Epithelial to Mesenchymal Transition (EMT) is a biological process characterized by the transition from immotile epithelial cells to motile mesenchymal cells. Though shown to be implicated in many biological processes, it has also been identified to enhance migration and invasion of cancer cells leading to metastasis. A class of microRNAs called “oncomiRs” plays a significant role in the regulation of malignant transformation and metastasis. In this review, the ability of different signaling pathways in controlling EMT through well-defined regulatory networks, and the role exerted by oncomiRs in regulating the specific signaling pathways like TGF-β, Wnt, Notch and Hedgehog in modulating breast cancer metastasis have been discussed with updated information. Further, this review focuses on the significance of up and down regulated microRNAs in the pathogenesis and progression of breast cancer and how such microRNAs could be treated as potential therapeutic targets to circumvent cancer. As a prospective strategy, we highlight the importance of circulating tumor cells (CTCs) and their derived microRNAs as prognostic indicators and cancer therapy monitoring tools.
Breast cancer is the second most common cancer among women and one of the leading causes of mortality worldwide. Most deaths are due to late diagnosis at metastatic stage as well as resistance to therapy (1). Recently, the phenomenon of Epithelial to Mesenchymal Transition (EMT) has been linked to tumor invasion, metastasis and drug resistance (2). The phenomenon of resistance in cancer cells can occur due to modulation of signaling pathways where the cells adapt themselves for self-sustenance. Secondary tumor is formed at the site of metastasis by a reverse process of EMT termed as MET, (Mesenchymal to Epithelial Transition) (Figure 1). A parallel change with loss of apico-basal polarity, degradation of cell- cell junctions, cytoskeletal rearrangement, upregulation of mesenchymal markers and simultaneous down regulation of epithelial markers is observed in oncogenic EMT (3)(Figure 2). The tight regulation of this sequential event is controlled by a set of transcriptional factors (TFs) like Slug, Twist, Snail, Zeb1/2 working together in the cellular cross talk which in turn helps the cell to gain attributes of invasiveness and migration (4). Thus, the event is mainly characterized by decrease in the expression of epithelial markers such as E-cadherins and δ-catenin with a simultaneous increase of mesenchymal markers such as Vimentin, N-cadherin, fibronectins and matrix metalloproteinases. It has been widely established that various signaling pathways play a crucial role in these two processes. This complex and multifunctional program is tightly regulated by the interplay of many complicated regulatory networks (Figure 3). Understanding this cross talk among networks might provide cues to target specific nodes of interaction so that effectively many of the detrimental steps of the metastasis pathways could be tackled simultaneously.
 Figure 1
Figure 1A schematic representation of Epithelial Mesenchymal Transition. Cancer cells detach from the basement membrane and intravasate to the nearby blood vessels as CTC and travel through blood vessels to secondary site in a process described as metastasis. They get lodged in different organs by a process termed as Mesenchymal Epithelial Transition (MET).
 Figure 2
Figure 2MicroRNA biogenesis and regulation. miRNA genes are transcribed by RNA pol II to generate primary transcripts called pri-miRNA. Further processing by RNA binding protein DGCR8 results in 85~nucleotide stem loop structures called pre-miRNA. After its cytoplasmic transportation by exportin, it is further processed by Dicer, RNase III enzyme to 22 nucleotide miRNA/miRNA* duplex. The mature miRNA with RISC (RNA induced silencing complex) binds to 3’UTR region of target mRNA to either undergo cleavage or translational repression.
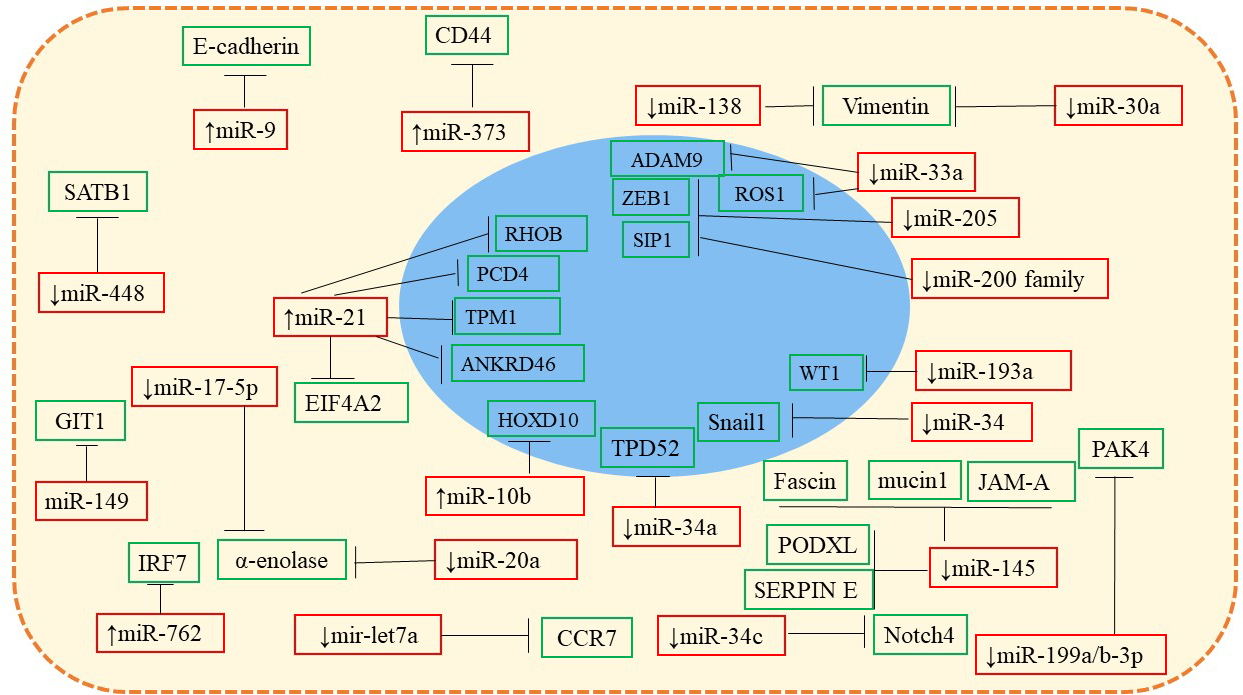 Figure 3
Figure 3Illustration of miRNAs: mRNA target network. MicroRNAs that are up and down regulated in breast cancer cells target EMT related genes in different pathways leading to metastasis. For example, upregulated miR-9 targets E-cadherin which in turn disrupts epithelial tight junctions and activates β-catenin cascade. Similarly, downregulation of miR-30a and miR-138 deregulate vimentin expression thus triggering EMT.
