1 School of Ecological Technology and Engineering, Shanghai Institute of Technology (SIT), 201418 Shanghai, China
2 Shanghai Key Laboratory of Protected Horticultural Technology, Forestry and Fruit Tree Research Institute, Shanghai Academy of Agricultural Sciences (SAAS), 201403 Shanghai, China
3 Department of Plant Physiology, Institute for Biological Research “Siniša Stanković” - National Institute of Republic of Serbia, University of Belgrade, 11060 Belgrade, Serbia
Abstract
Background: Mutant analysis remains one of the main genetic tools for
characterising unclarified gene functions in plants, especially in non-model
plants. Daylily (Hemerocallis spp.) is a popular perennial ornamental
plant grown worldwide. Analysis of daylily mutants can enhance understanding of
genes regulating the albino phenotype and improve the cultivar quality of
daylily. Methods: The natural albino mutant (Alb
Keywords
- carotenoid
- chlorophyll
- daylily (Hemerocallis spp.)
- drought tolerance
- natural albino mutant
- light saturation point
- RNA sequencing
Photosynthesis converts solar energy into chemical energy and relies on
photosynthetic pigments that absorb light energy. Albino mutations in plants
affect photosynthesis rate by reducing the content of chlorophyll and
carotenoids, which are the main photosynthetic pigments. Chlorophyll deficiency
is generally caused by the mutations of genes encoding enzymes that catalyse
biochemical reactions involved in chlorophyll metabolism or genes responsible for
changes in chloroplast ultrastructure [1, 2, 3]. Carotenoids belong to a large
pigment group and are categorised into carotenes, like
Daylilies (Hemerocallis spp.) are popular perennial ornamental plants. Most daylily cultivars are developed by sexual hybridisation [30], and their population amplification depends on division propagation. The American Daylily Association (https://www.daylilies.org/DaylilyDB/) has collected approximately 90,000 daylily cultivars. Mutant analysis is a widely used tool for characterising gene functions in plants including non-model plants, such as daylily, and great progress has been made in cloning genes from plant mutants in recent decades. Several studies have reported gene cloning using daylily [31, 32, 33, 34, 35, 36, 37]. Mapping-based cloning is an effective strategy for identifying causal mutations responsible for variations in a trait. In addition to mapping-based cloning, microarray [38], RNA sequencing (RNA-seq) [39], and mapping by sequencing [18, 40] are used in analysing the gene functions of mutants. Giving that the vegetative growth period of daylily is longer than 2 years, mapping-based cloning is not an ideal method for analysing the natural mutants of daylily. Thus, RNA-seq was performed to compare gene expression between wild-type and albino mutant plants of daylily in the present study.
To understand the genetic and physiological mechanisms underlying daylily albino mutations, we performed its functional characterization and determined the most likely genetic and physiological causes. Moreover, impacts on the physiological functions of the plant, such as photosynthesis, metabolism and tolerance to drought, were explored.
Hemerocallis Middebdorffii Trautv. & C. A. Mey and
Hemerocallis ‘black-eyed stella’ and ‘stella de oro’ were cultured in
experimental fields (Shanghai Institute of Technology, N: 30
The sections of daylily leaf tissue from albino mutants and wild-type plants were observed and recorded under a light microscope (ECLIPSE E200, NIKON, Japan).
