†These authors contributed equally.
Academic Editors: Amedeo Amedei, Edda Russo, Francesca Romano and Alessandra Fanelli
Background: SARS-CoV-2 is a positive-sense single-stranded RNA virus. It is enveloped by four structural proteins. The entry of the virus into the host cells is mediated by spike protein binding to the angiotensin converting enzyme 2 (ACE2) and proteolytic cleavage by transmembrane protease serine 2 (TMPRSS2). In this study, we analyzed the expression of the ACE2 receptor and TMPRSS2 in cases under investigation for SARS-CoV-2 infection. Methods: The study was carried out using the viral transport medium of consecutive nasopharyngeal swabs from 300 people under examination for SARS-CoV-2 infection. All samples underwent the SARS-CoV-2 transcriptase-mediated amplification assay (Procleix® SARS-CoV-2) to detect the virus. Immunocytochemistry was used in each sample to detect the presence of the SARS-CoV-2 nucleoprotein, the ACE2 receptor, and TMPRSS2. Results: An immunocytochemical study with monoclonal antibody against SARS-CoV-2 viral nucleoprotein showed positivity in squamous cells. ACE2 were not detected in the squamous cells obtained from the nasopharyngeal samples. Conclusions: SARS-CoV-2 predominantly localizes to squamous cells in cytology samples of patients with positive transcriptase-mediated amplification SARS-CoV-2 assay results. The immunocytochemical negativity for ACE2 evidenced in the present study could be related to the cellular heterogeneity present in the nasopharyngeal smear samples and could be related to variations at the genomic level. Our results suggest that SARS-CoV-2 might be present in the nasopharyngeal region because viral cell junctions are weaker. This facilitates viral concentration, infective capacity and migration to specific organs, where SARS-CoV-2 infects target cells by binding to their receptors and then entering.
SARS-CoV-2 belongs to the Coronaviridae family and is an enveloped positive-sense single-stranded RNA virus [1, 2, 3, 4]. Its genome consists of 14 open reading frames (ORFs), nine of which encode 16 nonstructural proteins (nsp 1–16). These form the replicase complex [5, 6]. The five remaining frames encode nine accessory proteins (ORFs) and four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N). Spike is the structural protein that mediates the entry of SARS-CoV-2 into host cells [7]. Its receptor-binding domain (RBD) mediates contact with a cell receptor, angiotensin-converting enzyme 2 (ACE2), and a polybasic S1/S2 cleavage site, which is proteolytically cleaved by cell cathepsin L and the transmembrane protease serine 2 (TMPRSS2) [1, 8, 9].
Different studies have revealed that ACE2 and TMPRSS2 are present in the enterocytes of the small intestine and the alveolar epithelial cells of the lung [10, 11, 12, 13]. They have also been shown to be present in arterial and venous endothelial cells and arterial smooth muscle cells, the colon, and the testis [10, 11, 12, 13]. The epithelial expression of ACE2 and TMPRSS2 and the fact that they are present in the vascular endothelium might explain the entry routes for SARS-CoV-2 and the main manifestations of COVID-19.
Oral and nasal mucosa and nasopharynx did not show expression of ACE2 or TMPRSS2 in superficial epithelial cells, suggesting that these tissues in the upper respiratory tract are not the main site of SARS-CoV entry. The presence of SARS-CoV-2 RNA in nasopharyngeal samples might be because the lower respiratory tract is infected [11]. In a previous study, we showed the presence of SARS-CoV-2 in squamous cells studied in the liquid-based cytologies of nasopharyngeal swabs from people under investigation for SARS-CoV-2 infection [14]. In this article, we investigate the presence of ACE2 and TMPRSS2 in the universal transport medium (UTM) used to investigate cases of SARS-CoV-2 infection. We also report on an immunohistochemical study of the viral nucleoprotein and viral RNA of SARS-CoV-2.
This is a prospective and descriptive cohort study conducted on cases under investigation (CUI) for SARS-CoV-2 infection at Sant Joan University Hospital, Reus, Spain between November 1, 2020 and December 30, 2020. The study protocol was reviewed and approved by the hospital’s ethics committee (registration number CEIm 204/2020). The research followed the principles of the Declaration of Helsinki 1964 and its subsequent modifications. We studied 260 consecutive cases who were positive for SARS-CoV-2 infection and 40 consecutive negative cases submitted to the molecular pathology unit of our pathology department (Fig. 1).
