Dengue is potentially a life-threatening arthropod-borne viral infection for which there are no known therapeutic agents till date. Early stage diagnosis of dengue infection is still lacking. Diagnosis is only made after severe manifestations and later stages of infection. Timely prognosis can prevent dengue related mortalities. The nucleic acid-based therapy has potential to emerge as a promising approach for early diagnosis and treatment of this viral infection. Many studies have been carried out suggested the regulatory role of ncRNAs thereby revealing the importance of protein-RNA and RNA-RNA interactions during infection. Various regulatory RNAs are either expressed by mammalian cells or generated by viral RNA have reported to play important roles in viral life cycle including dengue virus. Thus exploring host-virus interaction will pave the novel path for understanding the pathophysiology of febrile infection in dengue. Rapid advances in sequencing techniques along with significant developments in the field of RNA studies has made RNA therapeutics as one of the promising approaches as antiviral targets. The idea of RNA based therapies has been greatly backed by a Hepatitis C virus drug, Miravirsen which has successfully completed phase II clinical trial. In the present review, we will discuss the implications of different non-coding RNAs in dengue infection. Differential expression of small ncRNA may serve as a reliable biomarker of disease severity during different stages of infection and can also play regulatory roles in disease progression.
Dengue fever (DF) is a flu-like acute viral infection caused by single stranded positive sense RNA virus of the Flaviviridae family, which is an arthropod-borne/ transmitted virus. The dengue virus includes four serotypes viz. DENV-1, DENV-2, DENV-3 and DENV-4 [1, 2] which are mainly found in tropical and subtropical regions. Amongst them DENV-2 and DENV-3 are the ‘Asian’ serotypes originated from Southeast-Asia and are frequently associated with the severe disease accompanying secondary dengue infection [3]. These serotypes have approximately ~97% similarity at the gene level and are separated further by up to 25%–40% at the amino acid level [4]. Dengue mainly spreads through two principal mosquito vectors belonging to the Aedes genus i.e., Aedes aegypti and Aedes albopictus. Out of these two, Ae. aegypti is highly anthropophilic in nature and thus a competent vector well adapted even in an urban environment.
According to WHO, Dengue is the world’s fastest emerging pandemic prone viral disease. Before 1970, the dengue epidemic prevalence was reported in nine countries, but today the endemic has increased in more than 100 countries (Fig. 1). This upsurge has been mainly attributed due to an increase in population, global warming, immigration-emigration, urbanization, inefficient mosquito control and lack of health care facilities [5, 6, 7]. Recently a large dengue outbreak was reported in the year 2016 at global level, while a significant reduction was observed thereafter in the year 2017-18 post Zika outbreak period. Although the factors leading to this fall are still unknown. According to the latest report, sharp increase has been reported in the year 2019 especially with DENV-1 and DENV-2 serotypes. The majority of the cases were reported from tropical countries such as Philippines, Mexico, Nicaragua, Thailand, Malaysia and Colombia. In 2019, the first case of sexual transmission of dengue virus was reported in Spain. Other cases of sexual transmission has also been reported [8, 9].
 Fig. 1.
Fig. 1.European Centre for Disease prevention and Control (ECDC) report dated 16.07.2020 showing the number of dengue cases in the affected countries (https://www.ecdc.europa.eu/en/dengue). Reproduced with the permission from ECDC.
DENV infected/carrier mosquito interacts with multiple hosts during a single gonadotrophic cycle and thus confers multiple infections in order to complete a single blood meal. Once DENV is ingested by the mosquito, it establishes infection in the midgut from where the virus disseminates to other tissues. When the infection reaches salivary glands, transmission to humans takes place when mosquito feeds onto human blood for meal. Once the virus gains entry into the human host through skin, different immune responses leads to the progression of illness [1]. The detailed dengue life cycle has been depicted in the Fig. 2.