MicroRNAs are tiny non-coding RNA molecules that play a significant role in regulating the genes that influence proliferation and migration of cells and thereby, the tumor invasion and metastasis (5). Expression of miRNA levels are observed to vary across tumor types, grades and stages as compared to healthy tissues (6). Recent studies have also shown the involvement of microRNAs in EMT through careful modulation of oncogenic signaling pathways. Accumulating evidence show that microRNAs act as inducers and suppressors of transcription factors involved in EMT and thus influencing breast cancer progression. Overall, so far, there are 130 microRNAs reported to influence EMT directly (7). Role of miR-200 family has been well studied in several cancers. Enhanced expression of miR-200 family members was found to inhibit EMT through targeting ZEB family, while suppression of the same led to enhanced EMT leading to invasion and metastasis (8). Circulating tumor cells (CTC) are the cells shed from different stages of tumor into the peripheral blood in circulation. Since it can be isolated by liquid biopsy systems, these are considered to be predictive indicators for disease prognosis and therapy monitoring (9). Altered expression of specific microRNAs in CTC could provide a real time indicator of tumor progression and thus can be helpful in predicting/assessing the disease status (10). Thus, molecular characterization of CTC along with expression profiling of mRNAs and miRNAs in these cells could be used as an additional tool in deciding personalized therapy.
In this review, we provide an overview of the major signaling pathways mediated by TGF-β, Wnt, Notch and Hedgehog transcription factors with the involvement of microRNAs as regulators affecting these signaling networks during breast cancer metastasis via EMT. We also highlight the various isolation and enrichment techniques for CTC along with molecular characterization that could be employed as clinical parameters for diagnostic, prognostic and therapeutic monitoring of breast cancer.
For the cells to metastasize, a cascade of events such as down regulation of receptors for cellular adhesion, upregulation of factors for motility, and degradation of extracellular matrix by secreting proteases are necessary (11). Cross talk between tumor cells and their microenvironment through signal transduction, controls the expression of EMT inducing transcriptional factors which in turn determines the progression and physiology of the disease (12). Each factor has its own distinct expression pattern and role in tumor progression depending on the cell, tissue type, and the signaling pathways. Loss of E- cadherin, an epithelial junction protein is one of the major hallmarks of EMT. Slug, Snail and Zeb2/SIP directly repress transcriptional expression of E-cadherin, whereas Zeb1 and Twist act indirectly to orchestrate EMT (13). Induction of EMT in breast cancer involves the intricate cross talk between various molecules in the major signaling pathways like TGF-β, Wnt, Notch, Hedgehog, etc (14). MicroRNAs being the master regulators of gene networks, modulate oncogenic and tumor suppressor pathways by regulating the transcription factors involved in cell proliferation and thus bringing about dynamics in cell cycle regulation with several outcomes (15). Thus, it becomes imperative to understand the microRNA mediated regulation of the transcription factors that are involved in EMT.
TGF-β is a multifunctional cytokine growth factor which has a direct impact on breast cancer pathophysiology (16). It is one of the major players in cellular communication affecting wide range of cellular behavior including proliferation, differentiation, apoptosis, extracellular matrix formation (ECM) and immunoregulation (17). Being a tumor suppressor gene, it is also shown to have multifaceted role in cancer, depending on the tumor grade and stage. In early stages, this gatekeeper inhibits epithelial cell cycle by promoting apoptosis. But in later stages it is associated with increased tumor cell motility and invasiveness (18). TGF-β signaling cascade involves the binding of the ligand (TGF-β) to the TGF-β type II receptor. Recruitment and transphosphorylation of TGF-β type II receptor activates the receptor mediated adaptors Smad 2 and Smad 3 which in turn form complexes with Smad 4 (19). In addition, Smad independent pathway collaborates with various signaling cascades like phosphatidylinositol-3-kinase (PI3K) Akt, mitogen activated protein kinase (MAPK) and Rho family to regulate Snail and Slug activity (11).TGF-β induces desmoplastic and fibroblastic reactions which result in a dramatic change in tumor microenvironment. It helps in secretion of several extracellular matrix components (ECM) along with lysyl oxidase promoting metastatic progression (20). TGF-β plays a vital role in activating Snail which downregulates cadherin 16 and HNF-18 thereby inducing EMT. This in turn results in the downregulation of epithelial markers and the upregulation of mesenchymal markers with different phenotypic populations of cells (21). Annexin A1(Anx A1) an inhibitor of NF-κB, initiates TGF-β signaling and actin remodeling which results in elevated cell migration and invasion in breast cancer cells (22).
MicroRNAs play a major role in activation of TGF-β signaling cascade by upregulating/downregulating target proteins of signal transduction. Earlier studies have shown the downregulation of miR-200 family in TGF-β induced EMT cell line models (23). Over expression of miR-200 successfully prevented EMT, thereby inducing mesenchymal to epithelial transition (MET) and hence can be considered as regulators of epithelial identity (24). Cross talk between ZEB1/2 (TGF-β induced transcriptional repressors) and miR-200 family forms a double negative feedback loop regulating EMT. ZEB1/2 represses the promoter regions of miR-200b, -200a -429 and miR-220c/141 clusters and they in turn target 3’UTR of ZEB1/2 genes (25). Snail, an EMT inducer forms similar negative feedback loops with miR-34 family (26). The dynamics between these two loops (miR-34-SNAIL and miR-200-ZEB1/2) forms bistable cell switches of different phenotypes i.e. epithelial, partial EMT and mesenchymal within the same population of cells (27). TGF-β also upregulates several miRNAs like miR-181, miR-155,miR-10b, miR-30, miR-223 and miR-34 family thereby promoting EMT (28). Downregulation of miR-196a-3p led to increased neuropilin 2 levels which was required for TGF-β1 induced migration (29). Hsa-miR-155 helps in deregulation of the C/EBPβ which converts the TGF-β response from growth inhibition to EMT promoting metastasis (30). Increased plasma TGF-β1 levels in 23 breast cancer patients had a positive association with TGF-β signaling markers like parathyroid hormone related peptide (PTHrP) and interleukin 10 (IL-10) (31, 32). Earlier studies have shown the increased levels of CXCL1 along with TGF-β1 in blood, and resulted in poor prognosis and increased propensity to extended metastasis in breast cancer patients (33). A recent study of patients having estrogen receptor negative breast cancer reported the correlation between high TGF-β R-II expression and reduced survival indicating TGF-β as a major inducer of EMT (34).