Transmission electron microscopy analysis was performed according to the methods
described by Hashimoto et al. [42] with some modifications. In detail,
the leaf tissues of mutant and wild-type plant leaves were sampled, fixed and
pumped with 4% glutaraldehyde at 4 °C, stored at 4 °C
overnight and washed three times with 0.1 mol
The contents of chlorophyll and carotenoids were determined according to the methods described by Chen [43] with some modifications. The leaf samples of 20d daylily seedlings were ground using a pestle and mortar. Chlorophyll and carotenoids were extracted using a prepared extraction solution containing ethanol, acetone, and water in a ratio of 5:4:1 (v/v/v). The absorbance was measured by a microplate reader (Spark readers, TECAN, Swiss) at wavelengths of 470, 645 and 663 nm. The content of total chlorophyll, chlorophyll a (Chla), chlorophyll b (Chlb) and carotenoids was calculated according to the following formula:
The content of 5-aminolevulinic acid (ALA) was determined according to the
methods described by Dei [44] with some modifications [45]. Leaf
samples of 12-day-old daylily seedlings were ground and ALA was extracted by 4%
trichloroacetic acid (m/v). Approximately 5 mL of extraction solution, 2.35 mL of
sodium acetate (1 mol
The content of porphobilinogen (PBG) was determined according to the methods
described by Peng et al. [46] with some modifications. Leaf samples from
12 d old daylily seedlings were ground, and PBG was extracted by buffer
containing 0.1 mol
The contents of uroporphyrinogen III (urogen Ⅲ) and coproporphyrinogen III
(coprogen Ⅲ) were determined according to the methods described by Bogorad
et al. [47] with some modifications. Leaf samples of 12 d old daylily
seedlings were ground, and PBG was extracted by 0.067 mol
The content of protoporphyrin IX (proto Ⅸ), Mg-protoporphyrin IX (Mg-proto Ⅸ),
and pchlide were determined according to the methods described by Hodgins
et al. [48] with some modifications [46]. Leaf samples from 12 d old
daylily seedlings were ground after the addition of 25 mL of 80% alkaline
acetone. The extraction was centrifuged at 15000
Photosynthetic parameters were determined according to the methods described by
Iqbal et al. [45] with some modifications [49]. A portable
photosynthesis tester (CIRAS-3, PP SYSTEMS, USA) was used to determine
photosynthetic parameters. The fifth leaves from apical meristem of daylily were
selected for measurement in September and October 2022 (10:00–12:00 am).
Photosynthetic parameters including net photosynthetic rate (Pn), water use
efficiency (WUE), transpiration rate (Tr) and intercellular CO
The relative water content of the leaves was determined according to the methods described by Chen et al. [50] with some modifications. Daylily leaves with the same physiological status were treated under drought conditions, and weighed. Fresh weight was recorded as the initial weight. The saturated weights (Wt) were recorded after the samples being placed in distilled water for 2 hours to absorb water. After that, the dry weights (Wd) were recorded after the samples being dried at 80 °C for 48 h. The water content (RWC) was calculated according to the following formula:
Soil moisture content was determined according to the methods described by Wang
et al. [51] with some modifications. The wet weight (M1) of the soil was
acquired by randomly weighing the soil from three different positions in the pot.
Dry weight (M2) was recorded after the samples being dried at 80 °C for
48 h. The soil moisture (W) content was calculated according to the following
formula: (M1 – M2)/M2
Leaf samples from 12 d old albino mutant and wild-type plants were ground, and five sets of biological replicates of each sample were randomly sampled. Three sets with the good-quality samples were obtained for analysis. Total RNA was extracted after digestion by DNase. The mRNA was enriched by magnetic beads with oligo dT. Used mirVana™ miRNA ISOlation Kit, Ambion-1561 kit and Refrigerated centrifuge (ST16R, Thermo, USA), Gel imaging system (Tanon 2500, Biotanon Co., Ltd., Shanghai, China), UV spectrophotometer (NanoDrop 2000, Thermo, USA).
cDNA libraries were constructed using a VAHTS Universal V6 RNA-seq Library Prep Kit for Illumina sequencing according to the manufacturer’s instructions. The libraries were finally sequenced using an Illumina Novaseq 6000 platform. A total of 150 bp paired-end reads were generated. The transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China). Oligonucleotide adapters were removed using Trimmomatic software (version 0.36) [52], and clean reads were obtained after low quality bases and N bases were filtering out. Clean reads were then spliced through the paired-end method performed using Trinity software (version 2.4) to obtain transcript sequences [53]. The most extended sequence was selected as a unigene. The unigenes were annotated to the NR, COG/KOG, and Swissprot databases. GO annotation was acquired by mapping Swissprot ID to the GO term, and pathway information was acquired by comparing unigenes with the KEGG database [54].
