Academic Editor: Gustavo Caetano-Anollés
The role of Achromobacter species in lung disease remains unclear. The aim of this study was to characterize Achromobacter isolated from persons with cystic fibrosis and from other clinical samples. Whole genome sequences from 101 Achromobacter isolates were determined (81 from patients with cystic fibrosis and 20 from other patients) and analysed. Taxonomic analysis showed nine species including two putative novel species. Thirty-five novel sequence types were present. The most active agent was co-trimoxazole followed by imipenem, but Minimal Inhibitory Concentrations (MICs) were high. Acquired antibiotic resistance genes were rare. Their presence did not correlate with minimal inhibitory concentrations suggesting that other mechanisms are involved. Genes for proposed virulence factors were present in only some isolates. Two putative novel species were identified. The putative virulence properties of Achromobacter involved in infections are variable. Despite the high MICs, acquired resistance genes are uncommon.
The genus Achromobacter currently comprises 20 species.
Achromobacter species are mostly found in aquatic environments but may
also be present among the intestinal microbiota of healthy persons. Furthermore,
they may cause a range of human infections, in particular pulmonary infections in
persons with cystic fibrosis (CF) [1, 2, 3, 4]. In a Canadian and a French study,
respectively, 11% and 27% of persons with CF were tested positive for
Achromobacter by bacterial culture [5, 6]. Colonization and/or infection
may be persistent in persons with CF, but in some circumstances sputum can be
rendered culture negative following antibiotic therapy [6]. Nevertheless,
treatment is challenging due to both intrinsic and acquired resistance [2];
Achromobacter species encode OXA and AmpC type
Several genome assemblies have been reported, but these were limited to one to six isolates, which yields only limited insight in the occurrence of virulence genes and acquired antibiotic resistance [9, 10, 11, 12]. In this study we determined the whole genome sequences (WGS) of 101 Achromobacter isolates and report the diversity of the isolates, minimal inhibitory concentration for eight antimicrobial agents, and the presence of putative virulence genes. The isolates were obtained from CF, bronchiectasis (BE), and other diseases.
The aim of this study was to characterize the phylogenetics, antibiotic resistance, and virulence factors of clinical Achromobacter isolates based on whole genome sequencing.
A total of 101 Achromobacter isolates were analysed. These isolates had been cultured from respiratory samples of persons with CF (n = 81), respiratory samples of patients with other diseases (n = 13), blood cultures (n = 5), a patient with mastoiditis and a patient with otitis media. The isolates were recovered between 2003 and 2016 in four different countries: United Kingdom (n = 27), Spain (n = 27), the Netherlands (n = 46), and Australia (n = 1) (Supplementary Table 1).
Samples and patient data were collected in compliance with the Declaration of Helsinki ICH-GCP, the Declaration of Taipei regarding Health Databases and Biobanks, and with local and European regulations for collection and handling of patient data. Since the study concerned retrospectively collected anonymized patient data and bacterial strains, informed consent at the individual patient level was not required for this study. In addition, the Spanish and UK strains were collected in accordance with their local ethics guidelines and described in prior studies [13, 14]. In the Netherlands, use and analysis of bacterial strains with anonymized patient data does not require approval from Institutional Review Boards/Ethics Committees.
Isolates were initially identified by Matrix Assisted Laser Desorption/Ionisation and Time-Of Flight Mass Spectrometry (MALDI-TOF MS) using a Microflex with Biotyper software MBT-BDAL-5627 MSP library (Bruker, Germany) according to the instructions of the manufacturer.
Bacterial DNA was purified using the Qiacube with the DNeasy Blood & Tissue kit with the enzymatic lysis protocol (Qiagen, Carlsbad, CA). Library for sequencing with the MiSeq or Nextseq (Illumina, San Diego, CA) platforms were prepared with the Nextera XT library prep kit (Illumina) according to the manufacturers’ instructions. Contigs were assembled with SPAdes genome assembler v.3.6.2. with its default parameters and contigs shorter than 500 nucleotides were discarded [15]. Raw read sequences of all 101 isolates were uploaded to the NCBI’s SRA database under the BioProject ID PRJNA723829.
Fast-ANI, developed for fast alignment-free computation of whole-genome Average Nucleotide Identity (ANI), was performed to confirm the species assignments [16, 17]. A cut-off of 95% was used to define species [18, 19].
Multi-locus Sequence Typing (MLST) was performed using PubMLST with the scheme for Achromobacter xylosoxidans [https://pubmlst.org/general.shtml]. Novel alleles and sequence types from MLST were submitted to the PubMLST database [https://pubmlst.org/general.shtml]. The MLST-based Minimum Spanning Tree was generated with PHYLOViZ 2.0 using the GoeBurst algorithm [https://phyloviz.readthedocs.io/en/].
The evolutionary history of the OXA-type
The assembled contigs were analyzed for the presence of acquired resistance genes by ResFinder [last accessed October 28, 2019] from the Center for Genomic Epidemiology (DTU, Copenhagen, Denmark) [22].
Minimum Inhibitory Concentrations (MICs) of antimicrobial agents were determined
by the standard ISO broth microdilution method with frozen panels (Trek
Diagnostic Systems, Westlake, OH). The following antimicrobial agents
(concentration ranges) were tested: ciprofloxacin (0.03–32 mg/L); tobramycin
(0.125–128 mg/L); ceftazidime (0.25–256 mg/L); meropenem (0.06–64 mg/L);
imipenem (0.125–128 mg/L); aztreonam (0.25–256 mg/L);
trimethoprim/sulfamethoxazole (0.06–32 mg/L); and colistin (0.25–16 mg/L). The
MIC
WGS yielded an average of 247 contigs per isolate (range 109–714); the average
coverage was 51
MALDI-TOF, which is commonly used to identify isolates in routine diagnostic microbiology, identified 95 isolates as A. xylosoxidans, three as A. insolitus, and one as A. spanius. Two isolates were identified only to the Achromobacter genus level. Analysis of the WGS results confirmed 63/95 (66.3%) isolates as A. xylosoxidans. This was in agreement with the identification rates reported before with the used default MALDI-TOF database [2]. The remaining 32 A. xylosoxidans isolates were A. insuavis (n = 11), A. ruhlandii (n = 7), A. deleyi (n = 5), and two putative novel species, which were designated species1 (n = 8) and species2 (n = 1) in this manuscript. The isolate identified as A. spanius by MALDI-TOF was found to be A. deleyi by WGS. Two of the three isolates identified as A. insolitus by MALDI-TOF were confirmed by WGS, the third isolate was found to be A. aegrifaciens by WGS. The two isolates which were identified only to the genus level by MALDI-TOF were A. spanius and A. deleyi by WGS analysis (Supplementary Table 1). With the exception of the two novel species, all Achromobacter species have been reported previously in persons with CF [5].