Wnt signaling is mostly active during embryonic development and is related to cellular processes including cell differentiation, proliferation, maintenance of apicobasal polarity and migration (35). Aberrant activation of Wnt is usually associated with many human diseases including cancer (36). It has also been shown to play an important role in regulation of EMT through canonical (β-catenin) or noncanonical pathways (37). Canonical Wnt pathway recruits’ members like Wnt1 and Wnt3a which binds to FZD and LRP5/6 receptors, leading to the dissociation of destruction complex. This prevents β- catenin phosphorylation enabling it to translocate to the nucleus and activate downstream target genes like c-myc, CD44 etc. leading to uncontrolled proliferation of cells (38). Non-canonical pathways are independent of β- catenin and are divided into Wnt/Ca+2 and planar cell polarity (PCP) pathway which is usually initiated by Wnt5a and Wnt11 (39, 40). In Wnt/Ca+2 pathway, binding of Wnt ligands with the receptors activate the inositol trisphosphate through phospholipase C induction leading to stimulation of c-GMP factors and release of Ca+2 from the endoplasmic reticulum (41). In the planar cell polarity pathway, Wnt binds to the FZD receptors and its co receptors ROR and Ryk that activates Rho and Rac. This leads to activation of Rho-associated protein kinases (ROCK) and c-Jun N-terminal kinase (JNK), that plays a critical role in cellular polarization and migration through actin polymerization (42). Studies related to HER2 transgenic mice have revealed that breast cancer cells disseminate early in their transformation and both canonical as well as non-canonical pathways are the cause for this migration (43, 44). The aberrant expression of β-catenin in the nucleus or cytoplasm stimulates cell proliferation and it significantly correlates with the poor survival of breast cancer patients exhibiting an advanced stage of the cancer (45–47). Wnt pathway also up-regulates transcription factors like Slug and Twist which are prominent markers for EMT (48). The over expression of the co-receptors LRP5 and LRP6 of Wnt pathway is also implicated in mammary gland tumorigenesis and is suggested to be used as a major prognostic marker for detection of breast cancer patients (49).
MicroRNA dysregulation has been linked with hyperactivation of Wnt signaling that drives breast tumorigenesis (50, 51). OncomiRs were found to activate/inhibit canonical/non-canonical Wnt pathways by mutual feedback loop mechanisms. High levels of expression of miR-182 by β-catenin targeted MMP inhibitor RECK thereby increasing invasiveness (52). miR-374a overexpression inhibited the suppressors of Wnt pathway (PTEN and WIF1) resulting in an increased Wnt-mediated EMT and metastasis in breast cancer cells (53). miR-1 and miR-100 suppressed breast cancer progression by targeting the receptors FZD7 and FZD8 of the Wnt pathway (54, 55). miR-218 showed a positive feedback loop by targeting Wnt inhibitors (Sclerostin (SOST), SFRP2) resulting in increased osteoblast differentiation and metastasis of cancer cells from breast to bone (56). Different miRs like miR-22, miR-200b, miR-185-3p, miR-324-3p, miR-26a, miR-487b, miR-329, miR-410, miR-374b were found to inhibit different Wnt ligands of canonical pathway (57). miR-122 and miR-148a were shown to suppress WNT1 ligand which in turn reduced cellular proliferation and metastasis respectively (58, 59). Binding of Wnt antagonists like WIF, Dkk, sFRP, activates Wnt signaling. Certain miRNAs like miR-374a, miR-603, miR-1260b, miR-328 were shown to target these Wnt antagonists, thereby activating the pathway. Downregulation of miR-200 family promotes EMT and tumorigenesis through Wnt/β-catenin pathway. miR-220a inhibited cancer progression by either targeting 3’UTR of the β catenin or ZEB1/2 (60, 61). Other miRs which are studied beside miR-200 cluster that regulate β catenin levels are miR-214, miR-320, miR-101, miR-1826, miR-548b, miR-33a (57). Twenty-seven miRNAs predicted to be involved in the regulation of Wnt signaling pathway were identified as differentially expressed in locally advanced triple negative breast cancer (TNBC) (62, 63). Recent studies showed that increased expression of miR-218-5p led to bone metastases in TNBC patients (64). Hence, these putative miRNAs are the major determinants in altering the Wnt pathway to initiate EMT.
The formation of new blood vessels is very critical for the cancer cell migration and its metastatic spread. Tumor angiogenesis is regulated by several factors of which Notch is very crucial (65). High expression of Notch3 was reported to be associated with vessel formation and increased angiogenesis (66). Accumulating evidence suggest the deregulation of Notch in breast carcinogenesis (67). It also regulates various physiological processes including cell division, cell fate determination and migration that are involved in development and embryogenesis (68). The interaction between Notch receptors and their respective ligands triggers a cascade of sequential events like proteolytic cleavage to release Notch intracellular domain, its translocation to the nucleus, binding with the repressor thereby transcriptional activation of target genes that govern various cellular processes (71). Earlier studies have shown the involvement of Notch receptors as a primary site of integration of mouse mammary tumor virus (MMTV) (69). Over expression of Notch1 and Jag1 were mostly associated with poor prognosis in breast cancer patients (70). Induction of Slug mediates JAG1 activation of Notch in breast epithelial cells which in turn induces EMT (71). Constitutive activation of Notch1, Notch3, or Notch4 in mice led to formation of aggressive, metastatic breast tumors (67). Hey, a downstream target gene of Notch 1, activated Jagged-Notch-Slug signaling axis leading to an increase in breast cancer progression (71). Notch pathway was also found to promote EMT by mediating hypoxia stimulus which is mostly related to increased cell motility and invasiveness. Notch helps to indirectly stabilize Snail-1 by inducing Hypoxia inducible factor (HIF-1α) to lysyl oxidase (LOX) promoter (72). The interaction of Notch with other signaling pathways like Wnt and TGF-β bring about a dynamics which help in orchestrating EMT (73). LEF-1 a target gene of Wnt with Notch ligands Dll3 and Dll4 were detected simultaneously in array of 34 metastatic breast cancer (MBC) samples leading to its diagnosis (74).