Fragments per kilobase of exon model per million mapped fragments (FPKM) and unigene count were analysed in bowtie2 (version 2.3.3.1) [55] and eXpress software (version 1.5.1) [56]. The number of unigene reads in the samples were acquired by eXpress software. All the data were standardised by estimating SizeFactors, a function of DESeq package (version 1.18.0) [57]. The p-value and foldchange of the difference in gene expression between mutant and wild-type plants were calculated using nbinomTest software. The unigenes with difference value greater than 2 and p value less than 0.05 were selected for further analysis.
RNA-seq analysis was performed using Bioedit, NCBI, and Tair (https://www.arabidopsis.org/).
Six differentially expressed genes (DEGs) from RNA-seq data related to terpene,
carotenoids, and chlorophyll metabolism were randomly selected for quantitative
real-time PCR (qPCR) validation. Primers were designed using premier5 software
(Supplementary Table 1). The Prime ScriptTM RT reagent kit with gDNA
Eraser (TaKaRa) was applied to synthesise cDNA. qPCR was then performed according
to the methods described by Zuo [58]. HfUBQ and HfEF-1a were
used as internal reference genes [59]. The 2
Two-week-old sterile cultured self-pollinated progeny of daylily ‘Black-eyed stella’ were transplanted to a medium containing half-strength MS salts, 3% sucrose (v/v), and 10% PEG2000 (v/v) [60]. One-year-old daylily plants, which were cultured in pot with soil, were left unwatered for two weeks, whereas the control plants were regularly watered every two days. Relative soil water content was determined at the same time.
At least three independent biological replicates were performed for each experiment. Statistical analysis was performed with Excel 2010 and SPSS26. Figures were made using Origin8 software.
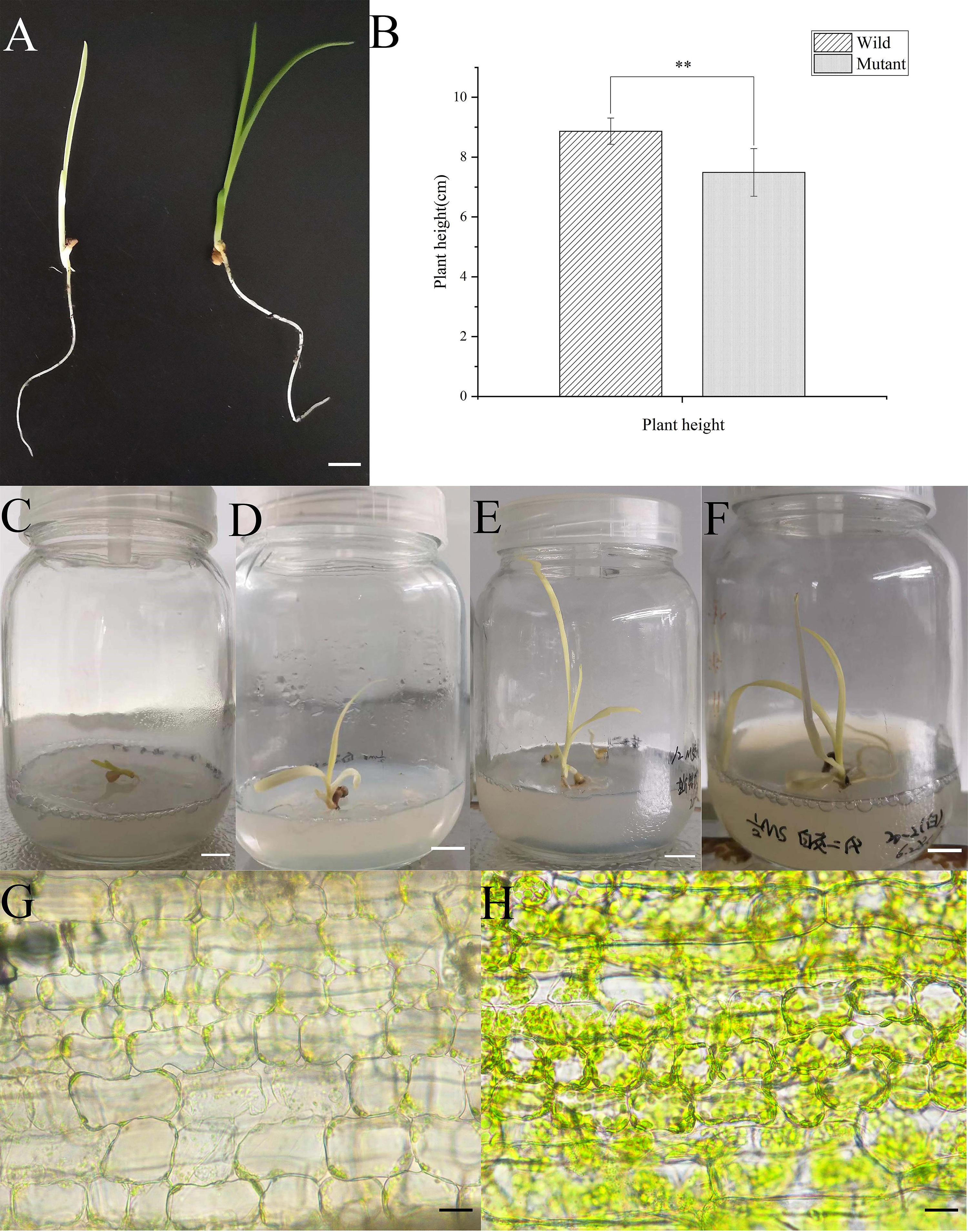
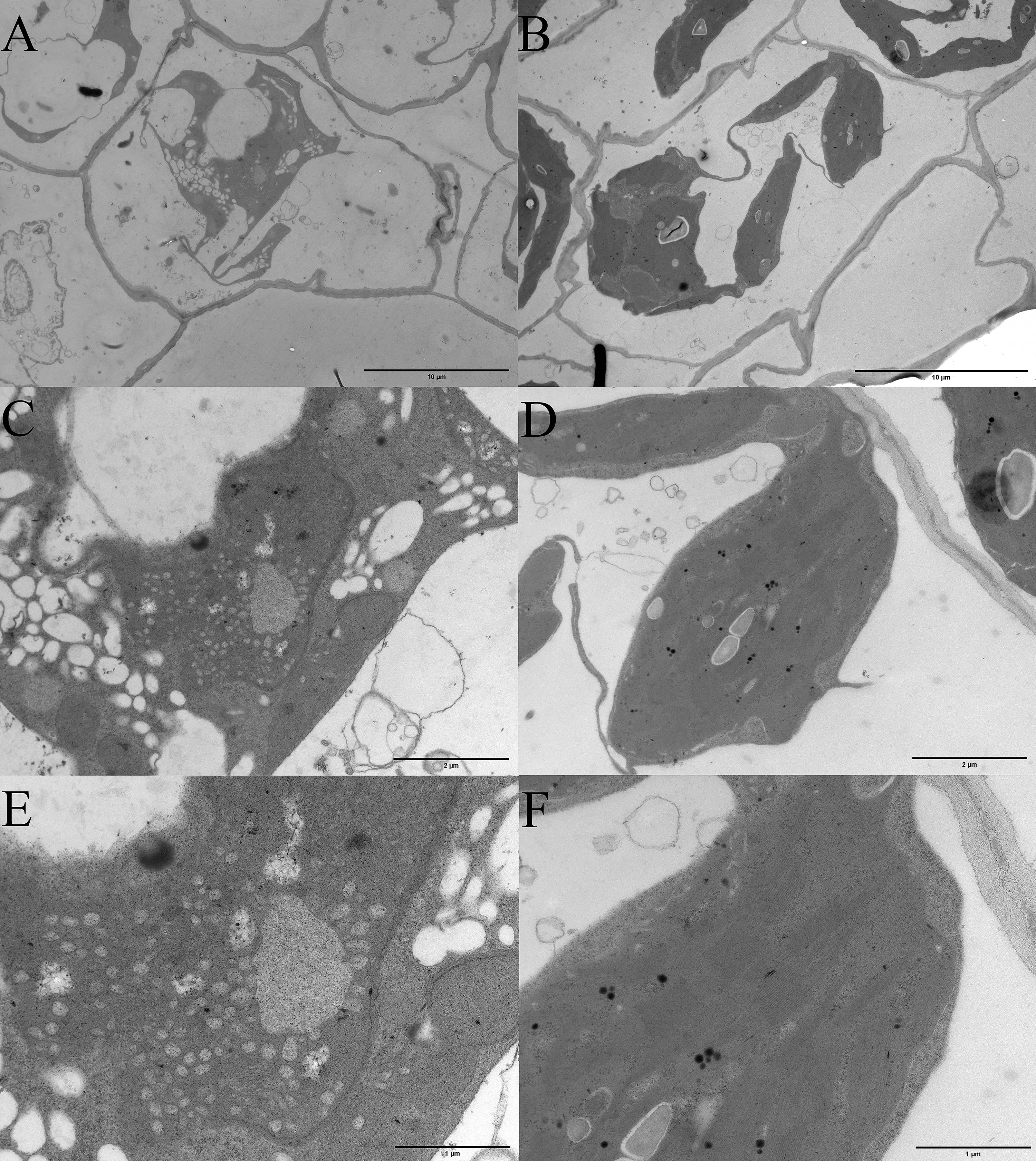
Albino seedlings were observed during the selection and development of new varieties from self-pollinated progeny of daylily ‘Black-eyed stella’ and one of its parents, ‘Stella de oro’ in May 2020. The leaf colour of the albino seedlings was light yellow (Fig. 1A). The albino seedlings displayed a dwarf phenotype (Fig. 1B), and survived for less than 15 days in soil. However, these seedlings survived longer when grown on the medium containing half MS salts and 3% sucrose under sterile culture conditions (Fig. 1C–F). The chlorophyll and carotenoid content of albino seedlings was determined and compared with those of wild-type plants. The results showed that the content of chlorophyll a (Chla), chlorophyll b (Chlb) and carotenoids decreased significantly compared with that of the wild-type plants (Supplementary Fig. 1). Albino phenotypes are generally attributed to changes in the metabolism of photosynthetic pigments and chloroplast development. Therefore, a microscopic analysis was performed to verify whether the chloroplast development of the mutant plants was altered. Extremely few chloroplasts were found in the albino mutant leaf cells (Fig. 1G,H). To better understand chloroplast development, transmission electron microscopy was used for investigating the morphology of mesophyll plastids. Both the number of thylakoids and the extent of stacking in the grana were significantly reduced in albino mutant plants (Fig. 2).
