- Academic Editors
Fluctuations in mechanical force vectors within living cells can substantially influence the behavior and functions of proteins. Tau protein can spontaneously be raptured and entangled in refolding under picoNewton compressive forces that are biologically available in a living cell: a hidden aggregation pathway due to stress and crowding. Our findings were achieved through a customized modification of atomic force microscopy (AFM) for single-molecule manipulation. This previously hidden phenomenon of proteins rupturing collectively while subsequently and spontaneously refolding into a complex entangled conformation, distinct from the Tau protein’s folded or unfolded states, could potentially explain the early-event initiation of the aggregation of the Tau protein seen in various neurodegenerative diseases. This article introduces our recent discovery of the missing Tau protein property that is of significant relevance to the Tau protein and neurodegenerative disease research and medical treatment, aiming to stimulate the collective observation and a new perspective on the Tau aggregation mechanism and disease mechanism studies.
Tau protein can undergo abnormal aggregation and form insoluble fibrils, a hallmark of several neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), and frontotemporal dementia. Literature has reported that increased oxidative stress and inflammation can promote the pathogenesis of Tau protein aggregation [1, 2, 3, 4, 5, 6, 7, 8]. Although the detailed mechanisms of the Tau protein aggregation are still not fully understood, it is generally believed that they involve the disruption of the Tau protein’s binding to microtubules, the induction of conformational changes, and the promotion of oxidative stress and inflammation [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12]. Oxidative stress and inflammation can increase the crowding in a living cell, which increases the local-environment mechanical force fluctuation amplitudes of the Tau proteins. Understanding the underlying reasons for the emergence of Tau aggregates and the impairment of their usual functions has been a primary area of investigation, aiming to enhance the development of improved diagnostics and therapeutic approaches [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14].
Recently, we have reported our discovery of a previously hidden property of Tau proteins: human neuronal Tau proteins can be spontaneously ruptured and entangled by picoNewton compressive force [15]. Under typical physiological conditions, it has been observed that the tertiary structure of human Tau protein can undergo abrupt and spontaneous rupture like a balloon being compressed. These tertiary-structural ruptures occur in response to biologically available picoNewton compressive forces. This suggests that even relatively low forces within the physiological range can cause significant structural changes and potentially impact the function and behavior of the Tau protein. Understanding the mechanical properties and vulnerabilities of the Tau protein under these conditions is crucial for comprehending its role in neurodegenerative diseases [15]. We made the experimental discovery by using a home-modified single-molecule atomic force microscopy (AFM) manipulation microscopic approach [15, 16, 17, 18, 19, 20]. Additionally, we have investigated the formation of an intertwined protein state that arises when proteins experience overcrowding-induced ruptures. These crowded proteins undergo simultaneous rupture and subsequently refold into an entangled folding state. This unique state differs from both the folded and unfolded states observed in individual Tau proteins. It represents a potential pathway or triggering mechanism for Tau protein aggregation, leading to bifurcation events [15].
AFM equipped with advanced picoNewton sensitivity and accuracy is a powerful and valuable approach for investigating protein conformation and fluctuation. AFM’s high sensitivity and accuracy allow for precise measurement and characterization of protein structures and dynamics at the nanoscale level. With AFM, it becomes possible to probe and manipulate individual protein molecules, examine their mechanical properties, and explore their conformational changes under various conditions, including the application of forces. This technique provides valuable insights into the behavior and stability of proteins, contributing to our understanding of their structure-function relationships and facilitating the study of biological processes at the molecular level [19, 20, 21, 22, 23, 24, 25, 26]. We have developed a new technical approach utilizing AFM that enables the manipulation of compressive forces on individual proteins under physiological conditions. This novel methodology allows for precise control and application of compressive forces directly onto a single protein molecule. By operating under physiological conditions, we can investigate the response of proteins to compressive forces in a biologically relevant context. This advancement provides a powerful tool for studying the mechanical properties, stability, and conformational changes of proteins, offering insights into their functional roles and potential implications in various biological processes.
In recent years, we have reported compressive force ruptures observed in several different protein molecules like Calmodulin, 6-Hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK), histone, Tau protein, and human nucleosome [15, 16, 17, 27, 28]. We have employed a combination of single-molecule fluorescence resonance energy transfer (FRET) microscopy and AFM to obtain precise insights into the conformational changes occurring during the rupture process. This innovative approach allows us to monitor the rupture process in real time, with sub-nanometer resolution [15, 16, 17, 18, 19, 20]. The collective data from our studies [10, 11, 12, 13, 14, 15] have revealed a strong correlation, providing evidence that the rupture process is both spontaneous and abrupt, driven by free energy [15, 16, 17, 27, 28].