Prior to this study, 485 Achromobacter STs had been reported. MLST analysis of this collection yielded 53 novel alleles and 35 novel sequence types (STs), indicating that only a portion of the genetic diversity had been assessed previously. A total of 61 STs were present among the isolates in this study. Novel alleles and STs were submitted to the PubMLST database [https://pubmlst.org/general.shtml]. A MLST-based Minimum Spanning Tree showed that the isolates clustered by species. The isolates of the different species differed by at least 6 or 7 loci. The majority of the A. xylosoxidans isolates were not closely related as only six pairs of single-locus variants were present, whereas 18 the other sequence types diverged by 2 or more loci (Fig. 1).
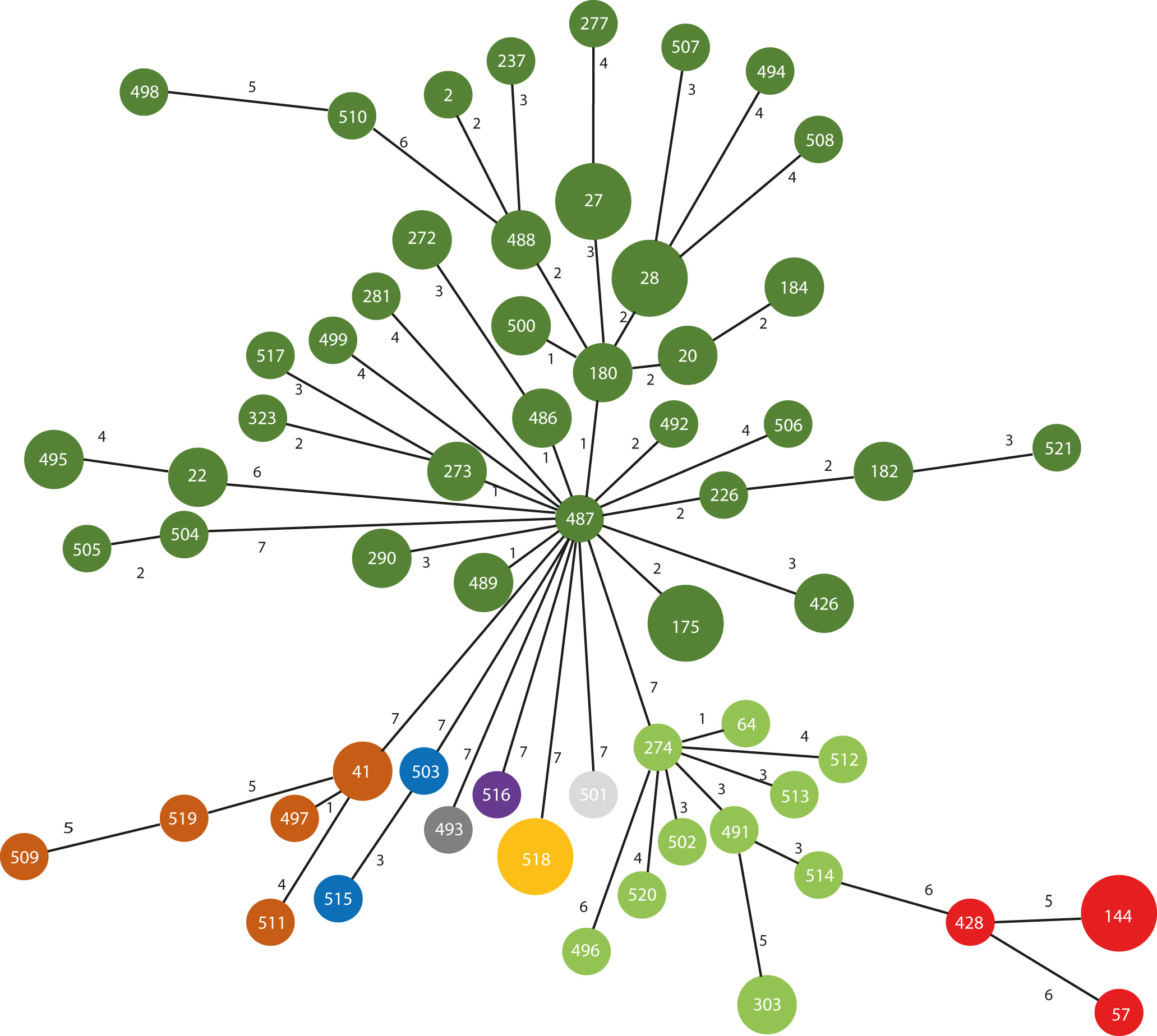 Fig. 1.
Fig. 1.MLST-based Minimum Spanning Tree of 101 Achromobacter isolates. Sequence types (STs) were based on seven housekeeping genes as determined by whole genome sequencing. The numbers in the nodes indicate the STs assigned by PubMLST (www.pubmlst.org). The numbers on the lines between the nodes indicate the number of loci differences between two STs. Distances between the nodes are not drawn to scale. Dark green: A. xylosoxidans; light green: A. insuavis; dark red: species 1; orange: A. deleyi; brown: A. ruhlandii; blue: A. insolitus; purple: A. spanius: dark grey: species2; light grey: A. aegrifaciens.