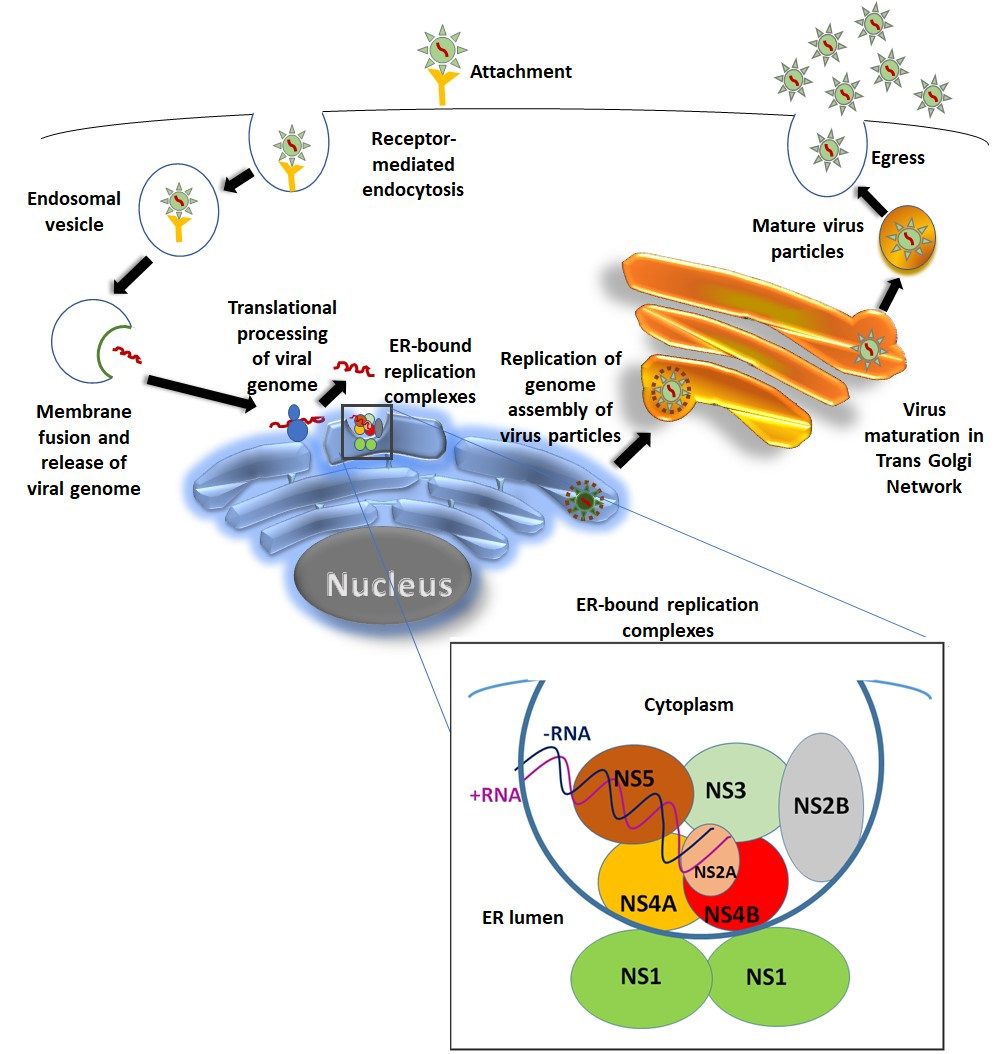 Fig. 2.
Fig. 2.DENV Life cycle. Virus gains entry into the target cell by getting attached to its surface receptor. Virus-receptor complex then gets internalized through receptor mediated endocytosis and forms the endosomal vesicle. Due to the acidic environment of endosome; endosomal membrane and viral membrane fusion takes place followed by release of RNA genome. The released RNA then goes to rough endoplasmic reticulum (RER) for genome translational processing and amplification. Flavivirus replication takes place on the surface of ER. DENV non-structural proteins together with some host proteins forms the replication complex. NS1 forms dimer in the ER lumen and aids other components of the replication complex on the other side of the ER membrane in replication process [21]. Further maturation and virus assembly takes place within the lumen of endoplasmic reticulum. Immature virus particle goes to the trans-golgi network for further maturation. Mature virus particles finally egress to the extracellular environment.
The genus Flavivirus includes more than 50 arboviruses. The significant human infecting flaviviruses includes dengue virus (DENV), Japanese encephalitis virus (JEV), West Nile virus (WNV), Zika virus (ZIKV), and tick-borne encephalitis virus (TBEV). The flavivirus genome translation is coupled to RNA replication. During translation, the replication complex formation takes place on the ER membrane. The replication complex formation is initiated by binding of NS3 and NS2A to the conserved regions of NS5 which works as RNA-dependent RNA polymerase (RdRp). Once translation is completed, NS2A, NS3 and NS5 of the replication complex binds to the 3’UTR of the genome. NS1 dimer formation takes place at the lumen side of the ER which aids the replication complex components on the cytoplasmic site of the ER (Fig. 2). The negative strand replicative intermediate forms during replication which is critical for priming positive strand RNA synthesis [10, 11, 12]. Limited coding capacity of flavivirus genome compels the virus to rely on molecular machineries of host cells for completion of its life cycle. The viral genome encodes a single polyprotein which is further cleaved into 10 mature proteins with the help of viral and host proteases. Various cellular factors both in humans and mosquitos play pivotal roles starting from the entry of virus into the host cell and up to its secretion. In humans DENV entry into target cells is mediated by structurally distinct group of membrane proteins such as glycosaminoglycans (heparan sulfate), C-type lectins (DC-SIGN (CD209), the mannose receptor (CD206) and immunomodulatory proteins (TIM/TAM receptors). Further, it also exploits the host system for trafficking, uncoating of structural proteins, translation of viral RNA, polyprotein processing, assembly, maturation and secretion of virus. Often these factors assist the viral replication through protein-protein interactions, protein-RNA interactions and viral-host RNA-RNA interactions [13, 14]. Various reports have extensively studied the interactions between DENV RNA and host proteins [4, 15, 16, 17]. Concurrently, several reported studies have suggested that, several types of ncRNA such as miRNAs, siRNAs, snRNAs, sfRNAs and long ncRNAs accomplish several critical roles in the replication of flavivirus including DENV. Most of these versatile RNA molecules are coded from genome of host cell. Various studies have further demonstrated the existence of ncRNA species derived from viral RNA as well [18, 19, 20].