Our experimental findings indicate that the Tau protein experiences a spontaneous rupture phenomenon triggered by a compressive force. This rupture occurs when the force applied reaches a specific threshold value, which we have accurately measured. Notably, the threshold force amplitudes align with the thermal fluctuation levels observed in living cells under normal physiological conditions. Moreover, the extent of rupture is contingent upon the speed at which the force is applied. In addition, our research has delved into the spontaneous and concurrent ruptures of multiple proteins when they are in proximity. This phenomenon leads to the formation of an entangled protein state, which could potentially serve as a mechanism or early event in the aggregation of Tau proteins [15].
Fig. 1A illustrates the characteristic force curve depicting the mechanical force applied to an individual Tau protein molecule in an aqueous environment and its corresponding response to an external compressive force [15]. As the force amplitude increases and reaches a specific threshold value, the protein reaches its breaking point and undergoes a spontaneous and abrupt rupture. This rupture event is captured by a sudden release of the compressive force, as shown in Fig. 1B. Additionally, Fig. 1C presents a distribution of rupture forces observed in relation to the varying degrees of compressive force loading [15].
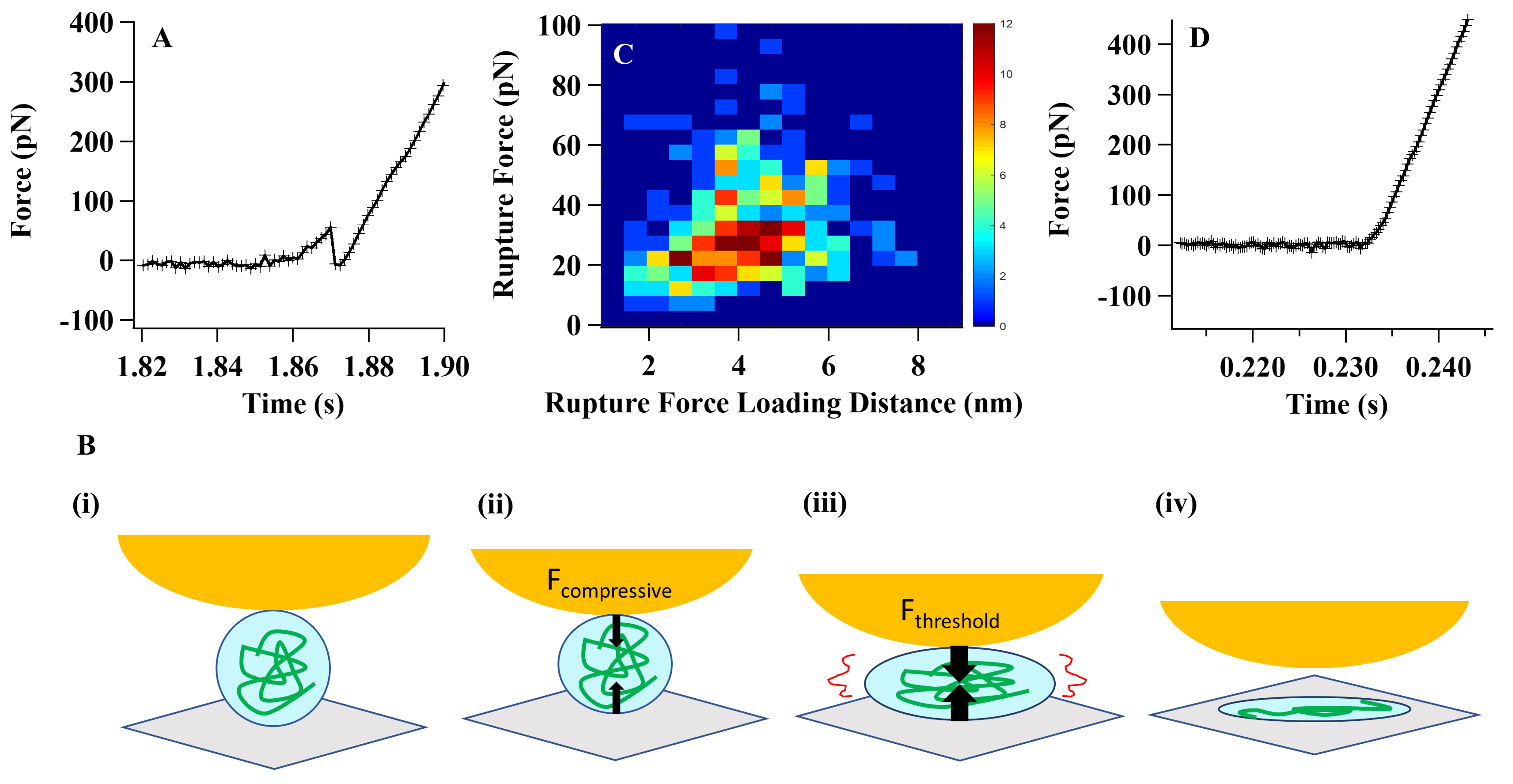 Fig. 1.