Notch pathway is also shown to be regulated by miRNAs and in turn activation of Notch signaling has been shown to downregulate the miR-221/222 expression thereby promoting the metastasis of ERα positive cell lines (75). The alteration in the miRNA expression pattern between the ER−ve and the ER+ve luminal subtypes are shown to be associated with migration and poor prognosis (76). Differential expression of miR-130b-3p demonstrated regression in cell migration and invasion and inhibition of MMP 9 by targeting DLL1 ligands (77). ZEB1 was also shown to initiate Notch signaling by stabilizing the expression of its receptors Jagged1, Maml2, and Maml3, through inhibition of the miR-200 clusters (78). miR-metastatic breast 34a was shown to downregulate Notch 1 pathway in breast cancer stem cells (79). miR-34 family that includes miR-34a, miR-34b, miR-34c regulates p53 and Notch pathway and is suggested to have a tumor suppressor role in various cancers (80). It directly modulates Notch1 and Notch 2 levels resulting in suppression of cell renewal properties and stemness in cancer stem cells (81). Overexpression of miR-199-5p led to decrease in population of cancer cells by blocking Notch pathway (82). miR-146a was shown to repress Notch signaling by directly targeting Numb, a Notch inhibitor in breast cancer (83). The contribution of Notch in EMT is evident and is considered as one of the most promising targets for inhibiting tumor progression and overcoming chemo-resistance (84).
The hedgehog (Hh) pathway is critical for stem cell development and maintenance apart from other biological processes like regulation of tissue homeostasis and regeneration during development (85). Recent studies underline the importance of tightly controlled Hh pathway activation for proper development of mammary glands (86). Hedgehog signaling starts with three different ligands, and two receptors Patched (Ptch 1) and Smoothened (Smo). In the absence of the Hh ligand, Ptch1 inhibits signal transduction by inhibiting Smo. Hh ligand binding activates Smo resulting in the activation of GLI transcription factors, which can function as either activators or repressors of transcription (87, 88). Increasing evidence implicate the role of Hh pathway in regulation of EMT by maintaining the plasticity of the different EMT phenotypes (89). Deregulation of Hh pathway is usually associated with multifarious human cancers leading to tumorigenesis, metastasis and chemo-resistance (90). Primary cilia are non-motile structure involved in tight regulation of different signal transductions. Claudin low subtype of breast cancer acquires stem cell like state by the growth of primary cilia through Hh signaling (91). Colavito et al. (2014) identified overexpression of GLI1 to be a critical determinant of breast cancer cells that have undergone EMT. Further they showed that Hh signaling is crucial for cancer stem cell behaviour and discovered a novel link between Nf-κB and GLI1 (92). Conditional expression of GLI1 in experimental mouse models developed mammary tumors leading to metastasis in bone, validating the role of Hh pathway in EMT-mediated breast tumorigenesis (93, 94).
Very little is known about the microRNAs involved in the regulation of Hh signaling in breast cancer metastasis. miR-1471 is known to be downregulated in breast cancer by modulating the expression of Shh thereby leading to inhibition of metastasis (95). miR-212 regulates cell proliferation and invasion by targeting Patched 1 of Hh signaling (96). miR-326 directly targets Smo and GLI2 hence negatively regulating Shh signaling that is involved in mesenchymal phenotype (97). miR-124 has been reported to be the downstream target gene of Hh signaling and GLI2/miR-124 axis is crucial for cellular proliferation (98). miR-138 is associated as a tumor suppressor gene repressing Smo expression and hence suppressing migration and invasion (99). Though Hedgehog pathway is mostly related to developmental process, its contribution to EMT and maintaining stemness of the breast cancer cells can’t be ignored.
MicroRNAs are termed as master regulators of gene expression since it controls about 30% of the genes in human (100). It is already known that microRNAs are important modulators of malignant transformation and metastasis and that altered signatures revealed in different cancer types are indicators of the same (101). Increasing experimental evidence for more than a decade established the role of microRNAs in cancer initiation and progression and such miRNAs are called oncomiRs which either act as oncogenes or tumor suppressors (102). Their role has been well established in controlling EMT and the reverse process MET through well-defined signaling pathways and other cellular components (103). Their over-expression/under-expression disturbs the patho-physiological balance that targets EMT modulators leading to disease initiation and progression. Thus, miR signatures from metastasized tissue can be used to detect the primary site of cancer origin (104) and also based on the expression levels, it can be used to predict prognostic status of the disease. Accordingly, microRNAs function as switch to directly / indirectly control EMT via various signaling pathways and downstream events which in turn play key role in metastasis.
Many microRNAs act as EMT regulators and their changed signatures have been well documented with the onset and progression of various cancers (105). A set of microRNAs including miR-200 clusters directly target EMT modulators for buffering transcriptional output in human mammary EMT models. Based on their up and down regulated expression pattern, it could be used as ideal indicators of the disease status. Increased miR9 levels downregulated E-cadherin expression by targeting CDH1 leading to initiation of EMT (106)(Tables 1-2). This loss of E-Cadherin, activated vascular endothelial growth factor (VEGF) leading to tumor angiogenesis (106). Elevated levels of miR-29a in breast carcinomas supressed tristetrapolin that disrupted epithelial polarity (107). miR-10b is a well-studied oncomiR in various types of cancers that promotes cell migration, tumor invasion and distant metastasis (108).
