- Academic Editors
†These authors contributed equally.
Background: Largemouth bass (Micropterus Salmoides) is an
economically important fish species in China. Most research has focused on its
growth, disease resistance, and nutrition improvement. However, the
sex-determining genes in largemouth bass are still unclear. The transforming
growth factor-beta (TGF-
The study of sex determination in animals is a challenging but critical task. The sex of diverse animals is determined by a hierarchical gene network, and is considered to be one of the most variable processes in evolution [1, 2]. Fish are at the intermediate stage of evolution between invertebrates and higher vertebrates. As such they show a much more diverse pattern of sex determination than higher vertebrates, exhibiting both genetic and environmental sex determination systems [3]. For example, arapaima (Arapaima gigas) [4] and tiger pufferfish (Takifugu rubripes) [5] have an XX/XY sex determination system, while greater amberjack (Seriola dumerili) [6] and half-smooth tongue sole (Cynoglossus semilaevis) [7] have a ZZ/ZW sex determination system. Barred knifejaw (Oplegnathus fasciatus) [8] has an X1X1X2X2/X1X2Y sex determination pattern. The sex of some fish is also affected by the environment [9, 10, 11]. Almost all types of vertebrate sex determination patterns have been reported in fish [12, 13, 14, 15, 16, 17].
In vertebrates, sex determination and differentiation are often highly plastic
and influenced by a range of different genetic and environmental factors [18, 19, 20, 21].
The transforming growth factor-beta (TGF-
Previous studies have reported that the TGF-beta signaling pathway is also involved in regulating the expression of cytochrome P450 family 19 subfamily A member 1 (cyp19a1) and forkhead box L2 (foxl2). For example, doublesex and mab-3 related transcription factor 1 (dmrt1) can directly inhibit cyp19a1 transcription in Nile tilapia. Overexpression of dmrt1 resulted in decreased expression of cyp19a1 and reduced serum estrogen levels in female tilapia, thereby leading to sex reversal [38]. Knockdown of amhy in the embryo of male Patagonian silverside (Odontesthes hatcheri) leads to the upregulation of cyp19a1 and foxl2 mRNAs and to ovarian development [39]. In orange-spotted grouper (Epinephelus coioides), foxl2 recombinant protein increases the expression of cyp19a1 mRNA, while foxl3 recombinant protein down-regulates the transcription of cyp19a1 but up-regulates the transcription of cyp11b, which is related to androgen synthesis [40]. In tilapia, foxl2 up-regulates the expression of cyp19a1 in vivo. In female fish, foxl2 mutation therefore results in sex reversal, decreased expression of cyp19a, and reduced serum 17ß-estradiol (E2) level [41, 42]. However, a systematic analysis of these 5 sex-related genes (amh, amhr2, gsdf, cyp19a1, and foxl2) has not yet been reported in largemouth bass.
Largemouth bass is one of the most economically valuable species in the Chinese aquaculture industry, with a production of over 600,000 tons in 2020. It presents a sexual growth dimorphism during early development, with females often living longer and reaching a larger size than males [43]. Our group recently published a draft genome assembly [44] and whole-genome resequencing [45] of largemouth bass. Several previous studies have reported that largemouth bass may have an XX/XY sex determination system [46, 47]. These basic studies are very helpful for us to study the sex-determining genes of largemouth bass.
Transcriptome is also an effective technique for identifying sex-determining genes. Several studies have in fact identified sex-related genes in various fishes such as in channel catfish [48], platyfish [49] and rainbow trout [50] through the use of transcriptome analysis. Transcriptomic comparisons could also allow prediction of new sex-related genes in largemouth bass. In the present study, we carried out detailed assessments of the sequences, gene structures, evolutionary traits, and gene expression patterns of 5 sex-related genes (amh, amhr2, gsdf, cyp19a1, and foxl2). These findings of this work would improve our understanding of the biological roles of these genes in largemouth bass. They should also contribute substantially to in-depth knowledge of sex-related genes in closely related species, such as smallmouth bass (Micropterus dolomieu).