The majority of the human genome is transcribed into non coding transcripts which itself do not expressed in the form of proteins but plays a critical role in regulating the expression of about 60% proteins coding genes. Fate of a particular mRNA is regulated by multiple ncRNA. Therefore, their aberrant expressions can be noticed for various human diseases [22]. Nevertheless, the field of ncRNA therapeutics is striding fast. For an instance, as soon as miRNA expression was linked with cancer, within a decade or two, MRX34 which is miRNA mimic to target hepatocarcinoma, entered into clinical trial [23]. Nonetheless, another drug miravirsen, antagomiR targeted against hepatitis C virus has also successfully cleared phase II clinical trials [24, 25]. Also, FDA approved its first ever RNAi based drug patisiran in 2018, to treat peripheral nerve disease (polyneuropathy) [26, 27]. At the end of the 2019, second siRNA based drug givosiran, also got approved to treat acute hepatic porphyrian (AHP) [28, 29, 30]. Distinguishing dengue from other febrile diseases is still challenging owing to its relatively common symptoms with other febrile diseases along with lack of its proper diagnosis. Treatment regimen for dengue is still complicated and there is still a lack of any licensed therapeutic agent [8]. Here, we briefly review about the roles and implications of various non-coding RNAs exhibited during DENV pathophysiology. Exploring these new strata of host-pathogen interaction will thus unravel future strategies as novel therapeutics in combating the menace of dengue infection. Many ncRNAs have been discussed below which are directly involved in DENV replication and pathogenesis. Although, ncRNA therapeutics is entirely based on a new concept that totally differs from conventional drug design approach. But the properties of these ncRNA are very much promising to be translated into therapeutics which one can witness through the already existing drugs [23, 25, 27, 30].
miRNAs are small (18–25 ntds long) noncoding RNAs, known to control the complex post transcriptional regulation of gene expression and can be derived either from host or viral RNAs. miRNAs are vital in various biological processes like cell development, apoptosis, cell proliferation, infection and inflammation [31, 32]. They also perform primary functions in the maintenance of B cell differentiation, Tregs lineage maintenance, regulating the differentiation of dendritic cells and macrophages via TLRs [33]. miRNAs have the novel therapeutic capability to manipulate the expression and function of various genes responsible for virus replication. They have also been reported to play critical roles in both virus replication and its inhibition. Dicer enzyme is primarily responsible for the generation of miRNA and siRNA. Various studies have suggested that endogenously produced miRNA have the potential to inhibit virus translation and replication [34, 35]. RNAi is a sequence dependent mechanism to process gene regulation by siRNA or miRNA. Both Dicer and Drosha are required for miRNA synthesis from dsRNA while only Dicer is required for siRNA production. During viral infection, viral RNA is recognized by miRNA/siRNA-RISC assembly which is then further targeted for silencing or degradation [36]. Exogenous viral genome is recognized by pattern recognition receptors (PRRs). These PRRs includes cytoplasmic retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), along with endosomal Toll-like receptor 3 (TLR3) and TLR7 [37, 38]. These receptors are crucial for innate immune response against virus which senses phosphate containing RNA in the cytoplasm and dsRNA [39]. Activation of these receptors induces type I IFN immune signaling. The IFN signaling is further amplified by JAK/STAT pathway that establishes systemic antiviral state by developing positive feedback loop in the surrounding cells. The binding of IFNs to the cell surface receptors leads to the activation of IFN stimulated genes like OAS/RNaseL and PKR. OAS recognizes viral dsRNA and tag it with 2’,5’-adenosine oligomers and it is then degraded by RNaseL. The resulting degraded remnants can further trigger IFN response through positive feedback loop. Another mechanism includes phosphorylation of translation factor eIF2a by PKR, causing global translational arrest finally leading to apoptosis of the infected cells [40, 41, 42, 43]. miRNAs or siRNAs generated as a result of antiviral immune response can also stimulate helicase RIG-1/MDA5 pathway which is suggestive that these proteins could lead to the conjuction of RNAi and IFN immune response [44, 45, 46, 47].