The five A. deleyi isolates were all recovered in the UK and belonged to a single ST; based on sequence alignments it is possible that they belonged to a single outbreak. Six of the eight novel species1 isolates belonged to ST144 and also appeared to be closely related; a definitive conclusion would require more knowledge of the population structure and mutation rates of this species. The two other isolates belonged to ST57and ST428.
OXA-family
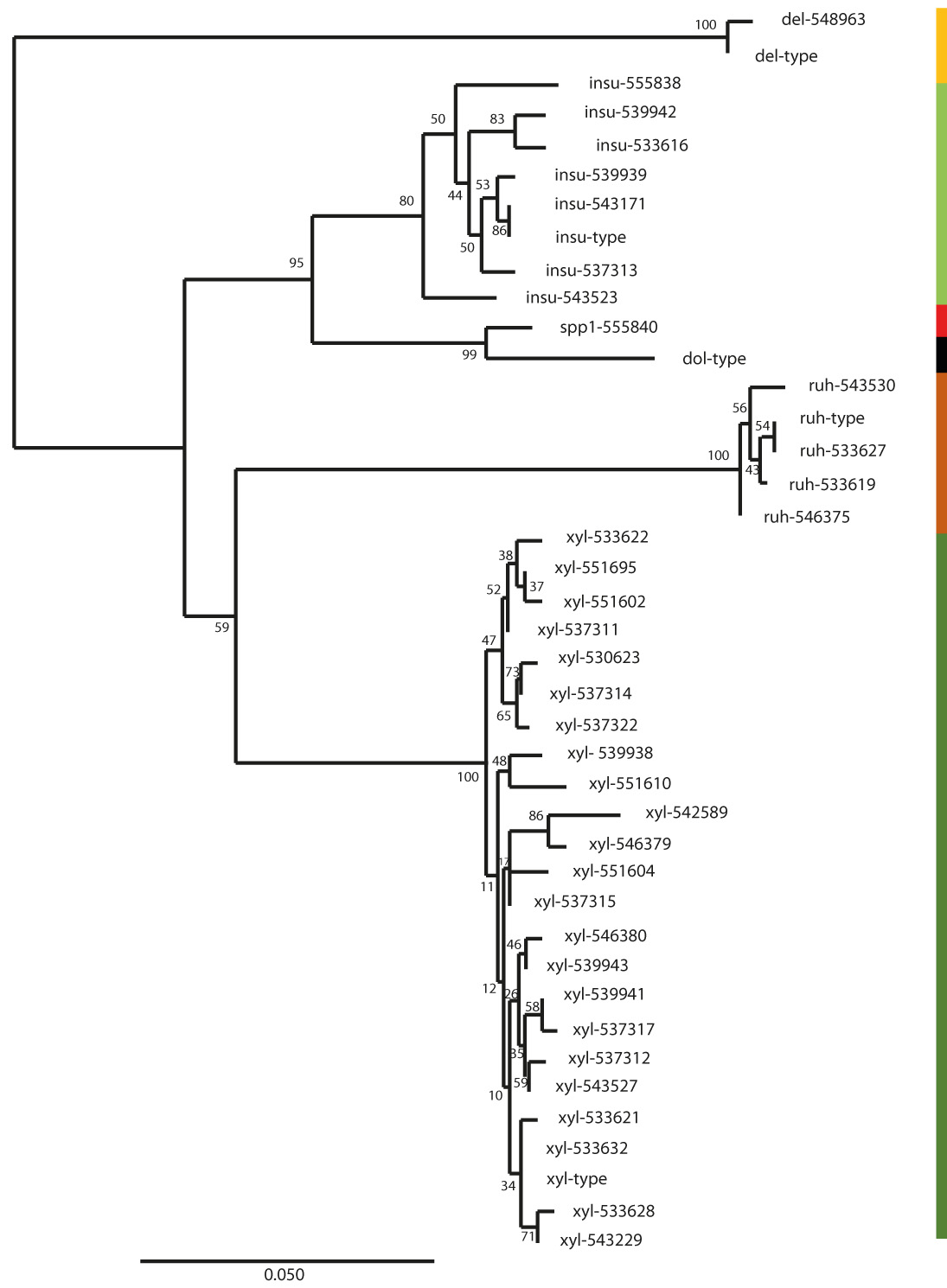 Fig. 2.
