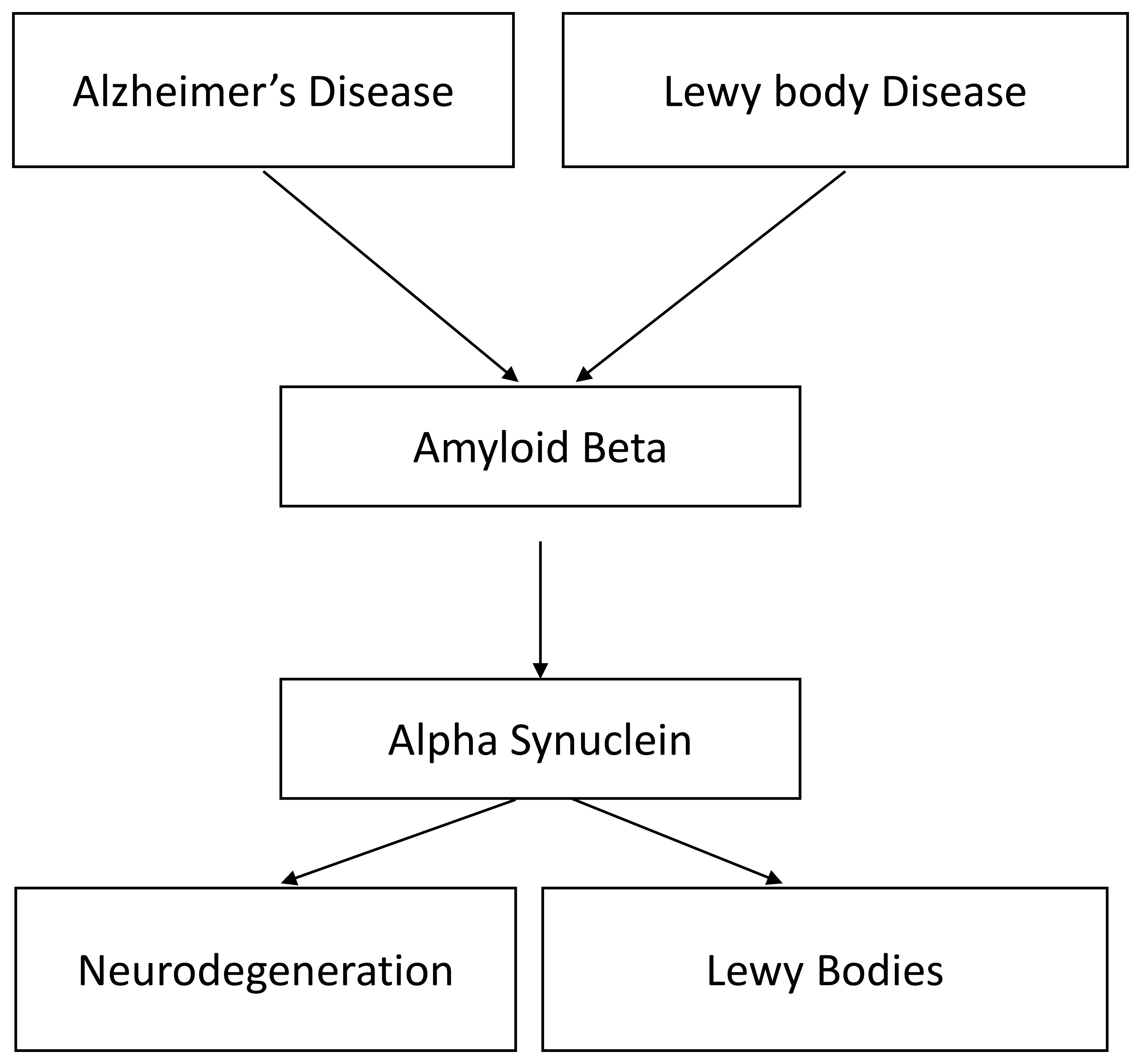
Background: Biomarker detection strategies have, in recent years, been
moving towards nucleic acid-based detection systems in the form of aptamers,
short oligonucleotide sequences which have shown promise in pre-clinical and
research settings. One such aptamer is M5-15, a DNA aptamer raised against human
alpha synuclein (