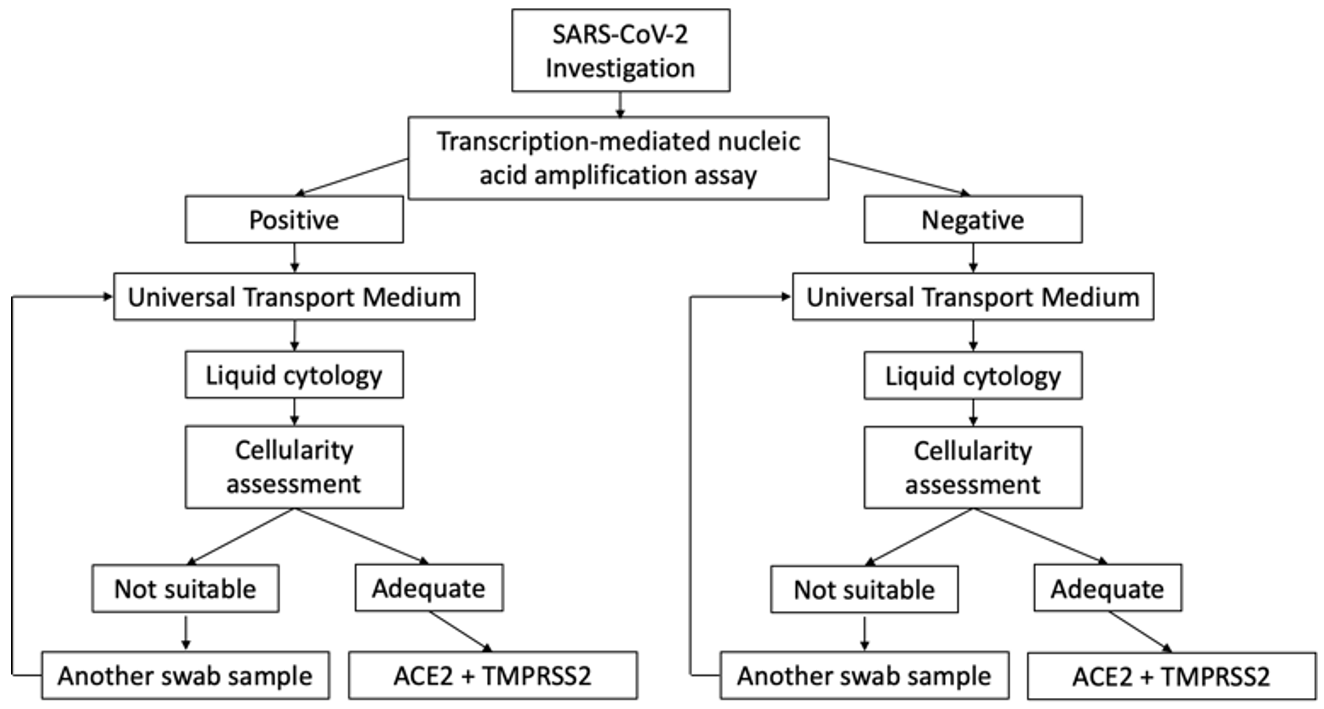 Fig. 1.
Fig. 1.Cases under investigation for SARS-CoV-2 infection involving nasopharyngeal swabs. Scheme of the selection of samples included in the study.
The Procleix® SARS-CoV-2 assay was used to detect infections
with swab specimens from the upper respiratory tract (oro/nasopharyngeal swab).
The assay uses transcription-mediated nucleic acid amplification (TMA) technology
and involves the amplification of the SARS-CoV-2 RNA target and the detection of
amplification products (amplicon) using the hybridization protection assay (HPA).