Related protein sequences for amh, amhr2, cyp19a1,
foxl2 and gsdf from zebrafish, large yellow croaker, and
European seabass were first downloaded from the USA National Center for
Biotechnology Information (NCBI). Next, whole genome sequences of 10
representative fishes (Table 1) were downloaded from the NCBI, and the genome
dataset of Mandarin fish (Siniperca chuatsi) was downloaded from China
National Gene Bank Database (CNGBdb). Nucleotide sequences for the 5 sex-related
genes were extracted from these genomes using BLAST v.2.2.26
(http://www.ncbi.nlm.nih.gov/blast) (e-value
| Species | Database | Assembly accession | Sex | Common name |
| Collichthys lucidus | NCBI | GCA_004119915.2 | Female | Big head croaker |
| Dicentrarchus labrax | NCBI | GCA_905237075.1 | Male | European seabass |
| Lateolabrax maculatus | NCBI | GCA_004023545.1 | Female | Spotted seabass |
| Lates Calcarifer | NCBI | GCA_001640805.2 | Male | Asian Seabass |
| Oreochromis niloticus | NCBI | GCA_001858045.3 | Female | Nile tilapia |
| NCBI | GCA_013350305.1 | Female | ||
| NCBI | GCF_000188235.2 | Female | ||
| NCBI | GCA_922820385.1 | Male | ||
| NCBI | SRA: ERR7448120 | Male | ||
| Perca flavescens | NCBI | GCA_004354835.1 | Male | Yellow Perch |
| Larimichthys crocea | NCBI | GCA_900246015.1 | Female | Large yellow croaker |
| Siniperca chuatsi | CNGB | CNP0000961 | Female | Mandarin fish |
| Danio rerio | NCBI | GCA_000002035.4 | Unknown | Zebrafish |
| Micropterus dolomieu | NCBI | GCA_021292245.1 | Male | Smallmouth bass |
| Micropterus salmoides | NCBI | GCA_014851395.1 | Female | Largemouth bass |
| NCBI | GCA_019677235.1 | Male |
NCBI, the USA National Center for Biotechnology Information; CNGB, China National Gene Bank.
A phylogenetic analysis was conducted using the downloaded or extracted amino acid sequences. Multiple protein sequences were aligned using the MUSCLE v.3.7 software (Roque Moraes Drive, Mill Valley, CA, USA) [51] with default parameters. Subsequently, a phylogenetic tree was constructed using IQTREE v.1.6.12 (Center for Integrative Bioinformatics Vienna, Vienna, Austria) [52] based on the maximum likelihood (ML) method and Jones-Taylor-Thornton (JTT+G4) model, with 1000 replicates for evaluation of the branch supports. Detailed locations of upstream and downstream neighboring genes to any sex-related gene on various chromosomes were compared.
One-year-old largemouth bass (five females: body length of 26.6
Sexually mature largemouth bass were harvested for culture in aerated tanks. The general natural spawning, fertilization, and husbandry procedures were performed as previously described [53]. Approximately 15–20 fry were collected at each time-point from 13 days post-fertilization (dpf) to 56 dpf (8 time points: 13, 18, 23, 28, 33, 45, 50 and 56 dpf). Fry were euthanized with MS-222 (20–30 mg/L for 2 mins) before sample collection. Each fry was cut into two portions (head and trunk), which were immediately frozen in liquid nitrogen and then stored at –80 °C until use. Male and female samples were distinguished using published molecular markers [43, 47] that identify male-allele carrying fish at early, undifferentiated stages (Supplementary Table 1).
Total RNA was extracted from each sample (total of 72 samples, 0.5–1 cm
cDNA libraries from three female and three male gonads were constructed as per
the manufacturer’s recommendations and then sequenced on an Illumina HiSeq 4000
platform (Illumina, San Diego, CA, USA) to generate 150-bp paired-end reads. Raw reads were
filtered with SOAPnuke v.1.5.6 (BGI, Shenzhen, China) [54] to remove adaptor
sequences, those reads with
qRT-PCR was performed to validate data generated by high-throughput sequencing. Tissue-specific expression patterns for the 5 sex-related genes were compared. qRT-PCR was carried out using gene-specific primers (Supplementary Table 1) designed by routine Primer6 software (PREMIER Biosoft International, San Francisco, CA, USA).
qRT-PCR was performed on an Applied Biosystems 7300 machine (Applied Biosystems,
Foster City, CA, USA). The amplification conditions were set as follows: 95 °C for
3 min and 40 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. All
qRT-PCRs were conducted with three biological replicates of three individuals.