Fig. 1.Single-molecule compressive force analysis of Tau proteins. (A)
The force curve presented here depicts the characteristic pattern observed during
the rupture of a Tau protein under a compressive force. The sudden decrease in
the force curve corresponds to the spontaneous rupture of the protein molecule.
To validate these spontaneous protein-tertiary structure rupture events, we
conducted a series of systematic control experiments [15, 27, 28]. (B) The
cartoon scheme below illustrates the process of Tau protein rupture under a
compressive force: (i) At the initial stage, there is no physical contact between
the atomic force microscopy (AFM) tip and the protein molecule. (ii) As the
contact is established, the molecule becomes squeezed under the applied
compressive force. The black arrows in the scheme represent the force vectors
acting on the molecule, the AFM tip, and the cover glass. (iii) As the
compressive force increases, it eventually reaches a threshold value.
Simultaneously, the molecular interaction weakens to a point where it can no
longer withstand the force. Consequently, the protein molecule undergoes rupture.
(iv) The final state depicted in the scheme represents the spontaneously ruptured
state of the protein. Please note that this is a simplified cartoon scheme
illustrating the concept and not an exact representation of the molecular
structure or the forces involved. (C) The distribution of threshold rupture force
was plotted against the rupture force loading distance using a force loading rate
of 3000 pN/s. This analysis aimed to examine the relationship between the applied
force and the corresponding threshold force required for rupture. (D) A typical
compressive force curve on a Tau protein under an Mg
The fundamental landscape of molecular biology and neuronal sciences is primarily based on quantitative chemical analysis of the concentration and number density of the molecules in the rate processes. Our reported results on the hidden protein properties strongly suggest a missing perspective to the scientific understanding: the basic physical forces can also play a critical role in molecular rate processes and dynamics in living cells. Such as in this case, the specific mechanical compressive forces can be generated from the stress and inflammation [1, 2, 3, 4, 5, 6, 7, 8] or other internal and external mechanical force sources, even including external brain trauma events, such as Tau aggregation and increased neuroinflammation in athletes after sports-related concussions, repeated-head nocking, and in traumatic brain injury patients [10, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39]. Cells are complex beyond simply a chemical test tube for molecules, involving mechanical force vector fluctuations. We are now seeing a complete picture of the molecular rate processes and dynamics in living cells in terms of understanding protein aggregation under protein crowding and mechanical force fluctuations.
We have found that the Tau protein rupture behaviors are highly sensitive to the electrolyte environment in the solution. The Tau proteins can have no rupture events (Fig. 1D) or have rupture events (Fig. 1A–C) under compressive force manipulation and analysis [15]. The diverse response behaviors observed under compressive force can be attributed to the structural confinement and the inherent rigidity of the protein’s structural nature. These behaviors are influenced by factors such as internal frictional forces and interdomain attraction forces, including hydrogen bonding and electrostatic forces. The structural confinement, along with these molecular interactions, contributes to the overall mechanical response of the protein to compressive forces [15, 16, 17, 40, 41, 42, 43, 44].
In our proposed comprehensive model, we classify protein responses under compressive force into two categories: “balloon” and “cotton ball”. The “balloon” type refers to proteins that may rupture when subjected to a compressive force, while the “cotton ball” type indicates proteins that undergo shape changes without rupturing. We suggest that by modifying the electrostatic environment through charge electric field manipulation, the rigidity of the protein can be softened, resulting in a more flexible form. This flexibility allows the protein to diffuse compressive forces, like a “cotton ball”, by releasing stress to the local environment and undergoing shape alterations [15].
We propose that there is a potential relationship between the electrostatic environment and the response of Tau protein under compressive force. This relationship could have significant implications for protein aggregation and tauopathy, a group of neurodegenerative disorders associated with the aggregation of Tau protein. By understanding and manipulating the electrostatic properties of the protein, we may gain insights into mechanisms underlying protein aggregation and potentially develop therapeutic approaches for Tau-related diseases [15, 43, 44].