Study by Bogerd & co-workers transfected the Dicer deficient cells with a
variety of RNA viruses including DENV. They did not observe any enhancement in
the replication level in the absence of miRNA and thus concluded that these
viruses have evolved resistant mechanisms for miRNA inhibition [48]. Cytokines
show synergistic or antagonist action for miRNA activation and inhibition and
thus regulate its target genes [49]. Studies carried out by Shahen et al. in 2018 suggested DDX3X (DEAD-Box Helicase 3, X-Linked) and PTEN
(Phosphatase and Tensin Homolog) to be the potential targets of DENV infection
regulated by M005 miRNA. DDX3X is a protein responsible for stimulating the IFN
promoter in DENV infected cells. It serves an important role in cell
proliferation regulation and thus can be used as a target to control the dengue
infection. Its upregulation can thus be used to treat DENV infection [50]. PTEN
suppression has also shown the potential antiviral activity against dengue
infection [51]. The suppression of PTEN inhibits autophagy which results in
decreased capsid protein expression and thus minimize the release of virus
particle [52]. There are various miRNAs which are found to affect the DENV life
cycle (Table 1). The examples include: (1) the insertion of miRNA recognition
element (MRE) for miR-122 (hepato specific) in the 3’UTR region of DENV-RNA
suppressed the viral replication in the transfected cells [53]; (2) the
incorporation of MRE for miR-142 (hematopoietic specific) in dengue RNA inhibits
viral replication in dendritic cells and macrophages, but not in
non-hematopoietic cell lineage [54]; (3) miR-30e* promotes interferon (IFN)
production through the NF-
| miRNA | Targets | Roles | Favors or suppresses DENV replication |
| M005 | DDX3X (Dead Box Helicase 3; X-linked) | Stimulates IFN promoter and thus regulates cell proliferation [50] | Suppresses |
| PTEN (Phosphatase and Tensin Homolog) | Cell growth; translational control and cytoplasmic shuttling [51]. | Favors | |
| miR-122 (Hepato-specific) | DENV genome | Insertion of miRNA recognition elements (MRE) in the 3’UTR of DENV genome [53]. | Suppresses |
| miR-142 (hematopoietic-specific) | DENV genome | Insertion of MRE for miR-142 in RNA genome suppresses viral replication in dendritic cells and macrophages [54]. | Suppresses |
| miR-30e* | NF-κB pathway | Promotes IFN production [55]. | Suppresses |
| Let-7c | BACH1 (Basic leucine zipper transcription factor 1) and HO-1 (Heme Oxygenase protein) | Let-7c directly binds to BACH1 (strong anti-inflammatory suppressor) and HO-1 (anti-oxidant) [56]. | Suppresses |
| miR 548g-3p | Stem Loop A (SLA) | miR 548g-3p binds to the SLA in the 5ˊ region of RNA genome of DENV [57]. | Suppresses |
| miR-133a | Polypyrimidine Tract Binding (PTB) | miR-133a interferes with PTB and thus suppresses replication [58]. | Suppresses |
| miR-146a | Viral replication | Dampens IFN production [64]. | Favors |
| Abbreviations: DENV, Dengue virus; IFN, interferon; miR, miRNA; MRE, miRNA recognition elements. | |||
Being vector, mosquitoes transmit numerous viral infections such as Dengue, West Nile, Chikungunya etc. [59]. Dynamics of mosquito transmitted viral diseases depends on interactions between vector and parasite [60]. miRNAs also play important roles in the viral cycle inside the vector. Blair and co-workers for the first time established the expression of small RNAs not only in tissues of female mosquitoes but also in cultured mosquito cells [61, 62]. In their study DENV-2 was administered orally into Ae. aegypti and subsequently cultured mosquito cells infected by DENV2. Both models reported the accumulation of small RNA derived from viral genome and virus specific small RNAs. Concurrent existence of these two small RNA increases virus replication and facilitate faster viral transmission [62]. In 2010, Wu et al. have reported the differential expression of miRNA’s in both Ae. albopictus DENV infected and uninfected cells [63]. In another study the researchers have demonstrated more than threefold over expression of miR-252 in the C6/36 cells infected with DENV. Over expressed miR-252 down regulated both the viral RNA copies and expression of E-protein (Yan et al., 2014). Campbell et al., 2014 investigated the modulation of microRNA levels in Ae. aegypti post-exposure to DENV-2 at 2, 4 and 9 days. Total of 35 miRNAs were modulated during the course of infection. By target prediction studies, they have identified that 365 mRNA targets contain miRNA-binding at their 3’UTR and the coding sequence. The gene products of target mRNA were associated with DENV replication and dissemination events including transport, transcription/translation and cytoskeletal/structural components. Tissue-specific miRNA expression was investigated in Ae. albopictus mosquitos infected with DENV New Guinea C strain. qPCR and Northern blot analyses suggested that miR-281 was over expressed in midgut tissue. Subsequent functional interventional studies have demonstrated that the overexpression of miR-281 supports the replication of DENV-2 [66]. Recently Su et al., 2019 investigated the differential expression of miRNAs in DENV-2 harboring Ae. albopictus mosquito. Results indicated that total of 46 miRNAs differentially expressed, among them 15 microRNAs were over expressed (at least two-fold) and two were down regulated (at least two-fold). Further, the transfection of miR-1767 and miR-276-3p enhanced virus replication but miR-4448 inhibited the replication of dengue virus replication in C6/36 cells [67].