We studied the 5 sex-related genes in the genomes of 11 representative bony
fishes. As in most species, largemouth bass was found to contain all the 5 genes
with a single copy, whereas gene loss or duplication was found in some other
species (Figs. 1,2,3). For example, two gsdf genes were identified in
European perch (Fig. 2B), which is consistent with previous reports [61]. An
earlier study reported a Y-specific duplicate of amh (referred to as
amhy) on the Y chromosome of male Nile tilapia [62]. However, only one
amh was identified in this published male genome assembly. This
inconsistency prompted us to reassemble a haplotype-resolved genome using
recently released HiFi reads (NCBI SRA: ERR7448120) in order to validate the
amh gene (Supplementary Table 2). Interestingly, one
amh gene was identified in haplotype X, whereas two amh
(amhy and amh
 Fig. 1.
Fig. 1.Characterization of a Y-specific duplication or
insertion of the amhy gene in the male Nile tilapia genome. (A) The
phylogenetic tree and synteny comparison of amh in various fishes. The
maximum likelihood (ML) tree (left panel) was constructed by IQTREE. Bootstrap values are shown on
branches. The right panel presents the synteny of the amh gene in each
fish species. (B) A synteny comparison between X and Y contigs. Note the
Y-specific insertion at the 10-kb region. (C) ClustalW alignments of
amh, amhy and amh
 Fig. 2.
Fig. 2.Comparison of gsdf among various fishes. (A) Phylogenetic tree (left) and synteny comparison (right) of gsdf. (B,C) ClustalW alignments of gsdf proteins in European perch and Nile tilapia. Abbreviations: aff1, AF4/FMR2 family member 1; cdk5rap2, CDK5 regulatory subunit associated protein 2; gtase9, beta-1,3-galactosyltransferase 9; klhl8, kelch-like protein 8; mmr1, macrophage mannose receptor 1; nup54, nucleoporin p54; ppef2, serine/threonine-protein phosphatase with EF-hands 2; ptges, prostaglandin E synthase; usp20, ubiquitin specific peptidase 20; nup188, nucleoporin 188.
 Fig. 3.
Fig. 3.Phylogenetic trees and synteny comparisons of amhr2 (A), cyp19a1 (B) and foxl2 (C) in various fishes. Abbreviations: cdcp7, cell division cycle-associated protein 7; copb, coatomer subunit beta; dmxl2, dmX-like protein 2; gldn, gliomedin; mp3k12, Mitogen-activated protein kinase kinase kinase 12; pcbp2, Poly(rC)-binding protein 2; pcph2, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta; pik3cb, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta; scg3, secretogranin-3; mrps22, 28S ribosomal protein S22; sp1, transcription factor Sp1; tnfaip8l3, tumor necrosis factor alpha-induced protein 8-like protein 3.
To elucidate the evolutionary history of each sex-related gene, we characterized and compared adjacent genomic regions of each gene locus in representative fishes. This synteny analysis indicated that amh, amhr2, cyp19a1, gsdf and foxl2 are conserved throughout the fish tree of life, with only slight modifications of upstream and downstream neighboring genes (Figs. 1,2,3).
The neighbors for amh are peak1 and oaz1 (upstream),
and dot1l and ell (downstream) (Fig. 1A). In Nile tilapia, the
synteny comparison of X and Y chromosomes confirmed a 10-kb Y-specific insertion
within the downstream region of amhy. Annotation of this insertion
revealed a single gene that is a duplicate copy of amhy (termed
amh
Most species have only one gsdf gene in their genomes (Fig. 2), although the European perch genome contains two gsdf genes located on the same chromosome (CAJNNU010000010.1) and sharing 85.71% sequence similarity, but with different neighboring genes (Fig. 2B). Interestingly, in Nile tilapia the protein sequence of gsdf (Fig. 2C) differs between the multiple male and female genomes (three females and two males; Table 1), suggesting its potential participation in sex determination. Compared to other species, the genus Micropterus (including largemouth bass and its relative smallmouth bass) had less conserved upstream and downstream regions around the gsdf gene, with mmr1 located on other chromosomes (Fig. 2A).
As a receptor for amh, amhr2 plays an important role in sex determination and sex differentiation [64]. A Y-specific insertion of the amhr2 gene (amhr2by) has been identified in yellow perch and is thought to be a candidate sex-determining gene [33]. As in zebrafish, amhr2 is lost in other cyprinid fishes [32], possibly due to chromosome rearrangement during fish evolution (Fig. 3A). Synteny analyses indicate that both cyp19a1 and foxl2 are conserved in the examined species (Fig. 3B,C). However, the foxl2 neighboring genes are less conserved in zebrafish, implying that it is more distantly related to other fish species.
qRT-PCR was performed to characterize the transcription patterns of amh, amhr2, gsdf, cyp19a1, and foxl2 in the liver, muscle, brain, and gonad (testis or ovary) tissues of mature individuals. All genes showed relatively higher transcription levels in the gonad, while low levels or no expression in the other tissues (Fig. 4). This result suggests that these genes are involved in gonad development and maintenance of gonad stability.