Detection is achieved by chemiluminescent markers. The probe is measured with a
luminometer and reported as relative light units (RLUs). All calculations are
determined by the Procleix® Panther System
Software® for each assay. For each sample, the light signal RLU
and the flashing signal RLU are determined. The RLU divided by the limit is
abbreviated as signal/limit (S/CO) in the report. Assay results are nonreactive
(S/CO
After Procleix® SARS-CoV-2 assay determination, the UTM was
processed for liquid-based cytology using the Thin Prep 5000TM LBC method
(Hologic Co., Marlborough, MA, USA). All the UTM material was fixed with the
hemolytic and preservative solution Cytolyt
The immunocytochemical staining protocol was previously validated with
anti-SARS-CoV-2 NP antibody, clone # 6F10 (BioVision
Incorporated®, CA, USA); anti-ACE2 antibody, clone SN0754
(GeneTex Inc®, Irvine, CA, USA); and 1:500 for anti-TMPRSS2
antibody, clone N2C3 (GeneTex Inc®, Irvine, CA, USA) in samples
obtained from UTM material processed by liquid-based cytology, with 10 positive
and 10 negative samples by RT–PCR and kidney, myocardium, and prostatic
adenocarcinoma. Additionally, we used cytokeratin AE1/AE3 to compare the
immunoreaction in positive and negative cases. Immunocytochemical staining was
carried out on all the samples processed for liquid cytology, which were placed
in a VENTANA® Benchmark ULTRA/LT automatic immunohistochemistry
processor (Ventana Medical Systems, Oro Valley, AZ, USA) using a previously
standardized protocol for SARS-CoV-2, ACE2, and TMPRSS2 detection that included
recovery solution pH 9 for 40 min at 100 °C and the
Optiview® DAB Immunohistochemistry Detection Kit
(VENTANA®). For incubation with the primary antibodies, after
they had each been reconstituted with 100 mL of distilled water, the following
dilutions were used: 1:1000 for anti-SARS-CoV-2 NP antibody, clone # 6F10
(BioVision Inc®, Milpitas, CA, USA); 1:500 for anti-ACE2
antibody, clone SN0754 (GeneTex Inc®, Irvine, CA, USA); and 1:500
for anti-TMPRSS2 antibody, clone N2C3 (GeneTex Inc®, Irvine, CA,
USA). The incubation took 32 min at 36 °C. Finally, the slides were
treated with diaminobenzidine, contrasted with Mayer’s hematoxylin, dehydrated
with alcohols at increasing concentrations, rinsed with xylol, and then examined
under an Olympus BX41 light microscope at 3.5
Viral medium from nasopharyngeal samples from 24 SARS-CoV-2-negative and 24
SARS-CoV-2-positive samples was added to 15 mL of Dulbecco’s phosphate buffered
saline (DPBS) (ThermoFisher Scientific Inc, Waltham, MA, USA) in a 50-mL tube.
The samples were then centrifuged at 3000
Denatured proteins (50
Viral media from nasopharyngeal specimens from five SARS-CoV-2-negative patients and five SARS-CoV-2-positive patients were processed as briefly described. The samples were centrifuged at 2000 rpm for 5 min and fixed in 2% glutaraldehyde in 0.1 mol/L phosphate buffer for three h at 4 °C. Then, they were fixed in 1% osmium tetroxide for 30 min and dehydrated in a series of acetone and soaked in a mixture of Epon-Araldite. Thin sections (60 nm) were stained with lead citrate, and images were taken at 80 KV using a JEOL JEM1011 microscope. For SEM, the fixed samples were dehydrated in an ethyl alcohol series, chemically dried with Amil-acetate, mounted in aluminum tubes, plated with gold, and photographed at 10 KV using a Scios 2 DualBeam scanning microscope (Thermo Fisher Scientific, Waltham, MA, USA).
Our analysis included 131 male and 169 female patients with a median age of 44.67 years (range, 4–98 years). Of the 260 cases that were positive for SARS-CoV-2, 196 were asymptomatic (75.38%), whereas 64 (24.62%) showed symptoms. The most frequent SARS-CoV-2 symptom was myalgia in 64 (100%) cases, followed by dry cough in 62 (96.86%), fever in 56 (87.50%), anosmia in 42 (65.63%), gastrointestinal symptoms in 40 (62.50%), odynophagia in 28 (43.75%), and dyspnea in 24 (37.50%). The mean hospitalization time in the COVID-19 patients was 32.33 days (range, 1–81). Nine (14.06%) patients required intensive care unit admission. All the cases that tested negative for the SARS-CoV-2 assay (40) were asymptomatic. Of those who tested positive, five (1.92%) died from COVID-19. Table 1 summarizes the results.