There are some studies which proves miRNA support for DENV replication, the examples are DENV increases the expression level of miR-146a, and thus supports the viral replication by dampening IFN production [64]. DENV infection changes the miRNA expression profile of PBMCs [68]. NS1 gene is mainly targeted by one of the functional miRNA/viral small RNAs encoded by DENV [69]. The miRNAs reported thus far are not comprehensive and are limited. Many other miRNAs have been reported elsewhere [70].
The clinical relevance of miRNA potential has further been explored by different studies carried out on patient samples. In 2020, Hapugaswatta et al. has proved this in a study in which they observed the relative expression of different miRNA and several putative target genes in peripheral blood cells (PBC) of 20 DF and 20 DHF patients. They observed significant up regulation of hsa-miR-150-5p in PBC of DHF patients compared to DF patients during acute phase of infection. Decreased expression of EZH2 gene (enhancer of zeste homolog 2) which is involved in histone methylation and thus transcriptional repression, was found to be down regulated during early stages of infection when no severe symptoms of developing severe manifestations were observed [71].
Similar kind of studies were also carried out by different groups, which studied miRNA transcriptomes in blood [72] and serum of dengue patients [73]. They reported different miRNA like miR21, miR146a, miR590, miR188, miR152, miR24, miR512, miR4640. The studies suggested these miRNA to be reliable and have the potential to be characterized as promising biomarker for dengue infection.
During severe DENV infection or DHF, cytokine dysregulation is observed, which results in cytokine storm. It is a phenomenon in which there occurs imbalance between pro-inflammatory and anti-inflammatory mediators. Cytokine dysregulation is observed during miR- 150 overexpression in DHF. Study by Chen et al. in 2014 observed the correlation of miR-150 and SOCS-1 (suppressor of cytokine signaling) expression which is crucial for regulating various immune related pathways. They confirmed that miR-150 overexpression suppresses SOCS-1 expression in DHF patients. Modulation of miRNA expression can potentially control abnormal immune responses during DHF disease severity [74].
A study carried out by Hussain and Asgari in 2014, identified microRNA like viral small RNA (vsRNA)-5 which were 23-nucleotide long. RNA viruses produces dsRNA by convergent transcription of their viral genomes. These dsRNA are long and perfectly complementary stem loop structures. Dicer creates pool of 22 bp dsRNAs which are structurally similar to miRNA duplex intermediate. One of its strands is then loaded into RISC to guide for complementary mRNAs. Since these sequences are completely complement to viral RNA, RISC binding results in the cleavage and degradation thus potentially decreasing viral replication [75, 76].
These vsRNA are also expressed by DENV2 in infected mosquito and mammalian cells. It has been found to be associated with the autoregulation in DENV2 replication by directly targeting the NS1 protein during late infection. It was mainly dependent on insect Argonaute 2 (Ago2) for its biogenesis. The prime feature of miRNA is its partial stoichiometric binding to the target site thereby leading to its efficient binding. miRNA is expressed in high levels unlike vsRNA which is expressed in vanishingly low amount. Inhibition of vsRNA leads to increase in DENV replication by 10,000-fold. Thus, this RNA works as a viral encoded repressor of DENV replication [69].
RNA interference (RNAi) is an RNA dependent gene silencing mechanism that silences the expression of genes by promoting degradation of RNA in a sequence specific manner which is mainly triggered by the double stranded RNA (dsRNA) in the cytoplasm of eukaryotic cells. The exogenously introduced dsRNAs during uncoating of viral RNA and the viral replication, are cut into 21–25 ntd length small interfering RNA (siRNA) by an enzyme called Dicer which has RNase III-like function. The siRNAs, described as the ‘hallmark’ of RNAi, form RNA-induced silencing complex (RISC) with other cellular components and thus leads to silencing of specific gene by introducing the cleavage of the homologous transcript [77, 78, 79]. RNAi is believed to be an effective endogenous mechanism for host cells for defense against virus attack [80] and is becoming an energizing field to develop anti-viral therapy including dengue virus.