| Status of the miRNA | miRNA | References |
|---|---|---|
| Upregulated in metastatic breast cancer cells | miR-9, miR-29a, miR-10b, has-miR-106b-5p, has-miR-7-5p, miR-1273g-3p, miR-21, miR-30b, miR-155, miR-106b, miR-450a, miR-148a, miR-150, miR-373, miR-762, miR-103, miR-23a miR-222, miR-23b, miR-24, miR-25 | (106), (107), (108), (109), (110), (104), (112), (116), (117), (197), (120) |
| Downregulated in metastatic breast cancer cells | miR-126, miR-355, miR-34, miR-34a, miR-200, miR-205, miR-200c, miR-30a, miR-33a, miR-138, miR-148a, miR-149, miR-199a/b-3p, miR-99a, miR-99a, miR-125b, miR, miR-130b, miR-448, miR-193a, miR-17-5p, miR-20a | (127), (129), (130), (131), (198), (126), (134), (133), (135), (137), (138), (139), (104), (142), (132), (141) |
| High levels found in serum | miRNA-106b, miR-103, miR-23a miR-222, miR-23b, miR-24, miR-25, miR-29a, miR-10b, miR-21, miR-125b, miR-145, miR-155, miR-19, miR-19a, miR-382, miR-181b, miR-1, miR-133a, miR-133b, miR-92a, miR-486-5p, miR-423-5p, miR-7, miR-185, let-7i, miR-16, let-7b, miR-93, miR-214, miR-16-2, miR-320a, miR-21, miR-18b, miR-103, miR-107, miR-652, miR-141, miR-200a, miR-200b, miR-200c, miR-210, miR-375, miR-801 | (116), (120), (121), (122), (113), (114), (124), (123) |
| Low levels found in serum | miR-223, miR-338-3p, miR-768-3p | (122), (123) |
| miRNA | Status | Target | Effect on breast cancer cells | References |
|---|---|---|---|---|
| miR-9 | Upregulated | E-cadherin | Loss of cell adhesion, activation of Wnt signaling, increase in VEGF levels | (106) |
| miR-10b | Upregulated | HOXD10 | Increased expression of pro-metastatic gene RHOC | (108) |
| miR-21 | Upregulated | PCD4, TPM1, EIF4A2, ANKRD46, RHOB | Increase in metastatic potential | (112), (115), (199) |
| miR-373 | Upregulated | CD44 | Increased migration and invasion | (117) |
| miR-762 | Upregulated | IRF7 | Invasion | (197) |
| miR-34c | Downregulated | Notch4 | Induction of EMT | (129) |
| miR-34 | Downregulated | Snail1 | Activation of EMT | (130) |
| miR-34a | Downregulated | TPD52 | Increased migration and invasion | (131) |
| miR-200 family and miR-205 | Downregulated | ZEB1, |
Loss of epithelial characteristics | (198) |
| miR-193a | Downregulated | WT1 | Increased metastatic potential | (132) |
| miR-30a, miR-138 | Downregulated | Vimentin | Increased expression of vimentin-mesenchymal protein in IDC | (134) |
| miR-33a | Downregulated | ADAM9, ROS1 | Increased metastatic potential | (133) |
| miR-138 | Downregulated | Vimentin | Increased expression of vimentin-mesenchymal protein. | (135) |
| miR-145 | Downregulated | Fascin, JAM-A, PODXL, SERPIN E, mucin-1 | Increased metastatic potential | (136) |
| miR-148a | Downregulated | WNT1, NRP | Increased metastatic potential in TNBC | (137) |
| miR-149 | Downregulated | GIT1 | Increased metastatic potential | (138) |
| miR-199a/b-3p | Downregulated | PAK4 | Increased migration and invasive capacity | (139) |
| miR-let-7a | Downregulated | CCR7 | Increased migration and invasive capacity | (140) |
| miR-17-5p, miR-20a | Downregulated | α-enolase | Increased migration and invasive capacity | (141) |
| miR-448 | Downregulated | SATB1 | EMT via activation AR-EGFR signaling | (142) |
Tumors that have metastasized to distant organs retain miR signatures found in primary tumors and these specific signatures seem to be from different metastasis locations. This attribute of miRNAs makes them ideal prognostic markers for cancers. It has been reported that breast tumors that metastasized to lungs and ovaries had dysregulated levels of miR-3201 (109). Patients having overexpression of miR-21 in breast tumor showed advanced stage of the cancer, lymph node metastasis with poor prognosis (110–112). Upregulation of miR-21 activated MAPK pathway in HER2/neu-positive breast cancer cells by downregulating programmed cell death (PDCD4). Knock-down of miR-21 suppressed cell growth and proliferation in xenograft models (112). Several studies have indicated that levels of circulating miR-21 had strong correlation with different cancer stages independent of oestrogen-receptor status (104, 113–115). miR-155 is one of the well-studied miRs in all types of cancers and is associated with lymph node metastasis in breast cancer. When miR-155 was knocked down in human breast cancer cell lines, there was an inhibition of growth in cancer cells by induction of cell arrest in G0/G1 phase of cell cycle with increased apoptosis (112). Elevated levels of Hsa-miR-106b, miR-30b, miR-450a, miR-148a, miR-150 and miR-323 were shown to be associated with large tumor size and lymph node metastasis (116). miR-373 and 520c are pro-metastatic miRs, promoting tumor cell migration and invasion by targeting CD44 (117).
MicroRNAs are also released into the peripheral blood as circulating nucleic acids or are present in CTC (circulating tumor cells) that originate from tumor tissues. (118). Their nuclease resistant hairpin loop structure makes them ideal and stable biomarkers for early and non-invasive detection of breast cancers (119). For example, miR-103, miR-23a, miR-29a, miR-222, miR-23b, miR-24 and miR-25 upregulated in both serum and breast cancer tissues (120) are candidates to be detected from the blood. Elevated levels of miR-10b, miR-21, miR-125b, miR-145, miR-155, miR-19, miR-19a, miR-382, miR-181b, miR-24, miR-1, miR-133a, miR- 133b, miR-92a, miR-486-5p, miR-423-5p, miR-7, miR-185, let 7i, miR-16, miR-214, miR-16-2, let 7b, miR- 93, miR-320a have been detected in serum of breast cancer patients (113, 121, 122). Higher levels of miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-210, miR-375, and miR-801 were seen in plasma of CTC-positive compared to CTC-negative metastatic breast cancer patients and healthy individuals (123). It has been opined/demonstrated that in patients with TNBC, miR-18b, miR-103, miR-107, and miR-652 levels could be used to accurately predict tumor relapse and overall survival of the patients (124). Analysis of miRNAs profiles form the CTC have opened new frontiers in the field of prognosis leading to clinical management of the disease.