 Fig. 4.
Fig. 4.Relative transcription of the five sex-related genes in
different tissues of adult largemouth bass. Data were normalized to
ß-actin. Values are expressed as relative gene expression (mean
amh, amhr2 and gsdf were predominantly transcribed in the testis and less in the ovary, whereas cyp19a1 and foxl2 were predominantly expressed in the ovary. The transcription levels of cyp19a1 and foxl2 were elevated during gonadal development, with much higher transcription levels observed in female (XX) gonads compared to male (XY) gonads at each of the 8 developmental stages and in the mature gonads. The transcription levels of amh, amhr2 and gsdf were elevated during gonadal development, with much higher levels in XY gonads than XX gonads at the 8 developmental stages and in the mature gonads (Fig. 5). The 5 sex-related genes showed significant differences in expression between males and females, especially at 56 dpf, suggesting this is a critical time-point for the initiation of sex differentiation. In adult gonads, the relative transcription of the 5 sex-related genes (Fig. 4) was consistent with the transcriptome data (Table 2).
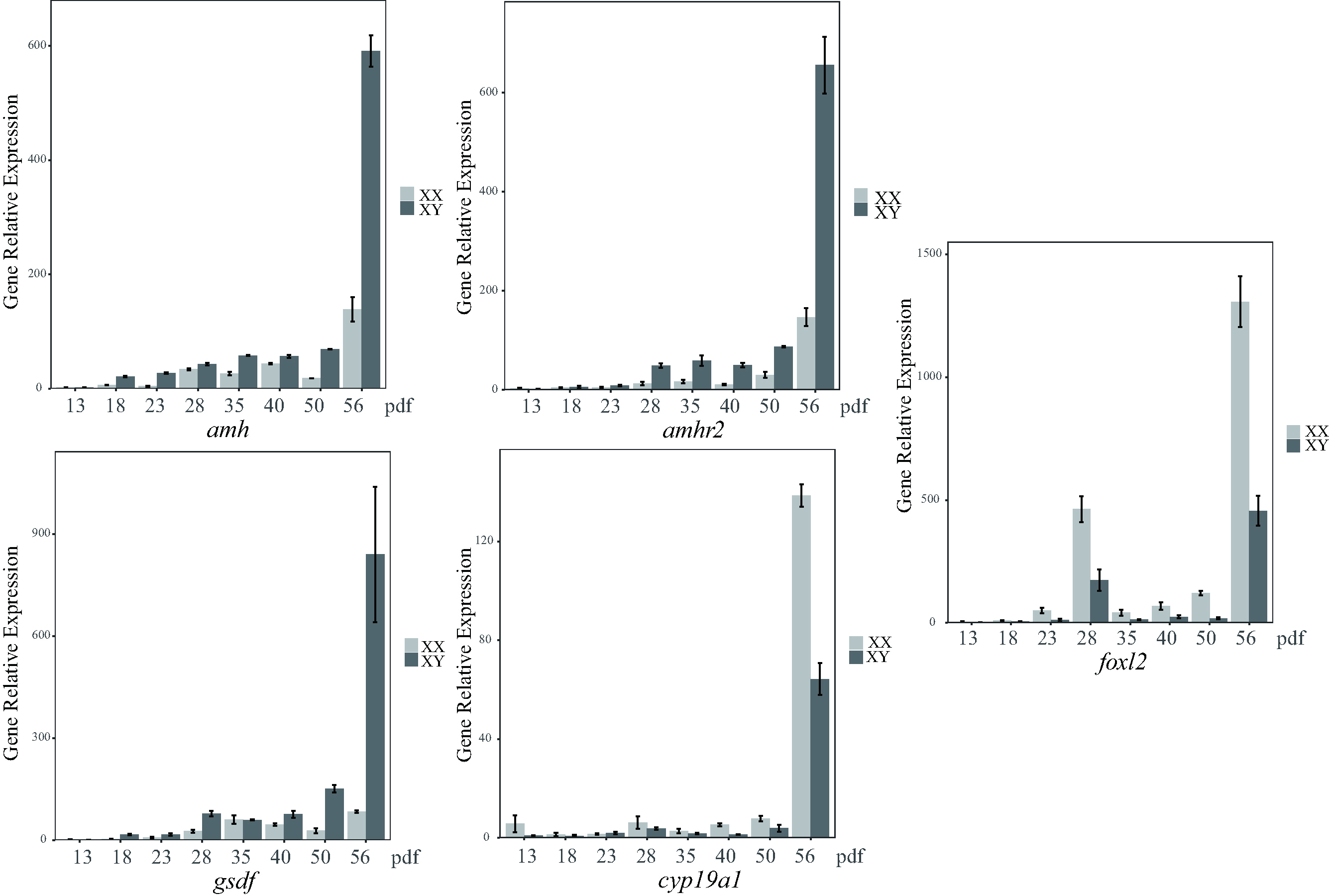 Fig. 5.