DENV generates intracellular dsRNA as an intermediate of their replication. This dsRNA can induce RNAi in the host cells. RNAi modulation could be the possible explanation for the mosquito’s non-pathogenic and persistent infections of DENV [62]. Studies by Vargas et al. in 2009 has shown that DENV2 does not completely evade RNAi, since silencing of genes like dcr2, r2d2, or ago2, which encode for the important sensor and effector proteins in the RNAi pathway, increased the virus replication in the vector and decreased the extrinsic incubation period required for virus transmission [62]. The dsRNA has been proven to mediate RNAi in mosquito C6/36 cells which leads to inhibition of DENV replication in cultured mosquito cells [61, 81]. The study carried out by Wu et al. in 2010 have shown that the synthetic siRNA against the DENV1 membrane glycoprotein precursor gene effectively inhibited DENV1 viral RNA replication and increased C6/36 cell survival rate which proves that siRNAs can be employed as potential targets in the treatment of dengue.
Long non-coding RNAs (lncRNAs) are long transcripts of more than 200 ntds and play crucial role in gene regulation. lncRNAs have been recognized to be associated with various biological processes including pathogenesis of diseases through diverse mechanisms. Its role and expression have been found to be associated with various liver specific disease [82, 83, 84]. Wang et al. in 2016, identified the expression profiles of lncRNAs in DENV1 and DENV2 infected L-02 cells to find the expression implication on transcription regulation and signal transduction during liver injury [85]. Mature miRNA are also related with RNAi and thus gene regulation. On the basis of their distribution and sequence similarity, lncRNA may serve as the precursors for miRNA and functions in the miRNA endogenous expression [86]. Their mutual interaction and that of miRNA with its target would lead to an insight of the role of lncRNA in gene regulation and thus will open novel perspectives in the research arena.
The differential expression of lncRNA was predicted by performing pre-miRNA prediction, lncRNA-miRNA co-expression and target gene analysis. In DENV1 infected group, lncRNA expression was found to be associated with biosynthesis, DNA/RNA related processes, estrogen signaling, sterol biosynthetic process and protein dimerization activity. In DENV2 infected group, their expression was found to be dominant in protein secretion, methyltransferase process, regulation of host cell cytoskeleton reorganization, small GTPase Ras family, inhibition of cell proliferation and induction of apoptosis [87], as depicted in Fig. 3.
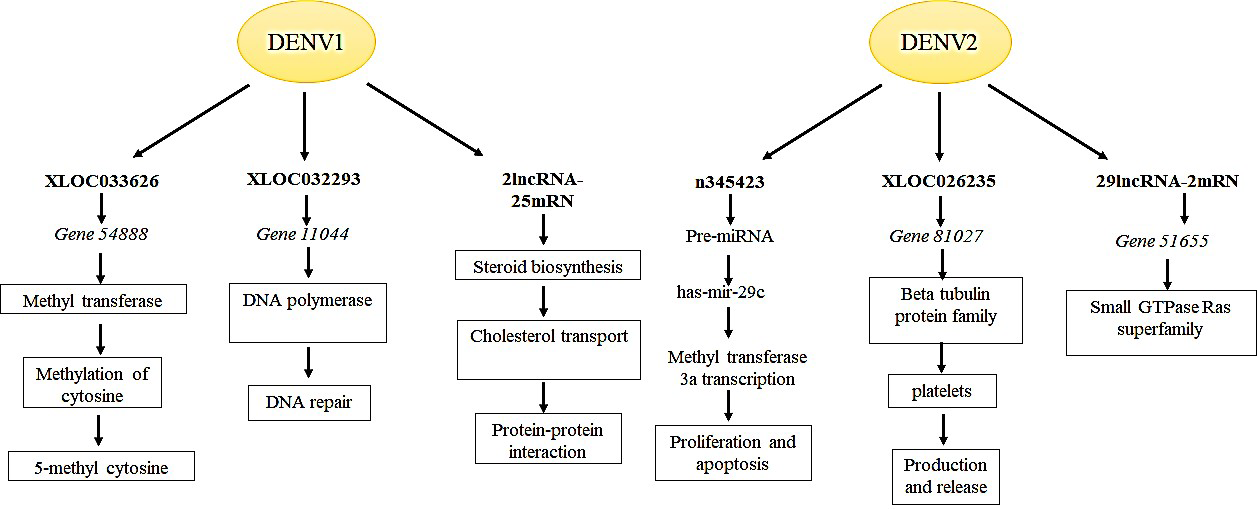 Fig. 3.
Fig. 3.Differentially expressed lncRNAs during DENV1 & DENV2 infection. lncRNAs found differentially enriched in DENV1 infection are primarily the genes involved in signaling; biosynthetic pathways; protein dimerization; RNA/DNA related processes. While lncRNAs associated with DENV2 infection are majorly involved in cytoskeleton organization; cell proliferation and apoptosis; methyltransferase processes and protein secretion. The Figure is based on the data from Wang et al. [87].