The microRNAs having a low expression during the diseased condition are those which are classified as tumor suppressing or metastasis suppressing miRNAs. There are several miRs whose expression is significantly low in breast tumors compared to healthy tissues. miR-200 cluster has been reported to be downregulated in breast cancer. Recently, Gregory et al. have demonstrated that a regulatory autocrine signaling axis of TGF-β/ZEB/miR-200 exists in breast cancer patients (24). In a complementary study, it has been demonstrated that the over expression of miR-200 members caused E-cadherin upregulation and inhibited EMT via targeting the transcription factors ZEB1/2 (125) and a decrease in invasion and migration potential of cancer cells was observed by reintroduction of miR-200c (126). Breast cancer cells lose expression of miR-126 and miR-355 while progressing into metastatic stage, however, a knock-in rescue assay with these microRNAs reduced cell proliferation and invasiveness (127). In some cases, downregulation of metastasis suppressor miRs occurs due to their epigenetic silencing by CpG island hypermethylation (128). Low expression of miR-34c was found in breast cancer stem cells due to hypermethylation of CpG islands leading to overexpression of Notch4, thus promoting EMT. Ectopic expression of miR-34c led to reduced self-renewal, EMT and migration of breast cancer stem cells (129). Decreased levels of miR-34 were seen in breast carcinomas due to loss of function or mutations in p53, which in turn suppressed Snail1 activity. (130). MiR-34a also inhibits EMT by directly targeting Tumor protein D52 (TPD52) (131).
A low expression of miR-193a compared to adjacent healthy tissues suppressed breast cancer proliferation and metastasis by targeting Wilm’s Tumor 1 (WT1) expression (132). On similar lines, a negative correlation observed between levels of miR-33a and lymph node metastasis by targeting ADAM9 and ROS1 genes (133). Decreased levels of miR-30a and miR-138 were also reported in invasive ductal carcinoma (IDC) and MBC patients respectively. Overexpression of this miR led to downregulation of vimentin, a major player of EMT progression (134, 135) (Tables 1-2). miR-145 regulates motility of breast cancer cells by targeting fascin, JAM-A, PODXL, SERPIN E, mucin-1 thus inhibiting tumor metastasis (136). miR-148a also acts as a metastasis suppressor in triple negative breast cancer by targeting WNT1 and NRP. When miR-148a was over expressed, metastasis was suppressed in vivo by reduction of cancer cell extravasation (137). Downregulation of miR-149 and simultaneous upregulation of G-protein-coupled receptor kinase-interacting protein 1 (GIT1) was observed in advanced breast cancer (138) and that led to the understanding that miR-149 is a metastasis suppressor gene and targets GIT1 transcripts.
Reduced expression of miR-99b, miR-125b, miR-205, miR-130b, miR-24, miR-99a and miR-199a/b-3p are reported in breast cancer cells having metastatic potential. miR-199a/b-3p arrests breast cancer cells in G1 phase and inhibits PAK4 which is known to have roles in tumorigenesis (104, 139). miR-let7a found in most of the cancers downregulates invasion propensity by targeting CCR7 (140). Reduction in the levels of miR-17-5p and miR-20a causes increase in α-enolase secretion, IL-8 and cyclin D1 which induces secretion of CK-8 that in turn activates plasminogen in promoting migration of cancer cells (141). miR-448 is downregulated in breast cancer cells subjected to chemotherapy and the suppression of the same promoted SATB1(Special AT-rich sequence binding protein1) expression which led to EMT via activation of AR-EGFR (Amphiregulin-Epidermal Growth Factor Receptor) and NF-κB signalling (142). Empirical evidences show that some microRNAs are released into various extracellular components under pathological conditions and studies have also established a correlation between circulating miRs and breast cancer status. (117). Interestingly, there are microRNAs whose levels are reduced in the serum of cancer patients compared to healthy individuals. For example, miR-223, miR-338-3p and miR-768-3p are downregulated in serum of breast cancer patients (123). Serum biomarker level changes during treatment and hence can be correlated with progression-free survival (PFS) and monitoring of disease status in metastatic breast cancer patients.
Since there is an inverse correlation between expression levels of microRNAs and their targets, they can be used as ideal therapeutic candidates for cancer therapy. MicroRNAs that are upregulated in cancerous conditions can be targeted by using antimiRs /miRNA sponges; while those that are downregulated or lost can be reintroduced using microRNA mimics that can complement the loss. Introduction of miR-33a, miR-149 and miR-145 as mimics reduced cancer cell proliferation (133, 138). miR-34a overexpression as mimics targeted TPD52, which resulted in less invasion and migration of breast cancer cells (131). Decreased vimentin levels and increased E-cadherin levels was observed when miR-138 was overexpressed in breast cancer cell lines (135). miR-199a/b-3p was used to target PAK4/MEK/ERK pathway resulting in reduced tumorigenesis (139). Anti-miRs of miR-373 and miR-762 suppressed migration of cancer cells in vitro suggesting that these miRs can be targeted to decrease invasiveness of breast cancer (117). Interestingly cancer cell sensitivity to chemical agents such as paclitaxel used in cancer therapy is increased by introduction of miR-200c by repression of class III beta tubulin (TUBB3) (126). Various DNA carriers such as nanoparticles, liposomes, dendrimers etc. are used to deliver these short nucleotides at the specific sites (143). BRCA1 deficient tumors were targeted by antimiRs of miR-21 as PNA (Peptide Nucleic acid) (112, 144). Effective knockdown of Twist expression was observed in vivo when PEI coated mesoporous silica nanoparticles was conjugated with siRNA (145). 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) based nanoliposomes were used to deliver EphA2 siRNA directly into the tumor cells with enhanced permeation (146).Thus, microRNAs could be potential futuristic molecules to target breast cancer. Though decisive and conclusive experimentation is still eluding, there is immense potential of miRNAs in cancer therapy as shown by some preliminary studies. There is still a long way to go for successfully translating this potential from the realms of laboratory to the clinical reality.
The process of dissemination of the cells from the solid tumor during metastasis has already been established in 1900s. Certain cells shed from the primary tumors, migrate early via blood stream and they are called circulating tumor cells (CTC). They serve as mediators of homing cascade in metastasis (147). Detection and evaluation of these cells in blood stream has an important implication in clinical applicability (148). Their presence in the peripheral blood is suggested to be an indicator of poor clinical outcome before and after chemotherapy (149). Also, it has been used to assess progression free survival and overall survival. CTC that undergo EMT, acquire partial mesenchymal phenotype which results in decreased epithelial gene expression and loss of cell contact from basement membrane (150). Therefore, it can be used as a biomarker to monitor cancer progression non-invasively and also decide the treatment regimen. However, it is limited by the methods of detection and isolation of the CTC as their numbers are as low as 1 CTC per billion normal blood cells (151). Therefore, improved detection techniques with cocktail of EMT markers and epithelial markers have been suggested for the isolation and enrichment of CTC (152). Analysis of blood samples from breast cancer patients revealed the presence of relevant number of CTC that correlates with different grades, stages and lymph node status of the cancer (153). Therefore, CTC hold a great promise in improving the current diagnostic and prognostic tools along with appropriate treatment regimen.