Fig. 5.Relative transcription levels of the five sex-related genes at
eight different developmental stages in the gonads of largemouth bass. Data were
normalized to ß-actin. Values are expressed as relative gene
expression (mean
| Gene | Female 1 | Female 2 | Female 3 | Male 1 | Male 2 | Male 3 | log |
| amh | 2.95 | 3.23 | 2.65 | 90.08 | 125.77 | 140.61 | 4.51 |
| amhr2 | 0.08 | 0.14 | 0.12 | 42.72 | 44.12 | 39.5 | 8.75 |
| gsdf | 14.27 | 7.68 | 4.33 | 2002.02 | 2679.3 | 2934.29 | 7.34 |
| cyp19a1 | 0.25 | 0.23 | 0.34 | 0.05 | 0.10 | 0.03 | –2.03 |
| foxl2 | 5.67 | 4.23 | 6.78 | 0.04 | 0.13 | 0.04 | –6.04 |
FPKM, fragments per kilobase of transcript per million mapped reads; FC, fold change.
A differential expression analysis was performed between testes and ovaries to screen for other sex-related candidate genes. A total of 15,962 differentially expressed genes (DEGs) were predicted. The testis showed 9072 up-regulated DEGs and 6890 down-regulated DEGs compared to the ovary (Supplementary Figs. 1,2). To identify genes potentially associated with sexual differentiation and gonadal development, we performed sex-related GO annotation and KEGG pathway analysis of these DEGs. This analysis revealed 7656 DEGs, of which 3636 were matched to 264 KEGG subcategories and to 52 GO terms (Supplementary Fig. 3). Many DEGs were predicted to be associated with sex-related GO terms, such as sex differentiation, gamete generation, acrosome assembly, fertilization, and spermatogenesis (Table 3). The transcription of many spermatogenesis-related DEGs, such as sperm surface protein Sp17 (sp17) and spermatogenesis-associated protein 22 (spata22), was significantly upregulated in the testis, whereas the transcription of some oogenesis-related genes, such as forkhead box protein N5 (foxn5) and bone morphogenetic protein 15 (bmp15), was markedly upregulated in the ovary.
| GO term | Annotation | Gene name | Log |
Sex biased |
| Gamete generation (GO: 0007276) | progesterone receptor | pgr | 5.31 | testis |
| membrane-associated progesterone receptor | mpgr | 1.07 | testis | |
| deleted in azoospermia like | dazl | 4.71 | testis | |
| Fertilization (GO: 0009566) | Sperm surface protein Sp17 | sp17 | 3.42 | testis |
| sperm acrosome membrane-associated protein 4 | samp4 | –15.5 | ovary | |
| Sex differentiation (GO: 0007548) | sex determining region Y box 9 protein | sox9 | 1.75 | testis |
| follistatin related protein | fsrp | 4.35 | testis | |
| doublesex and mab-3-related transcription factor 1 | dmrt1 | 12.76 | testis | |
| bone morphogenetic protein 15 | bmp15 | –6.67 | ovary | |
| doublesex- and mab-3-related transcription factor B1 | dmrtb1 | 14.20 | testis | |
| forkhead box protein N5 | foxn5 | –13.16 | ovary | |
| Spermatogenesis (GO:0007283) | kelch-like protein 10 | klhl10 | 8.92 | testis |
| spermatogenesis-associated protein 22 | spata22 | 4.23 | testis | |
| cilia- and flagella-associated protein 73 | cfap73 | 8.76 | testis | |
| coiled-coil domain-containing protein 103 | ccdc103 | 6.54 | testis | |
| meiosis expressed gene 1 protein homolog | meig1 | 7.08 | testis | |
| Acrosome assembly (GO: 0001675) | zona pellucida sperm-binding protein 3 | zp3 | –12.02 | ovary |
DEGs, differentially expressed genes; GO, Gene Ontology.