In a study by Pandey et al. in 2017, co-expression analysis of protein coding genes possibly regulated through deregulated lncRNAs was carried out. They observed reduced expression of NEAT1 (Nuclear Enriched Abundant Transcript 1) lncRNA in patients with dengue severe phenotype which can prove to be useful for diagnosis in monitoring the progression of disease. Also, an antithetical relationship was observed between NEAT1 and IFI27 (Interferon Alpha Inducible Protein 27) gene. Monitoring NEAT1 and IFI27 co-expression may provide the understanding about disease progression and thus can prove to be a potential biomarker [88].
The sub-genomic flavivirus RNAs (sfRNAs) are viral derived non-coding RNAs generated as a result of Xrn1 host exonuclease. Xrn1 exonucleases degrades the viral genome progressively from 5’ to the 3’ direction and halts at stable RNA structures, called as Xrn1 resistant RNA (xrRNAs). The xrRNAs are present at the 3’UTRs which are 300–500 ntds long. These sfRNAs has been hypothesized to be associated with the variation in 3’UTRs during host adaptation [89, 90, 91, 92, 93].
Different genotypes and serotypes are indicative of genetic variations which have its implications in deciding the viral virulence, viral fitness and epidemic potential [94, 95, 96]. Recent studies have demonstrated the adaption of different flavivirus including DENV in different hosts leading to diversification of the viral genome. Sequencing studies of DENV population isolated from different hosts, i.e., mosquitoes and humans, were carried out which showed the variations in 3’UTRs of viral genome extracted from DENV reared in different hosts [97]. The mutations which were mapped in the 3’UTRs of DENV genome extracted from mosquitoes were actually cleared off on transition to human cells [97].
Studies have found the significant role of sfRNAs in the evasion of cellular anti-viral response in different flavivirus as well as in DENV [42, 98, 99, 100, 101, 102, 103, 104]. Sequence variability at 3’UTRs and sfRNAs accumulation was found to be significant in deciding viral fitness as well as the epidemiological fitness [105]. Pompon and co-worker annotated the role of sfRNA in pathogenicity and transmission of DENV [106]. They examined the epidemiological fitness (EF) of the DENV by constructing phylogeny of isolates obtained over period of 20 long years (1981 to 2001) in Puerto Rico. In an epidemic in 1994 in Puerto Rico, PR-2B DENV-2 clade replaced the endemic PR-1 DENV-2 clade. Phylogeny data revealed that nucleotide changes in 3’UTR resulted in evolution of virus from low EF to high EF. Further functional assays confirmed 3’UTR of high EF strain up-regulate the sfRNA in salivary gland of Ae. aegypti mosquitoes [106]. Genome study analysis by Manokaran et al. in 2015 reported variations in NS1, NS3, NS5, and 3’UTRs regions of the two clades. The newly emergent clade showed high sfRNA:gRNA ratio during replication. Study by Finol et al. 2019 have discovered that, despite sequence divergence, structure of sfRNA is highly conserved [107]. The increased levels of sfRNA modulates innate immune response by showing the affinity towards tripartite motif 25 (TRIM25) in a sequence dependent manner. sfRNA binding to TRIM25 prevents TRIM25 deubiquitylation imperative for sustained and amplified expression of retinoic acid—inducible gene 1 (RIG-I)—induced type I interferon [47]. IFN renders uninfected cells resistant to viral infection, therefore the reduced IFN expression at early stages of infection could be of much importance for viruses as it would aid virus to replicate and spread in human host to reach the viremia levels for further arboreal transmission.
Study by Bidet et al. in 2014, identified three important host RNA-binding proteins (RBPs) namely G3BP1, G3BP2 and CAPRIN1 which play crucial role in host immunity by regulating IFN response during DENV infection. These three genes are required for the translation of interferon stimulated genes (ISGs), which play critical roles in innate immunity. But sub-genomic flaviviral RNA (sfRNA), binds to these RBPs and antagonizes their actions and thus the antiviral property gets inhibited [108].
Non-coding RNAs do not code for any protein but they significantly play varied regulatory roles in both cellular physiology and disease pathogenesis. Approximately, 95% of the human transcriptome constitutes for non-coding RNAs. Several studies have shown that non-coding RNAs are dysregulated and thus implicated in various processes leading to disease progression.