Fig. 2.The evolutionary history of the OXA-type
| ID | Species WGS | ST | Disease | Aztreonam | Ceftazidime | Ciprofloxacin | Colistin | Co-trimoxazole | Imipenem | Meropenem | Tobramycin | OXA |
Acquired antibiotic resistance genes |
| 537301 | A. aegrifaciens | 501 | CF | 16 | 8 | 0.5 | 8 | 2 | 0.5 | - | sul2 | ||
| 548963 | A. deleyi | 518 | CF | 8 | 8 | 0.25 | 2 | 1 | - | - | |||
| 548966 | A. deleyi | 518 | CF | 8 | 32 | 0.5 | 4 | 8 | - | - | |||
| 548967 | A. deleyi | 518 | CF | 16 | 32 | 1 | 0.5 | 4 | 8 | 128 | - | - | |
| 548969 | A. deleyi | 518 | CF | 8 | 32 | 0.5 | 4 | 8 | 64 | - | - | ||
| 548972 | A. deleyi | 518 | CF | 128 | 2 | 1 | 0.5 | 2 | 0.12 | 0.5 | - | - | |
| 548973 | A. deleyi | 518 | CF | 256 | 2 | 0.5 | 1 | 2 | 0.5 | - | - | ||
| 548974 | A. deleyi | 518 | CF | 256 | 8 | 8 | 0.5 | 0.25 | 0.5 | 2 | 4 | - | - |
| 534814 | A. insolitus | 503 | CF | 64 | 1 | 0.5 | 1 | 1 | - | aac(3)-IV, aadA11, aph(3”)-Ib, aph(4)-Ia, aph (6)-Id, tet(a), dfrA12, sul1 | |||
| 546376 | A. insolitus | 515 | CF | 64 | 2 | 16 | 4 | 0.5 | 1 | 0.12 | - | - | |
| 543171 | A. insuavis | 64 | other | 4 | 2 | 1 | 0.12 | 0.25 | 0.12 | 32 | + | - | |
| 537313 | A. insuavis | 274 | CF | 128 | 4 | 1 | 0.5 | 0.25 | 2 | + | aac(3)-IV, aph(4)-Ia, tet(A) | ||
| 533618 | A. insuavis | 303 | CF | 128 | 4 | 4 | 1 | 0.25 | 2 | 0.12 | 128 | + | - |
| 555838 | A. insuavis | 303 | CF | 8 | 1 | 1 | 1 | 4 | 2 | 16 | + | - | |
| 537308 | A. insuavis | 491 | CF | 256 | 4 | 8 | 2 | 1 | 1 | 1 | + | - | |
| 546381 | A. insuavis | 496 | other | 256 | 4 | 1 | 0.5 | 0.5 | 4 | 0.12 | 8 | + | - |
| 533616 | A. insuavis | 502 | CF | 8 | 32 | 2 | 4 | 2 | 0.5 | 32 | + | - | |
| 539942 | A. insuavis | 512 | CF | 128 | 4 | 2 | 1 | 2 | 2 | 64 | + | - | |
| 539939 | A. insuavis | 513 | other | 256 | 4 | 2 | 1 | 0.12 | 2 | 64 | + | - | |
| 537305 | A. insuavis | 514 | CF | 32 | 16 | 1 | 4 | 2 | 4 | + | - | ||
| 543523 | A. insuavis | 520 | other | 128 | 64 | 1 | 2 | 32 | 32 | + | - | ||
| 546373 | A. ruhlandii | 41 | CF | 0.5 | 1 | 4 | 32 | + | - | ||||
| 546375 | A. ruhlandii | 41 | CF | 1 | 0.5 | 4 | 32 | + | - | ||||
| 548953 | A. ruhlandii | 41 | CF | 32 | 2 | 0.5 | 4 | 32 | 128 | + | - | ||
| 548956 | A. ruhlandii | 497 | CF | 4 | 16 | + | - | ||||||
| 533619 | A. ruhlandii | 509 | CF | 32 | 128 | 8 | 32 | 1 | + | - | |||
| 533627 | A. ruhlandii | 511 | CF | 128 | 2 | 1 | 2 | 0.12 | 2 | 0.12 | 32 | + | - |
| 543530 | A. ruhlandii | 519 | CF | 4 | 8 | 2 | 8 | 4 | 32 | 2 | + | - | |
| 551600 | A. spanius | 516 | CF | 256 | 4 | 2 | 0.5 | 0.25 | 8 | 1 | 2 | - | - |
| 537315 | A. xylosoxidans | 2 | CF | 16 | 32 | 16 | 32 | 2 | 1 | + | aac(6’)-Ib3, aac(6’)-Ib-cr, sul1 | ||
| 533625 | A. xylosoxidans | 20 | CF | 16 | 2 | 1 | 0.25 | 2 | 4 | 64 | + | - | |
| 533630 | A. xylosoxidans | 20 | CF | 128 | 4 | 0.5 | 0.25 | 2 | 32 | 128 | + | - | |
| 542589 | A. xylosoxidans | 22 | other | 256 | 4 | 2 | 2 | 0.5 | 2 | 0.25 | 128 | + | - |
| 543228 | A. xylosoxidans | 22 | CF | 8 | 2 | 8 | 1 | 2 | 4 | 32 | + | - | |
| 537297 | A. xylosoxidans | 27 | CF | 16 | 8 | 8 | 16 | 2 | + | aadA1, sul1 | |||
| 537316 | A. xylosoxidans | 27 | CF | 8 | 2 | 64 | + | - | |||||
| 537320 | A. xylosoxidans | 27 | CF | 256 | 4 | 4 | 2 | 0.25 | 8 | 0.25 | 32 | + | - |
| 551605 | A. xylosoxidans | 27 | BE | 256 | 8 | 2 | 0.5 | 1 | 4 | 0.12 | 64 | + | - |
| 533615 | A. xylosoxidans | 28 | CF | 16 | 4 | 2 | 0.25 | 16 | 64 | + | - | ||
| 533617 | A. xylosoxidans | 28 | CF | 128 | 16 | 2 | 0.5 | 32 | 32 | 64 | + | - | |
| 533623 | A. xylosoxidans | 28 | CF | 64 | 8 | 8 | 8 | 64 | 32 | 64 | + | - | |
| 537310 | A. xylosoxidans | 28 | CF | 256 | 4 | 8 | 1 | 2 | 0.12 | + | ant(2”)-Ia | ||
| 537314 | A. xylosoxidans | 28 | CF | 128 | 2 | 2 | 2 | 0.5 | 8 | 0.5 | 16 | + | - |
| 533620 | A. xylosoxidans | 175 | CF | 256 | 4 | 2 | 1 | 2 | 0.12 | 16 | + | - | |
| 543529 | A. xylosoxidans | 175 | other | 128 | 4 | 2 | 1 | 2 | 0.12 | 32 | + | - | |
| 551604 | A. xylosoxidans | 175 | BE | 256 | 16 | 8 | 2 | 0.5 | 2 | 2 | 128 | + | - |
| 551607 | A. xylosoxidans | 175 | CF | 32 | 8 | 0.12 | 4 | 4 | + | - | |||
| 551608 | A. xylosoxidans | 175 | BE | 32 | 8 | 1 | 0.12 | 2 | 0.5 | + | - | ||
| 537303 | A. xylosoxidans | 180 | CF | 128 | 32 | 1 | 1 | 2 | 4 | + | - | ||
| 546371 | A. xylosoxidans | 180 | CF | 256 | 8 | 16 | 4 | 0.5 | 4 | 16 | + | - | |
| 537298 | A. xylosoxidans | 182 | CF | 128 | 2 | 2 | 8 | 1 | 8 | 0.25 | 128 | + | - |
| 537302 | A. xylosoxidans | 182 | CF | 128 | 1 | 2 | 2 | 0.25 | 8 | 0.25 | 32 | + | - |