As discussed earlier, microRNAs can be found free in blood as circulating tumor RNAs, with specific signatures of the grades and stages of tumors. These microRNAs are also present in the circulating tumor cells and a study by Tan.et.al (2019) established that there was a correlation in the expression pattern of microRNAs in the CTC with that of the unique expression pattern of microRNAs that was observed with the tumor cells in metastatic breast cancer patients. They further found that miR-106b detected from CTC of primary tumor could predict the prognostic potential of patients with MBC (154). Compared to the already discovered circulating miRNAs and tissue specific miRNAs, very less is known about the microRNAs present in the CTC. Deregulation of single miRNA can have profound effect on expression levels of hundreds of mRNAs leading to changes in actual target mRNAs’ regulatory functions in cancer development. Tumor specific miRNAs are the best set of miRNAs to profile and assess cancer types, grades and stages and thus are valuable tools for detecting the diseased condition. However, characterization of molecular signatures of CTC leading to miRNA and mRNA profiles can improve the currently available model towards personalized diagnosis and therapy (155).
In the scenario of metastatic disease settings, CTC has gained importance as a tool for therapeutic monitoring and stratification of patients. Though remarkable progress has been made in the technology-front for detection and enrichment of CTC from peripheral blood, the procedure remains still challenging. Identification and isolation of CTCs requires enrichment because of its heterogenous nature and low concentration in blood. Besides identification of CTC level in the blood, the need to characterize the CTC as a unique biomarker for therapy monitoring and prediction has been the focus of recent studies. Current technologies relies mostly on immunomagnetic and immunosorbent assay-based methods to isolate CTC (156). Surface based protein expression (EpCAM) and mRNA-based analysis (RT-PCR) are then used to characterize the cells. Because of these challenges both enrichment and characterization are often combined for enumeration of CTC in blood. Earlier studies have proved the variable expression of EpCAM levels in different breast cancer subtypes. Under this setting, both negative and positive markers like CD45, MUC-1, GA 73302, cytokeratins are currently employed for immunohistochemical analysis (157). The Ficoll and OncoQuick system works on density gradient centrifugation where CTC are separated from red blood cells and granulocytes. These are non-specific and CTC can also migrate between the layers losing their integrity (151, 158). The ISET (Isolation by Size of Epithelial Tumor cell) method employs filters of 8 μm thickness to separate the CTC from the blood cells.
Due to poor specificity and lower efficiency, these techniques have fallen out and immunological assay have taken their place. To increase the analyzed blood volume, an innovative method (Cell Collector) having EpCAM antibody activated wire, inside patients’ cubital veins for 30 minutes was done. Positive CTC having the antigen can be screened with high efficiency by this method (159). Microfluidic systems (DFF-chip, JETTA) combine hydrodynamic cell sorting and antibody-based selections for screening and isolation of these cells (160, 161). The only current gold standard method approved by FDA (Food and Drug Administration) for detection of CTC is the Cell Search CTC test (Veridex, Rarita NJ) (162). It is a cell spot analyzer which combines semi-automated isolation of EpCAM positive cells by magnetic nanoparticles and further characterization is done by immunofluorescent staining of different cytokeratins and CD45 to eliminate leukocytes. Cells having low EpCAM levels might escape from this analysis and hence reduce the efficiency of this technology (163). The alternate method is the immunomagnetic assay Adna Test Breast Cancer Select system (AdnaGen, Langenhagen, Germany). It uses magnetic beads coated with surface antigens (MUC-1, CD45, EpCAM) for isolation and enrichment of CTC (164). The detection rates of CTCs by this method are in the range of 69 to 73 % with less blood drawn from the patients (165). The IsoFlux CTC system is recently approved for CTC isolation and enrichment as it combines microfluidics with immuno-separation to achieve high end isolation with increased efficiency as standard methods. It uses a four microfluidic cartilage system to isolate CTC in less than 2 hours and requires only 7-10 ml of blood for analysis (166, 167).
Detection of miRNAs from CTC helps in providing better insights about real time evolution of tumor dynamics making it a potential alternative to tumor biopsy (168). After isolation and enrichment of CTC by EpCAM positive bead-based method, Markov et al. in 2016 selected a few miRNAs, miR-21, miR-146a, miR200 and miR-210 which were already established as markers for breast cancer invasion, migration and metastasis (169). To validate the same in enriched CTC, TaqMan qRT-PCR miRNA assay was performed on 55 patients’ samples with 20 healthy controls. miR-21 was found to be upregulated in both plasma and enriched EpCAM CTC in all the samples (170). Similar studies showed that CTC-positive patients have higher concentrations of circulating miR-200a-c and miR-210 than CTC-negative patients and controls (171–173). Thus, it can be inferred that the miRNA profiling from the CTC can be a promising candidate for the better assessment of the disease status (174). In another study, enriched CTC were isolated using Cell Search Profile kit from 50 patients of metastatic breast cancer for miRNA profiling before starting the chemotherapy regimen (175). After screening 436 miRNAs, 39 miRNAs were having ten times higher expression than normal cells. Moreover, 4 miRNAs were identified which could differentiate ER positive and ER negative tumors. miR-183 was identified as the “CTC specific” marker having high levels of expression in all patients with >5 CTCs/7.5 ml compared to those patients with <5 CTCs/7.5 ml (175). A novel ‘paper based miRNA expression profiling’ was carried out from CTC using cellulose paper based elute card (176). After identification and enrichment of CTC, they were spotted on to Micro Well Mini Tray and multiplexing was carried out. Selected miRNAs were profiled from 21 patients’ samples and 3 healthy controls. A fold difference of 2.3 was observed between the expression in CTC and CTC free samples. Moreover, decreased miR-28-5p and increased miR-221 levels was observed from samples drawn before and after the chemotherapy (176). Thus, characterization of CTC with their corresponding miRNA status would give an additional tool to monitor the therapeutic response in cancer patients.