| Patients | N | % | |
| Male | 131 | 43.67 | |
| Female | 169 | 56.33 | |
| Symptoms | 64 | 21.33 | |
| Myalgia | 64 | 100 | |
| Dry cough | 62 | 96.86 | |
| Fever | 56 | 87.5 | |
| Anosmia | 42 | 65.63 | |
| GI symptoms | 40 | 62.5 | |
| Odynophagia | 20 | 43.75 | |
| Dyspnea | 24 | 37.5 | |
| Asymptomatic | 236 | 78.67 | |
| ICU admission | 9 | 14.06 | |
| Hospitalized | 55 | 85.94 | |
| Lethal disease outcome | 5 | 1.92 | |
| Procleix® SARS-CoV-2 assay | |||
| Negative | 40 | 13.33 | |
| Positive | 260 | 86.67 | |
The immunocytochemical study of the SARS-CoV-2 nucleoprotein in the 260 positive samples showed that squamous cells were positive for granular cytoplasm. There were no signs of immunoreaction in ciliated respiratory-type cells, macrophages, or neutrophils (Fig. 2). The immunocytochemical study of 40 Procleix® SARS-CoV-2-negative samples showed that there was no cytoplasmic reactivity to SARS-CoV-2 nucleoprotein. The immunocytochemical analysis of the expression of the ACE2 receptor in the 300 samples studied (260 positive samples for SARS-CoV-2 and 40 negative samples for SARS-CoV-2), revealed no evidence of cytoplasmic reactivity in the cell component of the antibody (squamous cells, cylindrical cells or macrophages) obtained from nasopharyngeal samples. For the immunocytochemical study of TMPRSS2 a weak granular cytoplasmic immunoreactivity was observed for squamous cells, as well as a cross-reaction with bacteria. In certain cases, the immunocytochemical assessment of TMPRSS2 was difficult to interpret due to the presence of a large number of bacteria. In positive controls, granular cytoplasmic reactivity was demonstrated for both ACE2 and TMPRSS2 antibodies (Fig. 3).
 Fig. 2.
Fig. 2.Cases under investigation for SARS-CoV-2 infection involving
nasopharyngeal swabs. Relationship between the Procleix®
SARS-CoV-2 assay and immunocytochemistry study. An immunocytochemical study
showed no expression of ACE2 in epithelial cells (DAB, 20
 Fig. 3.
Fig. 3.ACE2 and TMPRSS2 immunocytochemistry study. The First
column shows kidney used as positive control. The rest of the columns show an
immunocytochemical study for ACE2 and TMPRSS2. No positivity is observed in
epithelial cells to ACE2 (DAB, 20
Western blot analysis of 24 SARS-CoV-2-negative and 24 SARS-CoV-2-positive samples showed the absence of ACE2 expression in both groups (Fig. 4). Regarding the TMPRSS2 analysis, the presence of two clearly defined bands between 55 and 46 kDa (Fig. 4) was evidenced for both groups, with a slightly higher intensity for the negative SARS-CoV-2 group. As a Western blot loading control, glyceraldehyde-3-phosphate dehydrogenase was used (Fig. 4).
 Fig. 4.
Fig. 4.Western blot study in SARS-CoV-2-negative and SARS-CoV-2-positive samples. No ACE2 expression is shown in 48 samples. TMPRSS2 shows bands between 55 and 46 kDa. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control.
Different sections of each sample were studied by both TEM and SEM. In the positive SARS-CoV-2 samples analyzed by TEM, we found the presence of virions in the extracellular compartment in relation to epithelial cells. Virions were observed in isolation and in small groups of variable measurements (Fig. 5). No intracellular viral particles were observed. In negative SARS-CoV-2 samples, we did not observe the presence of virions. In the SEM study of SARS-CoV-2-negative cases, the cytoplasmic surface was characterized by a smooth, regular appearance, and cytoplasmic folds were arranged in an orderly manner (Fig. 5a). In comparison, in the positive SARS-CoV-2 samples, the surface of the cytoplasmic membrane showed an irregular granular appearance, with irregularly arranged folds (Fig. 5b). Adhered to the plasma membrane, virions with a predominantly rounded shape were observed, although in some cases, slightly ovoid morphologies could be observed (Fig. 5b,c,d). Both in TEM and SEM, the variation observed in viral morphology could be attributed to a different state of virus activation or to degenerative changes resulting from the viral inactivation used.
 Fig. 5.
Fig. 5.Ultrastructural study. (a) SEM study of SARS-CoV-2-negative cases showing a cytoplasmic surface characterized by a smooth, regular appearance and cytoplasmic folds arranged in an orderly manner. (b) TEM study of SARS-CoV-2-positive cases showing a cytoplasmic surface characterized by a smooth, irregular appearance and virion particles (black arrows). (c) TEM study of SARS-CoV-2-positive cells showing extracellular granular material; no intracellular regions were observed. (d) Characteristic viral particles are shown (black arrows).
