) in the preoperative (Pre-op) and postoperative (Post-op) serum samples of HCC patients (n = 34).
To assess the diagnostic value of the three lncRNAs HOTAIR, BRM and ICR for HCC, ROC curve analysis was performed to determine their AUC, sensitivity, and specificity. AFP is widely used as a serum marker for HCC, hence this marker was analysed alone and in combination with the lncRNAs for the diagnosis of HCC. The three lncRNAs alone showed higher accuracy for identifying HCC compared to AFP (Fig. 3 and Table 4). Notably, lncRNA HOTAIR showed the highest AUC for distinguishing HCC patients from healthy controls, while lncRNA ICR showed better ability for distinguishing HCC from LC patients.
 Fig. 3.
Fig. 3.Diagnostic efficiency of individual serum lncRNAs orAFP or their combinations in HCC patients. (1) ROC curve analysis for the individual lncRNAs (a, b) or AFP (c) or their combinations in distinguishing HCC patients from healthy controls. (2) ROC curve analysis for the individual lncRNAs (d, e) or AFP (f) or their combinations in distinguishing HCC patients from LC patients. (3) ROC curve analysis for the individual lncRNAs or AFP or their combinations in distinguishing HCC patients with early BCLC stage (n = 22) from healthy controls and LC patients.
| Method | AUC (95% CI) | SEN (%) | SPE (%) | Cut-off | p-value | |
| HCC vs. HC | ||||||
| HOTAIR | 0.991 (0.976–1.000) | 96.7 | 95.0 | 0.49 |
||
| BRM | 0.983 (0.952–1.000) | 98.4 | 95.0 | 0.20 |
||
| ICR | 0.966 (0.925–1.000) | 98.4 | 85.0 | 1.76 |
||
| HOTAIR+BRM | 0.992 (0.979–1.000) | 96.7 | 95.0 | 0.597 | ||
| HOTAIR+ICR | 0.992 (0.979–1.000) | 91.8 | 100.0 | 0.863 | ||
| BRM+ICR | 0.985 (0.964–1.000) | 95.1 | 95.0 | 0.650 | ||
| HOTAIR+BRM+ICR | 0.994 (0.983–1.000) | 96.7 | 100.0 | 0.708 | ||
| AFP | 0.854 (0.774–0.934) | 65.6 | 100.0 | 16.75 | ||
| Three lncRNAs+AFP | 0.998 (0.994–1.000) | 98.4 | 100.0 | 0.641 | ||
| HCC vs. LC | ||||||
| HOTAIR | 0.811 (0.712–0.911) | 59.0 | 100.0 | 1.45 |
||
| BRM | 0.749 (0.621–0.877) | 55.7 | 94.7 | 0.83 |
||
| ICR | 0.850 (0.750–0.950) | 80.3 | 84.2 | 3.46 |
||
| HOTAIR+BRM | 0.850 (0.766,0.934) | 72.1 | 85.0 | 0.761 | ||
| HOTAIR+ICR | 0.855 (0.759–0.951) | 75.4 | 85.0 | 0.671 | ||
| BRM+ICR | 0.877 (0.786–0.968) | 82.0 | 85.0 | 0.647 | ||
| HOTAIR+BRM+ICR | 0.884 (0.807–0.960) | 65.6 | 94.7 | 0.849 | ||
| AFP | 0.788 (0.691–0.884) | 55.7 | 100.0 | 45.5 | ||
| Three lncRNAs+AFP | 0.955 (0.911–0.999) | 90.2 | 95.0 | 0.611 | ||
| BCLC (0–A) vs. LC+HC | ||||||
| HOTAIR | 0.848 (0.753–0.942) | 90.9 | 67.5 | 0.64 |
||
| BRM | 0.792 (0.682–0.902) | 90.9 | 60.0 | 0.25 |
||
| ICR | 0.894 (0.814–0.975) | 81.8 | 87.5 | 3.49 |
||
| HOTAIR+BRM | 0.873 (0.787–0.959) | 86.4 | 75.0 | 0.294 | ||
| HOTAIR+ICR | 0.892 (0.810–0.974) | 90.9 | 77.5 | 0.246 | ||
| BRM+ICR | 0.903 (0.827–0.980) | 81.8 | 90.0 | 0.358 | ||
| HOTAIR+BRM+ICR | 0.918 (0.849–0.988) | 86.4 | 85.0 | 0.338 | ||
| AFP | 0.778 (0.647–0.909) | 77.3 | 70.0 | 8.12 | ||
| Three lncRNAs+AFP | 0.955 (0.900–1.000) | 95.5 | 90.0 | 0.307 | ||
| AUC, Area under the curve; SEN, sensitivity; SPE, specificity; HC, healthy controls; LC, Liver cirrhosis; PLC, Primary liver carcinoma; AFP, Alpha fetoprotein; BCLC, Barcelona Clinic Liver Cancer. | ||||||
Based on the above results, we next used binary logistic regression to assess the diagnostic accuracy of lncRNA-based panels. As shown in Table 4 and Fig. 3, combination of the three lncRNAs resulted in better diagnostic power than the individual lncRNAs. The diagnostic accuracy of the three lncRNAs for HCC was further improved by combining with AFP, as indicated by the higher AUC values for distinguishing HCC patients from healthy controls and from LC patients (Table 4; Fig. 3(1c); Fig. 3(2c)). Thus, combination of the three lncRNAs with AFP resulted in the best diagnostic accuracy for HCC patients.