Evaluation of therapy response currently involves series of clinical tests, estimation of tumor biomarkers and various imaging techniques. CTC have emerged as an alternative for assessing prognosis and tumor burden before or after the treatment. The first relevant study on using CTC as a prognostic marker was done by Cristofanilli in 2004. Analysis of 177 metastatic breast cancer patients’ peripheral blood was done by using CTC and correlation studies with progression free survival (PFS) and overall survival (OS). Detection of at least 5 CTC in 7.5 ml blood was associated with shorter PFS and OS (median PFS: 2.7 vs. 7.0 months; OS: 10.1 vs. 18 months) (177). After serial analysis of CTC in patient’s serum a collective threshold of 5 CTC/7.5 ml blood was established as a prediction for good or poor clinical outcome in MBC patients. Although 2007 ASCO tumor marker guidelines clearly states that the measurement of CTC should not be used for treatment modifications and diagnosis, sufficient studies have proven that CTC are associated with an increased risk of disease reoccurrence (178). Reduction of cytokeratin 19 mRNA positive CTC during chemotherapy was an effective indicator of a clinical outcome (179, 180). In a serum based study of 267 patients, CTC count of 5 or greater in 7.5 ml blood was considered to be a strong prognostic value for PFS (HR 1.78) and OS (HR 2.33) (181, 182). The limitation of the above study was that the value of CTC could not be evaluated for meta-analysis because of the different treatment regimen followed with the study group itself.
Punnoose et al (2010) studied CTC count using Cell Search and CTC chip platforms for different MBC subtypes of patients. Lower count of CTC was observed in HER2 triple negative compared to ER positive cancer subtypes (183). This phenomenon may be due to low EpCAM expression and high percentage of EMT marker in basal like subtypes. A different approach has been used for assessment of HER2 expression in CTC by using immunohistochemistry and in situ hybridization (184). Another study of 254 patients with MBC indicated that 42% of HER2 positive tumor had HER2 negative CTC and 29% of HER2 negative tumor had HER2 positive CTC (185). Similar discrepancies of discordant HER2 status have been reported in early breast cancer studies (186). This indicated that the CTC have a phenotypic variability compared to the primary tumors (187, 188). Currently a multicenter study in Europe is underway for CTC of MBC patients with HER2 negative primary tumors and this might provide conclusive insights regarding the variability of HER2 phenotype (189). The overall survival analysis in a study of 468 patients with MBC was not predicted by CTC count in HER2 positive subtype, but their presence was found later in the same study after treatment with trastuzumab (190). As a follow up of this study, a neoadjuvant phase III NEO-ALTTO trial was set up in response to trastuzumab and lapatinib. No significant correlation was seen between CTC detection and complete pathological response by PET/CT scan (191). The phase III SUCCESS B trial for early HER2 positive primary tumor outcome was that if the patient’s blood had one or more detectable CTC with good staining, they were considered HER2 positive (192).
Proper randomized clinical trials are paramount for establishing the authenticity of any biomarker as indicators of the disease outcome. To assess whether the patients of MBC benefit from the therapy based on the CTC evaluation, a randomized phase III study was initiated by the Southwest Oncology group “SWOG S50500” (ClinicalTrials.gov identifier: NCT 00382018) (193). After the clinical evidence of disease progression, patients having > 5 CTC after one cycle of chemotherapy were randomized either to switch to new treatment or continue till tumor progression. Similarly, an ongoing CirCe01 trial evaluates and assesses the CTC count after an adjuvant therapy. If the response of CTC for the therapy was insufficient (the number of CTC did not come down), then the patients were switched early to a different treatment regimen (194). An existing STIC CTC METABREAST trial (ClinicalTrials.gov identifier: NCT01710605) was planned to check the aggressive therapy response in patients having high CTCs value (194). Patients were then organized randomly into groups based on their CTC count and clinicians’ choice. Patients with high CTC count (>5 CTC) were taken for chemotherapy whereas patients with low CTC counts were treated with endocrine therapy. The German DETECT V CHEVENDO trial (ClinicalTrials.gov identifier: NCT 02344472) is also set to evaluate the effect of dual anti- HER2 therapy (trastuzumab and pertuzumab) combining chemotherapy/ radiotherapy based on CTC levels (194). Bauer et al. 2018 investigated 574 breast cancer patients for tumor reoccurrence, two and five years after primary diagnosis in peripheral blood using FDA Cell Search system for CTC level detection (195). Later, in the follow up study, 8.2 % of CTC were detected after two years whereas no detection was observed after five years. Apart from CTC, serum tumor biomarkers (circulating tumor DNA and exosomes) have been considered for the monitoring of tumor progression in breast cancer. Dawson et al measured various biomarkers in thirty MBC patients and found that both CTC and circulating tumor DNAs had high levels and they were associated with low PFS and OS values (196). The detection and molecular characterization of CTC offer distinctive advantage to understand the etiology of metastasis which can serve as a liquid biopsy approach.
Breast cancer is highly heterogenous with various subtypes, each exhibiting different biological and cellular behavior leading to different clinical outcomes. Epithelial to mesenchymal transition (EMT) is emerging as one of the players of metastasis. Though the mechanism by which cell migrate and invade by EMT has been extensively studied, understanding the intricate cross talk between different signaling pathways and their regulators are necessary to bring about effective treatment regimen. MicroRNAs have completely transformed our comprehension of carcinogenesis because of their capacity to act as both tumor suppressors and oncogenes. Although many miRNAs have been found and identified as potent regulators of metastasis, most of them have not yet been characterized for potential clinical applications. However, the use of circulating microRNAs as diagnostic and prognostic serum markers appears to be inevitable for early breast cancer detection. The assessment and effectiveness of any prognostic tool is to evaluate the treatment effect. In this regard, the clinical impact emerging from CTC is due to its capacity to convey the information of tumour progression to clinicians based on its number or count. The transcriptome analysis of these designated cells can further help us in understanding the tumour biology in real time, thus giving impetus to formulate newer drugs for combating the hyper variability of tumour dynamics. However, careful monitoring has to be done for collection of blood along with tissue and their matched normals for proper correlative studies. With the latest advancement in microRNA research and with newer technologies being developed, an integrated model encompassing CTC, miRNA and mRNAs based detection and targeted therapies would emerge as a novel thernaostic approach for advanced breast cancer management.
Authors would like to acknowledge Dr. V. Dinesh Kumar for his critical remarks and help in editing this review article. AK is grateful to the Ministry of Human Resource Development, Govt. of India, for providing fellowship during this period.
miRNAs
MicroRNAs
deoxyribonucleic acid
Transforming growth factor
Wingless
Epithelial to Mesenchymal transitions
Circulating tumor cells
Triple negative breast cancer
Metastatic breast cancer