| 537309 | A. xylosoxidans | 184 | CF | 16 | 16 | 4 | 2 | 2 | + | aada2, ereA, tet(g), dfrA1, sul1 | |||
| 537312 | A. xylosoxidans | 184 | CF | 32 | 16 | 2 | 16 | 8 | 8 | + | sul1 | ||
| 546380 | A. xylosoxidans | 226 | CF | 256 | 2 | 4 | 4 | 0.25 | 2 | 0.25 | 128 | + | - |
| 537323 | A. xylosoxidans | 237 | CF | 32 | 16 | 1 | 32 | 4 | 8 | + | aac(6’)-IIc, ant(2”)-Ia, sul1 | ||
| 533614 | A. xylosoxidans | 272 | CF | 64 | 4 | 2 | 4 | 16 | 64 | + | - | ||
| 533622 | A. xylosoxidans | 272 | CF | 32 | 4 | 8 | 4 | 0.5 | 32 | + | - | ||
| 537304 | A. xylosoxidans | 273 | CF | 8 | 4 | 1 | 16 | 2 | 1 | + | aac(6’)-Ib3,aac(6’)-Ib-cr | ||
| 537324 | A. xylosoxidans | 273 | CF | 256 | 4 | 8 | 2 | 0.25 | 2 | 1 | + | aac(6’)-Ib-Hangzhou, aac(6’)-Ib-cr | |
| 543527 | A. xylosoxidans | 277 | other | 2 | 1 | 16 | 32 | 32 | + | OXA2, GES, aac(6’)-Ib3, ant(2”)-Ia, aph(3”)-Ib, aph(6)-Id, aac(6’)-cr, ereA, sul1 | |||
| 543233 | A. xylosoxidans | 281 | CF | 128 | 2 | 2 | 4 | 2 | 0.12 | 32 | + | - | |
| 533621 | A. xylosoxidans | 290 | CF | 8 | 8 | 1 | 2 | 2 | 64 | + | - | ||
| 543532 | A. xylosoxidans | 290 | other | 256 | 4 | 2 | 1 | 2 | 0.12 | 32 | + | - | |
| 537300 | A. xylosoxidans | 323 | CF | 256 | 16 | 8 | 1 | 0.5 | 1 | 1 | + | - | |
| 533624 | A. xylosoxidans | 426 | CF | 128 | 8 | 8 | 2 | 0.12 | 8 | 1 | + | - | |
| 539940 | A. xylosoxidans | 426 | BE* | 8 | 0.5 | 2 | 8 | + | - | ||||
| 543229 | A. xylosoxidans | 426 | CF | 256 | 4 | 2 | 4 | 2 | 4 | 2 | 64 | + | - |
| 533631 | A. xylosoxidans | 486 | CF | 128 | 4 | 2 | 1 | 0.12 | 2 | 0.12 | 128 | + | - |
| 533632 | A. xylosoxidans | 486 | CF | 128 | 2 | 2 | 1 | 0.12 | 2 | 0.12 | 128 | + | - |
| 533633 | A. xylosoxidans | 486 | CF | 128 | 4 | 2 | 1 | 2 | 0.12 | 64 | + | - | |
| 533628 | A. xylosoxidans | 487 | CF | 128 | 2 | 2 | 2 | 0.12 | 1 | 0.12 | 128 | + | - |
| 537322 | A. xylosoxidans | 488 | CF | 128 | 8 | 16 | 1 | 1 | 1 | + | - | ||
| 539941 | A. xylosoxidans | 488 | other | 256 | 8 | 4 | 2 | 2 | 0.12 | 128 | + | - | |
| 537317 | A. xylosoxidans | 489 | CF | 256 | 4 | 1 | 8 | 2 | 4 | 8 | + | - | |
| 537319 | A. xylosoxidans | 489 | CF | 256 | 8 | 8 | 2 | 2 | 2 | + | - | ||
| 551609 | A. xylosoxidans | 492 | CF | 256 | 2 | 8 | 0.5 | 0.5 | 1 | 0.25 | 8 | + | - |
| 546369 | A. xylosoxidans | 494 | CF | 8 | 16 | 1 | 0.5 | 2 | 0.5 | 128 | + | - | |
| 546379 | A. xylosoxidans | 495 | CF | 16 | 16 | 2 | 2 | 4 | + | - | |||
| 546382 | A. xylosoxidans | 495 | CF | 8 | 8 | 4 | 0.5 | 2 | 1 | 128 | + | - | |
| 533613 | A. xylosoxidans | 498 | CF | 4 | 16 | 2 | 1 | 1 | 8 | - | - | ||
| 551602 | A. xylosoxidans | 499 | other | 4 | 4 | 4 | 1 | 8 | 0.25 | 128 | + | - | |
| 539943 | A. xylosoxidans | 500 | other | 128 | 8 | 2 | 0.25 | 1 | 0.5 | + | - | ||
| 539944 | A. xylosoxidans | 500 | BE | 128 | 2 | 2 | 1 | 0.12 | 2 | 0.12 | 32 | + | - |
| 551610 | A. xylosoxidans | 504 | BE | 64 | 4 | 1 | 0.5 | 128 | 128 | + | aac(6’)-Ib3, aac(6’)-Ib-cr, sul1 | ||
| 539938 | A. xylosoxidans | 505 | other | 256 | 8 | 8 | 1 | 1 | 16 | 1 | 128 | + | - |
| 537299 | A. xylosoxidans | 506 | CF | 32 | 8 | 1 | 2 | 16 | 32 | + | - | ||
| 537311 | A. xylosoxidans | 507 | CF | 256 | 8 | 32 | 2 | 4 | 1 | 8 | 128 | + | - |
| 537296 | A. xylosoxidans | 508 | CF | 128 | 16 | 4 | 4 | 4 | 16 | 64 | + | - | |
| 543230 | A. xylosoxidans | 510 | CF | 128 | 2 | 2 | 16 | 0.25 | 2 | 0.12 | 8 | - | - |
| 537318 | A. xylosoxidans | 517 | CF | 8 | 4 | 16 | 0.12 | 1 | 1 | + | - | ||
| 543526 | A. xylosoxidans | 521 | other | 256 | 4 | 4 | 2 | 0.5 | 2 | 0.25 | 64 | + | - |
| 546367 | species1 | 57 | CF | 128 | 16 | 2 | 8 | + | sul1 | ||||
| 533626 | species1 | 144 | CF | 128 | 2 | 16 | 1 | 1 | 1 | 1 | 2 | + | - |
| 548957 | species1 | 144 | CF | 128 | 2 | 1 | 1 | 1 | 2 | 0.12 | 16 | + | - |
| 548959 | species1 | 144 | CF | 16 | 1 | 4 | 8 | + | - | ||||
| 548961 | species1 | 144 | CF | 128 | 4 | 2 | 1 | 0.5 | 2 | 0.12 | 16 | + | - |
| 551603 | species1 | 144 | other | 256 | 8 | 4 | 4 | 0.25 | 4 | 0.25 | 32 | + | - |
| 555840 | species1 | 144 | CF | 16 | 8 | 1 | 2 | 1 | 128 | + | - | ||
| 533629 | species1 | 428 | CF | 128 | 2 | 2 | 1 | 0.12 | 1 | 0.12 | 32 | + | - |
| 533612 | species2 | 493 | CF | 64 | 4 | 4 | 1 | 8 | 16 | - | - | ||
| *bronchiectasis. | |||||||||||||
MICs for individual isolates are reported in Table 1. The most active agent was co-trimoxazole followed by imipenem. The MIC-distribution of the isolates in this study has been reported previously [23].