The DEGs enriched in the sex-related KEGG pathways included the TGF-beta signaling pathway (ko04350), steroid hormone biosynthesis (ko00140), FoxO signaling pathway (ko04068), steroid biosynthesis (ko00100), estrogen signaling pathway (ko04915), Wnt signaling pathway (ko04310), and ovarian steroidogenesis (ko04913) (Table 4). Interestingly, the transcription levels of five cyp members (cyp2k1, cyp1a1, cyp2u1, cyp2j2 and cyp2f2) were significantly upregulated in the ovary, whereas the transcription levels of cyp20a1 and cyp27c1 were significantly upregulated in the testis. Four bmp members (bmp1a, bmp7, mbp8a and bmp15) were preferentially expressed in the ovary.
| KEGG pathway | Annotation | Gene name | log |
Sex biased |
| TGF-beta signaling pathway | bone morphogenetic protein 7 | bmp7 | –9.01 | ovary |
| bone morphogenetic protein receptor type-1A | bmpr1a | –1.85 | ovary | |
| bone morphogenetic protein 8A | bmp8a | –12.6 | ovary | |
| bone morphogenetic protein 15 | bmp15 | –6.67 | ovary | |
| anti-Mullerian hormone | amh | 4.51 | testis | |
| follistatin related protein | fsrp | 4.35 | testis | |
| follistatin-related protein 3 | fsrp3 | 5.56 | testis | |
| transforming growth factor beta-2 | tgfb2 | 5.53 | testis | |
| FoxO signaling pathway | insulin receptor | ir | 2.77 | testis |
| growth differentiation factor 9 | gdf9 | –6.51 | ovary | |
| epidermal growth factor-like protein 7 | egfl7 | 3.8 | testis | |
| transcriptional regulator Myc-B-like | mycbl | –4.07 | ovary | |
| transcription factor Sp1 | sp1 | –1.68 | ovary | |
| Estrogen signaling pathway | heat shock cognate 71 kDa protein | hsc71 | –1.64 | ovary |
| SHC-transforming protein 2 | shc1 | –11.4 | ovary | |
| Ovarian steroidogenesis | scavenger receptor class B member 1 | scarb1 | –6.05 | ovary |
| cytochrome P450 2K1 | cyp2k1 | –8.89 | ovary | |
| estradiol 17-beta-dehydrogenase 1 | hsd17b1 | –5.67 | ovary | |
| cytochrome P450 1A1 | cyp1a1 | –3.5 | ovary | |
| cytochrome P450 2U1 | cyp2u1 | –7.25 | ovary | |
| cytochrome P450 2J2 | cyp2j2 | –2.89 | ovary | |
| cytochrome P450 2F2 | cyp2f2 | –4.0 | ovary | |
| steroidogenic acute regulatory protein | star | 5.67 | testis | |
| insulin-like growth factor 1 | igf1 | 6.79 | testis | |
| cytochrome P450 20A1 | cyp20a1 | 1.5 | testis | |
| cytochrome P450 27C1 | cyp27c1 | 4.33 | testis | |
| Steroid hormone biosynthesis | steroid 11-beta-hydroxylase | cyp11b | 13.29 | testis |
| steroid 17-alpha-hydroxylase/17,20 | cyp17a1 | 7.42 | testis | |
| estradiol 17-beta-dehydrogenase 8 | 17 |
2.17 | testis | |
| Wnt signaling pathway | frizzled-3 | fzd3 | 4.3 | testis |
| frizzled-6 | fzd6 | 6.5 | testis | |
| frizzled-7 | fzd7 | 3.1 | testis | |
| transcription factor 7 | tcf7 | 15.3 | testis |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Most of the master sex-determining genes or candidate genes discovered recently in various fishes belong to the TGF-beta signaling pathway. This pathway appears to play a critical role in the process of sex determination and sex differentiation in diverse fishes. For example, amh is the sex-determining gene for three-spined stickleback (Gasterosteus aculeatus), mandarin fish and Black rockfish (Sebastes schlegelii) [65, 66, 67]. gsdf for sablefish [68], and bmpr1bby for Atlantic herring (Clupea harengus) [69]. TGF-beta signaling pathway may also be able to influence sex determination by inhibiting aromatase activity [70]. Knockdown of the sex determination gene amhy in male Patagonian silverside can activate the expression of downstream foxl2 and cyp19a1a. These are important genes in the female signaling pathway and can lead to increased estrogen synthesis, thereby causing sexual reversal from male to female [39]. However, the detailed mechanisms involved in sex determination, sex differentiation, and gonadal development in largemouth bass have thus far remained unclear.