The spontaneous rupture of proteins under compressive force is a complex
phenomenon influenced by various local factors, including the
hydrophilic-hydrophobic force field of the protein molecules and their
surrounding environment. According to statistical thermodynamics and the
Boltzmann distribution, molecular state-to-state change processes with activation
energy barriers below 10 k
In crowding, proteins that undergo simultaneous rupture have multiple possible outcomes. They can either refold back to their native states or into an entangled state, as both pathways are energetically viable. When a protein is ruptured, it can exist in a metastable state, where it remains temporarily before undergoing refolding back to its original conformation. This refolding process typically occurs within a few milliseconds. However, based on our experimental observations, we have also noted instances where the protein can adopt different conformational states, potentially remaining in a nonnative state for an extended period [15, 16, 17].
Under conditions of molecular crowding, such as the accumulation of Tau proteins on neuronal microtubules, fluctuations in compressive force can trigger rare events involving the simultaneous ruptures of multiple protein molecules. These simultaneously ruptured proteins can then spontaneously fold into an entangled aggregation state. This formation of the protein-entangled state may act as a nucleation event, initiating the early stages of Tau protein aggregation and subsequent fibril formation. Within the context of biological processes, the formation of the entangled state can serve as a critical bifurcation point in the pathways leading to Tau fibril formation. Under the influence of biologically relevant compressive forces, Tau protein molecules can undergo rupture in crowded environments, giving rise to the protein-entangled state. Eventually, this leads to the loss of protein activity and detachment from the microtubule surface, ultimately contributing to the degeneration of the microtubules.
Inflammation and oxidative stress have been suggested to play a role in triggering or promoting Tau protein aggregation [1, 2, 3]. In neurodegenerative diseases, including AD, chronic inflammation is often observed in the brain and is associated with the accumulation of abnormal Tau protein aggregates [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12]. For example, recent research has compellingly shown that mechanical stretching can trigger phosphorylation-dependent Tau misplacement into dendritic spines, leading to associated synaptic dysfunction. This dysfunction is thought to result from the presence of soluble Tau oligomers. It is important to note that Tau protein aggregation and inflammation can influence each other based on the molecular interplay perspectives in the literature [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12]. Our discovery that Tau protein aggregation can be triggered by compressive force fluctuations may provide a new and previously missing perspective for understanding the pathways and the mechanism of Tau protein aggregation. Beyond that, the inflammation and oxidative stress can increase the crowding in a living cell, which in turn increases the mechanical force fluctuation amplitudes around the Tau proteins. Such inflammation and oxidative stress can also alter the protein’s local environment of solvation, osmotic pressure, domain surface tension, as well as the liquid-liquid phase separation in living cells [45, 46]. Additionally, inflammation-induced oxidative stress and the release of reactive oxygen species can create a local environment for the Tau protein to be sensitive to compressive force fluctuations and get ruptured and aggregated.
It is noteworthy that Tau, an intrinsically disordered protein, exhibits such behavior. It has been demonstrated that Tau can exist in various conformational states, with some being more compact than others [47, 48, 49]. The spontaneous tertiary-structural rupture of the protein at the threshold compressive force provides further evidence that protein fluctuation dynamics, including intermolecular hydrogen bonding, inter-domain interactions, friction with solvent molecules, and inter-domain frictions, can lead to structural confinement capable of withstanding compressive forces in the order of picoNewton, as applied by the AFM tip apex [27, 28, 50]. In a biological neuronal system, the Tau protein raptures and aggregates at their bounding to the microtubule, which may significantly limit the Tau protein’s structural flexibility. This inherent structural confinement may have significant implications for understanding Tau protein aggregation.
In summary, our observations have revealed the occurrence of sudden and spontaneous ruptures of Tau proteins when subjected to compressive forces within a range of approximately 5 pN to 125 pN. Notably, these force amplitudes fall within the biologically relevant range of forces observed in living cells [15, 51]. Furthermore, our research has uncovered a close relationship between the structural response of proteins under compressive force and the local electrostatic environment. We have specifically investigated the phenomenon of multiple-protein ruptures leading to the formation of entangled structures in crowded environments when subjected to compressive forces. These findings highlight the intricate interplay between mechanical forces, protein structure, and the surrounding electrostatic environment in governing the behavior of proteins under compression in complex biological systems. The results of our experiment offer compelling evidence of entanglement interactions occurring between refolded Tau proteins following simultaneous and spontaneous paired protein ruptures in crowded environments. Importantly, these experimental conditions are achievable under physiological conditions, such as the presence of Tau proteins on neuronal microtubules. The findings reported in our study may contribute to uncovering a previously missing piece of the puzzle regarding the origin of Tau protein aggregation, a phenomenon closely linked to neurodegenerative diseases.
HPL designed the research study and wrote the paper. HPL contributed to editorial changes in the manuscript. HPL read and approved the final manuscript. HPL had participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
HPL acknowledges the support of this work by the Ohio Eminent Scholar Endowment.
This research was supported by the Ohio Eminent Scholar Endowment.
The author declares no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