Only 14 isolates carried known acquired antibiotic resistance genes. One
A. xylosoxidans isolate carried nine resistance genes including a
GES-type Extended-Spectrum
Despite infections by Achromobacter spp. being mostly limited to persons with CF and immunocompromised hosts, many virulence factors have been proposed for these species [12, 27]. The role of many of these (putative) virulence factors in infection remains to be elucidated. Some of these factors, such as O-antigens and capsules, may play a role in disease, but are also just basic bacterial structures. For our analysis, based on the encoding sequences, the following virulence factors were studied: hlyA, yqfA1, and yfqA2, and regions 1, 6, 23, and 24, defined by Li et al. [26]. Proposed factors without a defined role in virulence were excluded from the analysis [12].
Region 1, encoding a type III secretion system that has been implied in pathogenicity in other bacteria [27], was absent in some A. xylosoxidans isolates (n = 12, 19.0%), one A. ruhlandii isolate, and all isolates of A. aegrifaciens, A. insolitus and A. spanius. The region 1 sequences in the other isolates were species-specific, but in A. xylosoxidans two variants were present. A ~5 kb region of variant 1 was replaced by a ~6.5 kb sequence. Annotation showed that the assigned gene functions of both sequences were similar.
Region 6, encoding genes for dTDP-rhamnose synthesis, an O-antigen component, was present in only 19 isolates and 6 of these belonged to the same ST. The sequences could be divided into seven variants, which clustered into four groups. One variant, present in four A. insuavis isolates, lacks the gene for CDP-glucose 4,6-dehydratase. An additional 25 isolates encoded only the glucose-1-phosphate cytidylyltransferase (Table 2, Ref. [12]).
| ID | Species WGS | ST | Region1 |
Region6 |
Region13 |
hlyA | yqfA1 | yqfA2 | Region23 | Region24 |
| 537301 | A. aegrifaciens | 501 | - | - | - | - | + | + | - | - |
| 548963 | A. deleyi | 518 | + | +/- | MSF, LysR | + | + | + | - | - |
| 548966 | A. deleyi | 518 | + | +/- | MSF, LysR | + | + | + | - | - |
| 548967 | A. deleyi | 518 | + | +/- | MSF, LysR | - | + | + | - | - |
| 548969 | A. deleyi | 518 | + | +/- | MSF, LysR | - | + | + | - | - |
| 548972 | A. deleyi | 518 | + | + | MSF, LysR | + | + | + | - | - |
| 548973 | A. deleyi | 518 | + | - | MSF, LysR | + | + | + | - | - |
| 548974 | A. deleyi | 518 | + | +/- | MSF, LysR | - | + | + | - | - |
| 534814 | A. insolitus | 503 | - | - | - | - | + | + | - | - |
| 546376 | A. insolitus | 515 | - | +/- | - | + | + | + | - | - |
| 543171 | A. insuavis | 64 | + | + | + | - | + | + | + | + |
| 537313 | A. insuavis | 274 | + | + | + | - | + | + | + | + |
| 533618 | A. insuavis | 303 | + | + | + | - | + | + | - | + |
| 555838 | A. insuavis | 303 | + | - | + | - | + | + | - | + |
| 537308 | A. insuavis | 491 | + | + | + | - | + | + | + | + |
| 546381 | A. insuavis | 496 | + | - | + | - | + | + | - | + |
| 533616 | A. insuavis | 502 | + | - | + | - | + | + | - | + |
| 539942 | A. insuavis | 512 | + | - | + | - | + | + | - | + |
| 539939 | A. insuavis | 513 | + | + | + | - | + | + | + | + |
| 537305 | A. insuavis | 514 | + | + | + | - | + | + | + | + |
| 543523 | A. insuavis | 520 | + | +/- | + | - | + | + | + | + |
| 546373 | A. ruhlandii | 41 | + | +/- | no MSF & LysR | + | + | + | - | - |
| 546375 | A. ruhlandii | 41 | + | +/- | no MSF & LysR | + | + | + | - | - |
| 548953 | A. ruhlandii | 41 | + | +/- | no MSF & LysR | + | + | + | - | - |
| 548956 | A. ruhlandii | 497 | + | +/- | no MSF & LysR | + | + | + | - | - |
| 533619 | A. ruhlandii | 509 | + | + | no MSF & LysR | + | + | + | - | - |
| 533627 | A. ruhlandii | 511 | + | + | no MSF& LysR | + | + | + | - | - |
| 543530 | A. ruhlandii | 519 | - | +/- | no MSF & LysR | + | + | + | - | - |
| 551600 | A. spanius | 516 | - | +/- | no MSF & LysR | + | + | + | - | - |
| 537315 | A. xylosoxidans | 2 | +/- | - | + | + | + | + | - | - |
| 533625 | A. xylosoxidans | 20 | - | - | + | + | + | + | - | - |
| 533630 | A. xylosoxidans | 20 | - | - | + | + | + | + | - | - |
| 542589 | A. xylosoxidans | 22 | +/- | - | - | + | + | + | - | - |
| 543228 | A. xylosoxidans | 22 | +/- | - | - | + | + | + | - | - |
| 537297 | A. xylosoxidans | 27 | + | - | + | + | + | + | +truncated | - |
| 537316 | A. xylosoxidans | 27 | + | - | - | + | + | + | + | - |
| 537320 | A. xylosoxidans | 27 | + | - | - | + | + | + | + | - |
| 551605 | A. xylosoxidans | 27 | + | - | - | + | + | + | + | - |
| 533615 | A. xylosoxidans | 28 | +/- | - | + | + | + | + | - | - |