We conducted an immunocytochemical study of the universal transport medium for the determination of transcription-mediated nucleic acid amplification of SARS-CoV-2 in 300 patients admitted to our institution for suspected SARS-CoV-2 infection, Wuhan-Hu-1/2019 (MN908947) strain according to epidemiological reports of the health authority. The strategy recommended by the CDC for diagnosing patients with COVID-19 is to test samples from the respiratory tract to determine whether one or several nucleic acid targets specific to SARS-CoV-2 are present [15]. A nasopharyngeal sample is the technique of choice [16]. This study on the viral transport medium of people being investigated for SARS-CoV-2 infection shows cytoplasmic immunoreactivity for the viral nucleoprotein in squamous cells. We reported this finding in a previous publication by our group [14], and it confirms that SARS-CoV-2 is present in nasopharyngeal samples from patients being studied for COVID-19.
An enveloped virus, SARS-CoV-2 has virions that are round, sometimes pleiomorphic and approximately 80 to 120 nm in diameter [4, 5, 6, 7, 17]. It contains positive-strand RNA and has the largest genome (approximately 30 kb) of all RNA viruses [4, 5, 6, 7, 17]. The RNA genome is complexed with the basic nucleocapsid (N) protein so that a helical capsid is formed within the viral membrane [17]. This membrane contains at least three viral proteins: spike (S), a type I glycoprotein that creates the eplomers on the virion surface and gives the virus its crown-like morphology; a membrane (M) protein, which has a short N-terminal ectodomain and a cytoplasmic tail and spans the membrane three times; and a small, highly hydrophobic membrane protein (E) [17]. SARS-CoV-2 attaches to specific cellular receptors via the spike protein, which triggers a conformational change in the protein. This leads to fusion between the viral and cell membranes and causes the nucleocapsid to be released into the cell [17]. Two integral proteases of the renin–angiotensin system enable SARS-CoV-2 to enter the cell: ACE2 and neutral aminopeptidase (aminopeptidase N [APN]) [18, 19, 20].
In our working hypothesis, we propose that if SARS-CoV-2 viral particles are present in squamous cells and surface receptors, such as ACE2 and TMPRRS2, are necessary for cell infection, these receptors should be found on the surface of the squamous cells. However, the immunohistochemical study and the Western blot analysis did not demonstrate the presence of ACE2 in the squamous cells of nasopharyngeal swab samples. ACE2 receptor expression has been identified in the nasopharynx, lungs, intestine, kidney, and testis [10, 21, 22]. Different works have shown that the ciliated columnar cells of the nasopharyngeal region shows expression of ACE2 receptor [21, 22]. However, Hamming et al. [10] showed that tissues of the upper respiratory tract, such as oral, nasal mucosa, and nasopharynx, do not show ACE2 expression on the surface of epithelial cells. These differences may be due to the cell population studied and the methodology used. In our study, we did not show immunocytochemical expression to ACE2 receptor in the squamous cells of the nasopharyngeal smears. In addition, the ACE2 receptor is key to producing the cellular internalization of SARS-CoV-2 [23]. Our results would indicate, that in the squamous cells obtained from the viral transport medium, SARS-CoV-2 internalization does not occur due to the absence of the ACE2 receptor in this cellular component. Thus, these results raise questions about the interactions between SARS-CoV-2 and epithelial cells in the nasopharyngeal region.
Recent work on the interaction between SARS-CoV-2 and host cells has shown that the amount of ACE2 messenger RNA (mRNA) in the lungs of infected mice increased significantly in comparison to the control group. This indicates that SARS-COV-2 upregulates the ACE2 receptor and increases the internalization replication and dissemination of the virus [24]. As has been demonstrated both in vitro and in vivo, one way in which the ACE2 receptor can be upregulated is by gene stimulation of the ACE2 receptor via a human interferon [25]. Our study failed to demonstrate cellular protein expression of the ACE2 receptor in squamous cells of the nasopharyngeal region, which suggests that cell mechanisms are not involved in upregulating the ACE2 receptor in this region and that squamous cells are a reservoir that maintains SARS-CoV-2 because after four weeks, 26.3% of the RT–PCR test samples were still positive [26].