Dengue is an arboreal disease caused by flavivirus for which there is no licensed therapeutic yet. Early detection of infection with no warning signs is crucial for proper disease management. Patients with no warning signs may later develop severe infection that can alleviate disease related complications which may prove to be fatal. In view of this, recent ongoing research studies are mainly focusing on timely prognosis, DENV replication and host immune invasion. Taking into account the various regulatory roles played by ncRNAs, these have been hypothesized to be clinically important. The altered expressions of ncRNA during pathophysiology of a disease marks their potential for the putative biomarkers and the drug targets. Here, we have reviewed certain examples from the literature that highlights different regulatory roles that various ncRNAs could play during DENV infection. These regulatory roles can either be host dependent or host restricted. In the present review article, we have highlighted the significance of various miRNAs which either support or inhibit the DENV replication. miRNA therapies can be targeted for (a) supporting immune system by targeting IFN production like miR-30e*, (b) suppressing cytokine production by targeting miR-150, (c) inhibiting DENV genome replication by targeting miR-122 which aids in DENV genome replication. The same miRNA is crucial for HCV replication against which antgomiR-122 has been developed, that is Miravirsen which has successfully cleared phase II clinical trials. Lastly, (d) by supplementing the miRNA mimics which are involved in DENV suppression like miR-133a, let7c, etc. Moreover, miR146a is the only miRNA which is expressed during different flaviviral infections and also inhibits viral replication by promoting IFN production [109, 110, 111, 112]. Therefore, miR-146a has the potential to serve as the easy and direct approach to develop new antiviral against multiple infections. MRX34, therapeutic tumor suppressor mimic clinical trial I was halted due to immune perturbations. But now its role have also been found in induction of IFN responsive genes for type I IFN. miR-34 mimics can have the potential to combat DENV infection [113, 114]. vsRNA, a miRNA like ncRNA works as a viral repressor and thus inhibits DENV replication. lncRNAs associated with DENV infection have also been reported with immense potential to serve as a biomarker or a target for therapeutics. dsRNA which forms as an intermediate during DENV amplification can functions as miRNA/siRNA and thus induce RNAi in the host cell which leads to viral genome degradation.
On the contrary, DENV has evolved ways to counteract host antiviral response. It
uses its NS proteins to target various signaling molecules and pathways that
leads to antiviral response. Type I IFN mediated immune response is the main
antiviral mechanism which is mediated by PRR recognition of viral genome. It
engages NS5 to prevent genome methylation [115], NS3 and NS4a/b inhibits RIG-I
cascade, NS2a and NS4b blocks RIG-I/Mitochondrial antiviral signaling and thus
IFN
sfRNAs, a DENV encoded non-coding RNAs evade host anti-viral response machinery. sfRNAs are involved in immune evasion which inhibits IFN mediated signaling cascade. These observations highlight the potential of various ncRNA which can be further exploited as a standalone or as a conjunct therapy in the treatment of dengue. Also, for an effective treatment, a proper diagnosis is the prerequisite, a role perfect for ncRNAs which can even serve as potential biomarker for diagnostics.
Non-coding RNA based therapies have slowly but steadily emerged in the last few years. It has unveiled many directions for unravelling the nexus between host-pathogen interaction which can arm us with novel therapeutic strategies in the field. While at one level it is important to understand the impact of DENV infection on host ncRNA regulation mechanism, it is also desirable to explore pathogen generated ncRNAs and its implications on various infection aspects. Further research is thus required to analyze the differential gene expression during ncRNA regulation and further observe its implication on DENV replication. Integrating this information can give us a better insight about host response to infection which can then finally result in bench to bedside translation. Due to advancements in high-end computational capabilities and next-generation sequencing, ncRNA based therapeutics have been flourishing for the last decade. Altogether, in the near future we will see the utility and application of non-coding RNAs in personalized therapies as well.
Not applicable.
DM, KKP, RKYS, SB, and BV contributed to conception, design, analysis, and drafting of the manuscript. JK, YR, LS, and RKS, contributed to data collection and manuscript drafting. All authors approved the final version of the manuscript for submission.
We acknowledge our laboratory members for their help and discussion. We also thank anonymous reviewers for their excellent criticism of the article.
This work was supported to BV from DST; Govt. of India (grant EEQ/2018/000838) and AIIMS; New Delhi (grant A-651). DM and LS are supported by fellowship from Council of Scientific and Industrial Research (CSIR); and University Grants Commission (UGC); GOI respectively.
The author declares no conflict of interest.
DENV, Dengue virus; DF, Dengue fever; DHF, Dengue hemorrhagic fever; IFN, interferon; JEV, Japanese encephalitis virus; lncRNA, long non coding RNAs; miR, miRNA; MRE, miRNA recognition elements; PBS, peripheral blood cells; PKR, Protein Kinase R; PTEN, Phosphatase and Tensin homolog; sfRNA, Sub-genomic flavivirus RNAs; UTR, untranslated region; vsRNA, Viral small RNAs; WHO, World health organization; WNV, West Nile virus; ZV, Zika virus.