In the 22 HCC patients (22/61, 36%) with early-stage disease (BCLC stage 0+A), the diagnostic accuracy of the combined panel of three lncRNAs and AFP was also significantly better compared to each lncRNA alone or to AFP alone (Table 4, Fig. 3).
These data suggest that a combined panel comprising three lncRNAs and AFP could be a potential biomarker for HCC.
The prognostic value of the lncRNAs HOTAIR, BRM and ICR was evaluated in 47 HCC patients (survival time 19~25 days, median 23 days) by Cox regression analysis. Serum levels of the lncRNAs HOTAIR, BRM and ICR were significantly correlated with HCC prognosis (Table 5).
| Variables | HR | 95% CI | p |
| HOTAIR level (low, high) | 3.311 | 1.697–15.726 | 0.032 |
| BRM level (low, high) | 2.090 | 1.120–10.892 | 0.038 |
| ICR level (low, high) | 1.437 | 1.003–4.338 | 0.006 |
| HR, Hazard Ratio; CI, Confidence Interval. | |||
Accurate early diagnosis and prediction of outcome for HCC patients remains a major clinical challenge worldwide. Previous studies have implicated the lncRNAs HOTAIR, BRM and ICR in liver cancer and LCSCs. However, the clinical value of these lincRNAs in the management of HCC patients has yet to be explored. The present study is the first to assess the diagnostic value of the serum lncRNAs HOTAIR, BRM and ICR in HCC patients. Elevated serum levels of the three lncRNAs were found to be accurate diagnostic markers for HCC, as well as prognostic biomarkers for these patients. A significant decrease in the serum level of HOTAIR and ICR was observed following surgical resection in HCC patients. Hence, the lncRNAs HOTAIR and ICR may be useful markers for monitoring the outcome of surgical resection in HCC patients. Another novelty of this study was the finding that combination of the three lncRNAs with the classical tumour marker AFP resulted in improved accuracy for HCC diagnosis.
An increase in the HOTAIR level has been reported to promote the malignant transformation of normal liver cells, including normal liver stem cells (NLSCs), via epithelial-mesenchymal transition (EMT) [17]. Hence, the serum level of HOTAIR may be useful for the early diagnosis of HCC. Indeed, elevated levels of HOTAIR have been observed in many cancer types and a high level could therefore facilitate their diagnosis and perhaps also predict poor many cancer types including HCC [18, 19, 20, 21]. In these aspects, our data are consistent with the previously published data [22] in that elevated serum HOTAIR level significantly correlated with TNM stage, BCLC stage, extrahepatic metastasis, vascular invasion, portal vein tumour thrombus, and tumour size.
The oncogenic role of BRM has been reported in multiple cancers including colorectal [23], ovarian [24] and liver [14]. Previous studies have also shown that the lncRNAs BRM and ICR were highly expressed in liver CSCs and HCC tumours [14, 15], and that ICR could regulate liver CSC properties and contribute to PVTT development [15]. In particular, BRM promotes the self-renewal of liver CSCs and initiates tumour propagation via YAP1 signalling, while the serum level of BRM was positively correlated to the disease severity of HCC patients [14]. In the current study, an increased serum level of BRM was significantly correlated with advanced TNM stage, BCLC stage, vascular invasion, portal venous invasion, and tumour size. Furthermore, the serum level of the three lncRNAs decreased significantly following surgical resection of HCC tumours. These data provide support for the potential application of three lncRNAs (HOTAIR, BRM and ICR) for the early diagnosis, prediction of survival outcome, and monitoring of disease progression in HCC patients. A novel finding of our study was that combination of these three lncRNAs with the classical HCC marker AFP further increased the diagnostic value of the lncRNAs. Additional studies in large patient cohorts are needed to validate our findings.
Considering the known oncogenic roles of the lncRNAs studied here, we speculate they could also serve as therapeutic targets for HCC. For example, an RNA-based dominant negative molecule that counteracts HOTAIR function (HOTAIR-SNAIL-binding domain, HOTAIR-sbid) has been shown to inhibit mobility, invasiveness and EMT in HCC cells [25]. The potential application of these lncRNAs as therapeutic targets for liver cancer warrants further investigation.
A major limitation of this study was the small number of patients analysed and the short follow up time. Further studies in larger cohorts of patients should be conducted to confirm our findings. These should include patients with various pre-malignant conditions (e.g., hepatitis, fatty liver disease), as well as patients who have been treated with surgery or other approaches (e.g., regional ablation).
Increased serum levels of the lncRNAs HOTAIR, BRM and ICR could be used in a panel of markers for early diagnosis of HCC. Combining these lncRNAs with AFP improves the diagnostic accuracy of HCC even further, while also providing a useful tool for monitoring therapeutic effects in HCC.
All data generated or analysed during this study are included in this published article. Additional data/files would be available from the corresponding author upon reasonable request. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
ZHL, KYX, and JFB designed and revised the manuscript. ZHL wrote the manuscript and drew figures. ZHL, XQS, and YOY prepared the patient samples. LQ, and KYX provided assistances with revising the manuscript. ZYL, JSH, and XHR conducted data collection. LBM, FL, YW, and AF conducted data analysis. All the authors read and approved the final version of the manuscript.
The protocal regarding human serum samples was approved by the Hangzhou Xixi Hospital and an agreement was signed in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all patients and healthy donors (Approval NO. 20181228Y22).
We thank anonymous reviewers for excellent criticism of the article and all the authors in the reference list.
This work has been supported by the Natural Science Foundation of Zhejiang Province (LSY19H030002), the project of Hangzhou Science and Technology Bureau (20181228Y22), the project of Yuhuan Science and Technology Bureau (2018075).
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