In our current study, evolutionary and collinear analyses revealed that the 5 sex-related genes (amh, anhr2, cyp19a1, gsdf and foxl2) were generally conserved in 11 representative teleost fishes. However, some genes were found to be duplicated (amh, amhr2 and gsdf), mutated (gsdf) or lost (amhr2) in several fish genomes (Figs. 1,2,3). In certain Perciformes species, some of these genes (amh, amhr2 and gsdf) were reported to be replicated or altered to become the master sex-determining genes [67]. Therefore, most sex-determining genes may be derived from their orthologous genes (usually sex-related genes) by altering the structures of encoded proteins.
In male Nile tilapia, for example, an extra amh
Compared with other fishes, largemouth bass and smallmouth bass are conservative in that they contain all five sex-related genes studied here, without duplication, loss, or differences in gene features between males and females. Nevertheless, having no differences at the gene level does not rule out that they are sex-related, since many studies have reported that differences at the expression level can also determine a fish’s sex. For example, the sex-determining gene in channel catfish, breast cancer anti-resistance 1 (bcar1), shows similar male and female genome sequences. However, a comparative transcriptome analysis revealed that sex-specific isoform expression through alternative splicing may underlie the sex determination processes [71]. Based on our comparative transcriptome data of gonads from adult largemouth bass, we found no new alternative splicing transcripts for the five sex-related genes, suggesting that their transcript sequences were also conserved in the mature gonads. Interestingly, the sex-determining region (~1.7 Mb) in largemouth bass was previously reported on Chr7. However, this large non-recombinant region of the Chr7 does not contain any obviously potential master genes for sex determination [43]. In our current study, the 5 sex-related genes were distributed on different chromosomes of the largemouth bass genome (i.e., not just on the Chr7). This finding suggests that the identification of sex-determining genes in largemouth bass remains difficult.
To further investigate the 5 sex-related genes, we conducted expression analyses of adult tissues and gonads at different developmental stages using transcriptome sequencing and qRT-PCR. The 5 sex-related genes displayed significant differences in transcription between the ovary and testis at different developmental stages and in adult gonads (Figs. 4,5). The gsdf gene in particular exhibited the highest transcription level in males (XY) (Table 2 and Fig. 4), consistent with those findings reported in Nile tilapia [32]. Overexpression of gsdf in the developing gonads of medaka and Nile tilapia converted XX individuals into functional males [72, 73]. Furthermore, knockout of gsdf in XY fish resulted in male-to-female sex reversal [37, 74], suggesting that gsdf could be involved in male sex differentiation, sex maintenance, and testis development in medaka and Nile tilapia. Both amh and amhr2 exhibited higher transcription levels in the testis than the ovary, implying that these genes may play a role in testis development in largemouth bass.
During the natural sex reversal process of ricefield eel (Monopterus albus), increased expression of amh and decreased expression of dosage-sensitive sex reversal (dax1) in the ovaries are important for the activation of testis development. High expression of amh and low expression of dax1 are necessary for the maintenance of testis functions [75]. In contrast, cyp19a1 and foxl2 are expressed at higher levels in the ovary than in the testis. In olive flounder (Paralichthys olivaceus), foxl2 may play an important role in ovarian differentiation by maintaining cyp19a1 expression and antagonizing the expression of dmrt1 [76]. These results indicate that foxl2 and cyp19a1 promote ovarian development and male-related gene expression. In largemouth bass, the expression of the 5 sex-related genes was significantly different between adult male and female gonads, and during gonadal development. amh, amhr2 and gsdf were predominantly expressed in the testis, whereas cyp19a1 and foxl2 were mainly transcribed in the ovary. These differences indicate the genes may have important roles in the process of sex differentiation and the sex stability of largemouth bass. At 86 dpf in Senegalese sole (Solea senegalensis), the follicle stimulating hormone receptor (fshr) can activate other gonadal marker genes (such as amh and cyp19a1) to promote gonad differentiation [77]. In the present study, amh, amhr2, gsdf, cyp19a1 and foxl2 were significantly expressed in the testes and ovaries on 56 dpf. We therefore speculate that differentiation between males and females in largemouth bass starts at about the 50th day after fertilization.