| 533617 | A. xylosoxidans | 28 | +/- | - | + | + | + | + | - | - |
| 533623 | A. xylosoxidans | 28 | +/- | - | + | + | + | + | - | - |
| 537310 | A. xylosoxidans | 28 | +/- | - | + | + | + | + | - | - |
| 537314 | A. xylosoxidans | 28 | +/- | - | + | + | + | + | - | - |
| 533620 | A. xylosoxidans | 175 | +/- | - | + | + | + | + | - | - |
| 543529 | A. xylosoxidans | 175 | +/- | - | + | + | + | + | - | - |
| 551604 | A. xylosoxidans | 175 | +/- | - | + | + | + | + | - | - |
| 551607 | A. xylosoxidans | 175 | +/- | - | + | + | + | + | - | - |
| 551608 | A. xylosoxidans | 175 | +/- | - | + | + | + | + | - | - |
| 537303 | A. xylosoxidans | 180 | - | - | + | + | + | + | - | - |
| 546371 | A. xylosoxidans | 180 | - | +/- | + | + | + | + | - | - |
| 537298 | A. xylosoxidans | 182 | +/- | - | + | + | + | + | - | - |
| 537302 | A. xylosoxidans | 182 | +/- | - | + | + | + | + | - | - |
| 537309 | A. xylosoxidans | 184 | +/- | +/- | + | + | + | - | - | - |
| 537312 | A. xylosoxidans | 184 | +/- | +/- | + | + | + | + | - | - |
| 546380 | A. xylosoxidans | 226 | + | - | + | + | + | + | - | - |
| 537323 | A. xylosoxidans | 237 | +/- | - | + | + | + | + | - | - |
| 533614 | A. xylosoxidans | 272 | +/- | - | + | + | + | + | - | - |
| 533622 | A. xylosoxidans | 272 | +/- | - | + | + | + | + | - | - |
| 537304 | A. xylosoxidans | 273 | +/- | - | + | + | + | + | - | - |
| 537324 | A. xylosoxidans | 273 | +/- | - | + | + | + | + | - | - |
| 543527 | A. xylosoxidans | 277 | + | - | no MSF & LysR | + | + | + | + | - |
| 543233 | A. xylosoxidans | 281 | +/- | - | + | + | + | + | - | - |
| 533621 | A. xylosoxidans | 290 | + | +/- | + | + | + | + | - | - |
| 543532 | A. xylosoxidans | 290 | + | +/- | + | + | + | + | - | - |
| 537300 | A. xylosoxidans | 323 | +/- | - | + | + | + | + | - | - |
| 533624 | A. xylosoxidans | 426 | +/- | - | + | + | + | + | - | - |
| 539940 | A. xylosoxidans | 426 | +/- | +/- | + | - | + | + | - | - |
| 543229 | A. xylosoxidans | 426 | +/- | - | + | + | + | + | - | - |
| 533631 | A. xylosoxidans | 486 | +/- | - | + | + | + | + | - | - |
| 533632 | A. xylosoxidans | 486 | +/- | - | + | + | + | + | - | - |
| 533633 | A. xylosoxidans | 486 | +/- | - | + | + | + | + | - | - |
| 533628 | A. xylosoxidans | 487 | - | - | + | + | + | + | - | - |
| 537322 | A. xylosoxidans | 488 | - | - | + | + | + | + | - | - |
| 539941 | A. xylosoxidans | 488 | - | +/- | + | + | + | + | - | - |
| 537317 | A. xylosoxidans | 489 | - | +/- | + | + | + | + | - | - |
| 537319 | A. xylosoxidans | 489 | - | + | + | + | + | + | - | - |
| 551609 | A. xylosoxidans | 492 | +/- | +/- | + | + | + | + | + | - |
| 546369 | A. xylosoxidans | 494 | + | - | no MSF & LysR | + | + | + | + | - |
| 546379 | A. xylosoxidans | 495 | +/- | - | - | + | + | + | - | - |
| 546382 | A. xylosoxidans | 495 | +/- | - | - | + | - | + | - | - |
| 533613 | A. xylosoxidans | 498 | - | - | - | + | + | + | - | - |
| 551602 | A. xylosoxidans | 499 | +/- | - | + | + | + | + | + | - |
| 539943 | A. xylosoxidans | 500 | +/- | - | + | + | + | + | - | - |
| 539944 | A. xylosoxidans | 500 | +/- | - | + | + | + | + | - | - |
| 551610 | A. xylosoxidans | 504 | + | + | + | + | + | + | - | - |
| 539938 | A. xylosoxidans | 505 | + | + | + | + | + | + | - | - |
| 537299 | A. xylosoxidans | 506 | + | - | no MSF& LysR | + | + | + | + | - |
| 537311 | A. xylosoxidans | 507 | + | + | no MSF & LysR | + | + | + | - | - |
| 537296 | A. xylosoxidans | 508 | + | - | no MSF & LysR | + | + | + | - | - |
| 543230 | A. xylosoxidans | 510 | - | - | - | + | + | + | - | - |
| 537318 | A. xylosoxidans | 517 | + | +/- | + | + | + | + | - | - |
| 543526 | A. xylosoxidans | 521 | - | +/- | + | + | + | + | - | |
| 546367 | species1 | 57 | +/- | - | + | - | + | + | - | + |
| 533626 | species1 | 144 | +/- | + | + | - | + | + | - | + |
| 548957 | species1 | 144 | +/- | + | + | - | + | + | - | + |
| 548959 | species1 | 144 | +/- | + | + | - | + | + | - | + |
| 548961 | species1 | 144 | +/- | + | + | - | + | + | - | + |
| 551603 | species1 | 144 | +/- | - | + | - | + | + | - | + |
| 555840 | species1 | 144 | +/- | +/- | + | - | + | + | - | + |
| 533629 | species1 | 428 | +/- | + | + | - | + | + | - | + |
| 533612 | species2 | 493 | + | + | no MSF & LysR | + | + | + | - | - |
Region 13, involved in activation of hemolysin, was present in 49 A. xylosoxidans isolates as well as all A. insuavis and species1 isolates and absent in nine A. xylosoxidans isolates including four from ST27, two from ST22 and ST496 each. The two A. insolitus isolates and the single A. aegrifaciens isolate also lacked this region. Five A. xylosoxidans isolates lacked the gene for the MSF transporter and the LysR family regulator. This was also the case for the A. ruhlandii, A. spanius, and the species2 isolate. Interestingly, the A. deleyi isolates encoded only the MSF transporter and the LysR family regulator (Table 2).