Another discovery that has been key to understanding the SARS-CoV-2 infection mechanism is the role played by transmembrane serine protease 2 (TMPRSS2), a cell-surface protein expressed by the epithelial cells of some tissues, including those in the aerodigestive tract [12, 23, 27]. SARS-CoV-2 critically depends on TMPRSS2 if it enters the host and spreads. The first step that enables it to enter the host cell is when the spike protein attaches to ACE2, which is expressed in respiratory epithelial cells. In the second step, the spike protein is cleaved, and the virus is internalized. This second step depends on proteases in the host cell, particularly TMPRSS2 [12, 23, 27, 28]. Therefore, both proteins are necessary to produce SARS-CoV-2 infection. In our study, regarding TMPRSS2, we were able to show the presence of this protein by Western blot analysis, and the immunocitochemical study showed its expression in squamous cells and bacterial elements, studied from the universal viral medium of nasopharyngeal smears in patients under investigation for SARS-CoV-2. In a work developed by Huang et al. [29], the authors were able to show that in the oral region, the target cells for the replication of SARS-CoV-2 are the minor salivary glands and basal cells of the mucous epithelium. Furthermore, the coexpression of ACE-2 and TMPRSS2 in the same cellular components is not frequent [29]. Variability in the expression of ACE2 and TMPRSS-2 is demonstrated in the nasal region. Two cell groups have been described in this region, goblet cells and cilliated cells, with the highest expression of ACE2 in the upper respiratory system [27]. This expression has been demonstrated by scRNA-seq both in nasal brushes and in biopsies [30]. These findings possibly imply that surface cells might serve as a viral reservoir in the oro/nasopharyngeal region, and contain virus from secretory cells with high expression of ACE2 and TMPRSS-2.
Variation in the expression of ACE2 and TMPRSS2 has also been shown in relation to certain comorbidities. Thus, in asthmatic patients, the presence of AEC2 in the nasal epithelium would be low, and TMPRSS2 would be high in the bronchial region [31]. In smoking patients, an increase in both proteins is evidenced at the level of the respiratory tract, which could be associated with more severe symptoms of SARS-CoV-2 infection within this group of patients [31]. Finally, age might have implications for the expression of ACE2 and TMPRSS2, as it has been shown that in children, the expression of ACE2 and TMPRSS2 in the upper and lower respiratory tract is low compared to that in adults [32]. All these variables might have implications in the protein determination of AEC2 and TMPRSS2 in addition to the different antibodies and the techniques (IHC, ISH) used for their demonstration.
One possible explanation for why SARS-CoV-2 is present in squamous cells obtained from nasopharyngeal smears is the functional consequences of virus–host interactions. Molecular interactions can also occur with attachment factors or coreceptors, which are often found on the cell surface as carbohydrate structures (i.e., lectins and sialic acids). They might serve only to bind SARS-CoV-2 particles, helping to concentrate viruses on the cell surface [18, 33]. Unlike attachment factors, SARS-CoV-2 receptors actively promote cell entry. In our ultrastructural study, we demonstrated virions in close relationship with the cytoplasmic membrane and in the extracellular region. These findings could confirm the viral concentration without cellular internalization. However, the binding of SARS-CoV-2 to the cell surface and its subsequent internalization could depend on other alternative receptors, such as CD147 (basigin), a highly glycosylated transmembrane protein that is robustly expressed in blood cells [34, 35, 36, 37, 38]; CD26 can also bind to SARS-CoV-2 according to recent structural studies [39]. Other receptors that are potentially used by SARS-CoV-2 are amino peptidase N (ANPEP; a receptor for human and porcine coronaviruses), ENPEP and glutamyl aminopeptidase 29, and DC-SIGN and neuropilin-1 [40, 41].
A limitation of our study is the alterations due to the preservation of the material, since the universal viral transport medium does not adequately preserve the different cellular elements. Thus, squamous cells are more resistant, while columnar cells and goblet cells are more labile, which can cause alterations in the immunocytochemical expression of this cellular component.
In summary, we have shown that SARS-CoV-2 mainly localizes to squamous cells in patients who are positive for SARS-CoV-2 transcription-mediated nucleic acid amplification. SARS-CoV-2 might be located in the nasopharyngeal region because of the presence of nonspecific receptors, which facilitate viral accumulation and the contagious state of the disease. The fact that ACE2 receptor is not found in the squamous cells of nasopharyngeal region seems to suggest that this region is not associated with replication or virulence. Further studies are required if the role of the nasopharyngeal region in SARS-CoV-2 infection is to be clarified.
ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease serine 2; S/CO, signal/cutoff; ICC, immunocytochemistry study.
DP, JG, and KBP designed the research study. JG, CG, BP, AH, HC, LD and MG performed research. DP, KBP, and FR analyzed the data. DP, KBP, JG and FR wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
The study protocol was reviewed and approved by the Hospital’s Ethics Committee (registration number CEIm 204/2020). Written informed consent was obtained from each subject and followed the principles of the Declaration of Helsinki 1964 and its subsequent modifications.
The authors thank John Bates for his assistance in the revision and grammar correction of English.
This research received no external funding.
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