In the present study, we also performed comprehensive transcriptomic analyses of the testis and ovary in largemouth bass. Differential expression analysis allowed us to identify multiple sex-biased genes that may be associated with steroidogenic hormones, ovarian steroidogenesis, and sex differentiation. In particular, several additional members of the TGF-beta signaling pathway were identified that showed significantly different expression between the testis and ovary. These include bmp7, bmpr1a, bmp8a, fsrp, fsrp3, tgfb2, ir, gdf9, SMAD family member 1 (smad1), smad2, smad6b, smad7, inhibitor of DNA binding 3 (id3), repulsive guidance molecule BMP co-receptor a (rgma), and activin A receptor type 1 (acvr1). A comparative transcriptome analysis of yellowfin bream (Acanthopagrus latus) identified several genes in the TGF-beta signaling pathway that were differentially expressed, including bmp1-8, smad1-8, follistatin and transcription factor e2f4 [78]. id2 and id3 are members of the dominant-negative, basic helix-loop-helix transcription factor family. The id2bbY gene is a duplicated copy of the autosome id2b gene on the Y chromosome, which has been reported as a sex-determining gene in arapaima [4]. Gonadal transcriptome analyses of Channa argus and C. maculate showed that the transcription level of id2 in the testis was significantly lower than that in the ovary [79]. The id3 expression level was also significantly different between the male and female gonads of largemouth bass, suggesting that it may be critical for gonad development in this fish.
Dmrt and the transcription factor Sox (sox) are well-known sex-related gene families in various animals. Dmrt1 plays an important role in sex determination, male germ cell differentiation, and sex maintenance in diverse mammals, birds, and fishes [42, 80, 81, 82]. Moreover, dmrt2-5 are important for testis development in many fish species [83, 84, 85]. Our results showed that dmrt1, dmrtb1, dmrta1, dmrt2 and dmrt3 were highly expressed in largemouth bass testis (Table 4). The sox family has multiple roles in several biological processes, including gonad development and sex determination. sox3 is a sex-determining gene in Indian ricefish (Oryzias dancena) [72], and sox9 cooperates with sox8 to protect the adult testis from male-to-female genetic reprogramming and complete degeneration [86, 87]. High expression levels of sox2, sox5, sox7, sox9a and sox10 in the testis, and of sox3, sox11 and sox19b in the ovary suggests that sox genes may have complex roles in the sex differentiation of largemouth bass. Interestingly, similar results were reported in yellowfin bream [88].
This study characterized 5 sex-related genes (including amh, amhr2, cyp19a1, foxl2 and gsdf) in largemouth bass and in 11 other teleost representatives, including their transcription profiles. Phylogenetic and comparative genomics analyses provided additional evidence for validating the orthologs of these genes. Transcriptome sequencing and qRT-PCR revealed distinct transcription levels for these sex-related genes in various tissues of largemouth bass and at different stages of development. In summary, this study provides a comprehensive overview of these sex-related genes in largemouth bass. These interesting data establishes a basic foundation for in-depth functional analyses of these genes in relation to sex determination and sex differentiation.
The transcriptome raw reads were deposited in NCBI BioProject database under accession number PRJNA973811.
QS and XYe conceived and designed the research. XZ, CS, ZR, YH, XL, HL and XYou performed data analyses. XZ, ZR, CH, JX, XW and CS prepared samples and data. XZ and CS wrote the manuscript. QS, XYe, XL, HL, YH and XYou revised the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
All the experiments on fishes were conducted in accordance with the specific guidelines on the care and use of animals for scientific purposes as outlined by the Institutional Animal Care and Use Committee (IACUC) of the Pearl River Fisheries Institute, Chinese Academy of Fishery Sciences (CAFS), China. The IACUC approved this study under the CAFS project “Breeding of LMB-2019”.
We thank the anonymous reviewers and editors for their careful editing and helpful comments on this manuscript.
The research was supported by National Key Research and Development Program of China (no. 2022YFE0139700), and Special Program for Science Technology Innovation and Industrial Development of Shenzhen Dapeng New District (nos. KJYF202101-02 and KJYF202101-13).
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