The annotation of the A. xylosoxidans type strain (accession number NZ_LN831029) showed the presence of three different hemolysins encoded by the hlyA, aqfA1 and aqfA2 genes. The first encoded a 3296 amino acid protein. However, this region has also been annotated as an agglutinin with additional hypothetical proteins. The opposite strand encoded a putative isopropyldehydratase large subunit. Of note, A. xylosoxidans isolates do not show hemolysis on sheep blood agar. The hlyA region was absent from A. insolitus, A. insuavis, A. spanius, species1, and A. deleyi with one exception (Table 2). The region, which was also present in A. ruhlandii, appeared to be variable; a thorough assessment of this variability was not possible, because the assembly of these sequences was highly fragmented for most isolates (despite good genome coverage for sequencing). The aqfA1 gene was present in all isolates, except for one A. xylosoxidans isolate, and also the aqf2 gene was present in all isolates, except for one A. xylosoxidans. The presence or absence of the hemolysin activating region did not match with the presence or absence of any of the putative hemolysin genes; this suggests either issues with the annotation or a complex regulation of expression.
Region 23, involved in lipopolysaccharide biosynthesis, was present in 15 isolates including six A. insuavis isolates and with 3 isolates belonging to A. xylosoxidans ST27. The nearly 22 kb region was truncated after approximately 15.6 kb. The function of the lacking part is unknown. Region 24 encoding capsule production was present in all A. insuavis and species1 isolates, but not in any of the other species (Table 2).
It should be noted that expression of virulence factors was not confirmed in vitro or in vivo and that virulence features are only predicted on the bases of sequence analysis.
The 20 non-CF isolates consisted of 15 A. xylosoxidans (75%), four A. insuavis (20%) and one species1 isolate (5%) (Table 1). The CF-isolates appeared to be more diverse: 48 A. xylosoxidans (59%), seven A. insuavis, A. deleyi, A. ruhlandii and species1 (each 8.6%), two A. insolitus (2.5%), and one A. aegrifaciens, A. spanius and species2 (each 1.2%). Comparison of the average MICs for CF isolates and non-CF isolates showed that the MICs for aztreonam, imipenem and tobramycin were approximately two times higher for non-CF isolates (non-CF vs CF:153.6 and 84.1 g/L, 11.0 and 5.5 mg/L, and 58.4 and 34.5 mg/L, respectively), whereas the MICs for ceftazidime, ciprofloxacin, colistin, co-trimoxazole, and meropenem were 1.4-2.9 higher for CF isolates (CF vs non-CF: 19.3 and 12.9 mg/L, 8.9 and 3.1 mg/L, 2.0 and 1.4 mg/L, 2.4 and 1.0 mg/L, 5.0 and 3.4 mg/L) (Table 1). No relevant differences were observed in the presence of putative virulence factors.
In conclusion, only 63/95 (66.3%) of the isolates were correctly identified using routine MALDI_-TOF identification probably indicating a lack of well-typed Achromobacter isolates in the standard database. Two putative novel species were identified. Isolates from persons with CF appeared to be more diverse. Despite the high MICs the presence of acquired resistance genes is uncommon, although some isolates harbored several acquired resistance genes. The average MICs for CF isolates were lower for aztreonam imipenem, and tobramycin, but higher for ceftazidime, ciprofloxacin, colistin, co-trimoxazole, and meropenem. The putative virulence genes of Achromobacter involved in infections or colonization are variable, but no difference in putative virulence factors were observed.
CF, cystic fibrosis; ESBL, Extended-Spectrum
ACF, MDA, MMT, JSE RC and MBE designed the research study. BB-T, MvW, JFM performed the research. ACF and JRB analyzed the data. ACF, JRB, MBE wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement number [115721-2], resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies in kind contribution. Spanish CF isolates were recovered from projects supported by Instituto de Salud Carlos III of Spain (grants PI12/00743 and PI15/00466) and cofinanced by the European Development Regional Fund (A Way to Achieve Europe program; Spanish Network for Research in Infectious Diseases grant REIPI RD12/0015). MD-A was partially supported by Fundación Francisco Soria Melguizo (Madrid, Spain).
The authors declare no conflict of interest.