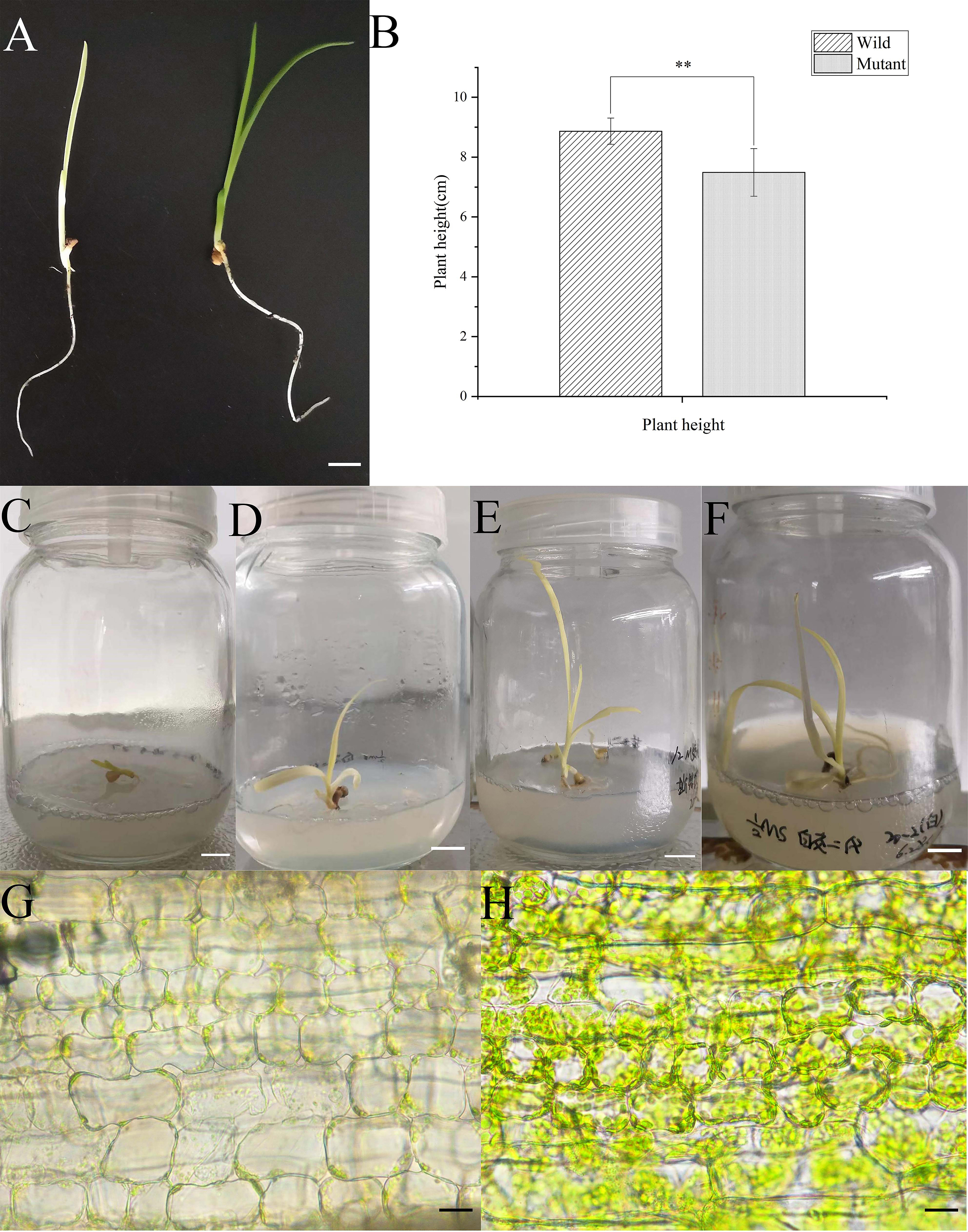 Fig. 1.
Fig. 1.Comparison of the morphological and characterization of the
plastid between albino and wild-type seedlings of daylily. (A) Seedlings of 15 d
old wild-type (right) and albino mutant (left) seedlings cultured in the soil.
(B) Height of seedlings of 15 d wild-type (left) and albino mutant (right)
seedlings cultured in the soil. (C–E) Seedlings of albino mutant culture on the
media containing half MS salts and sucrose under sterile conditions. (G,H) Light
microscopy of the chloroplast of the leaf cells of albino mutant (G) and
wild-type seedlings (H). The bars in (A–F) represent 1 centimeter, and in (G,H)
represent 20 micrometers. Mean values
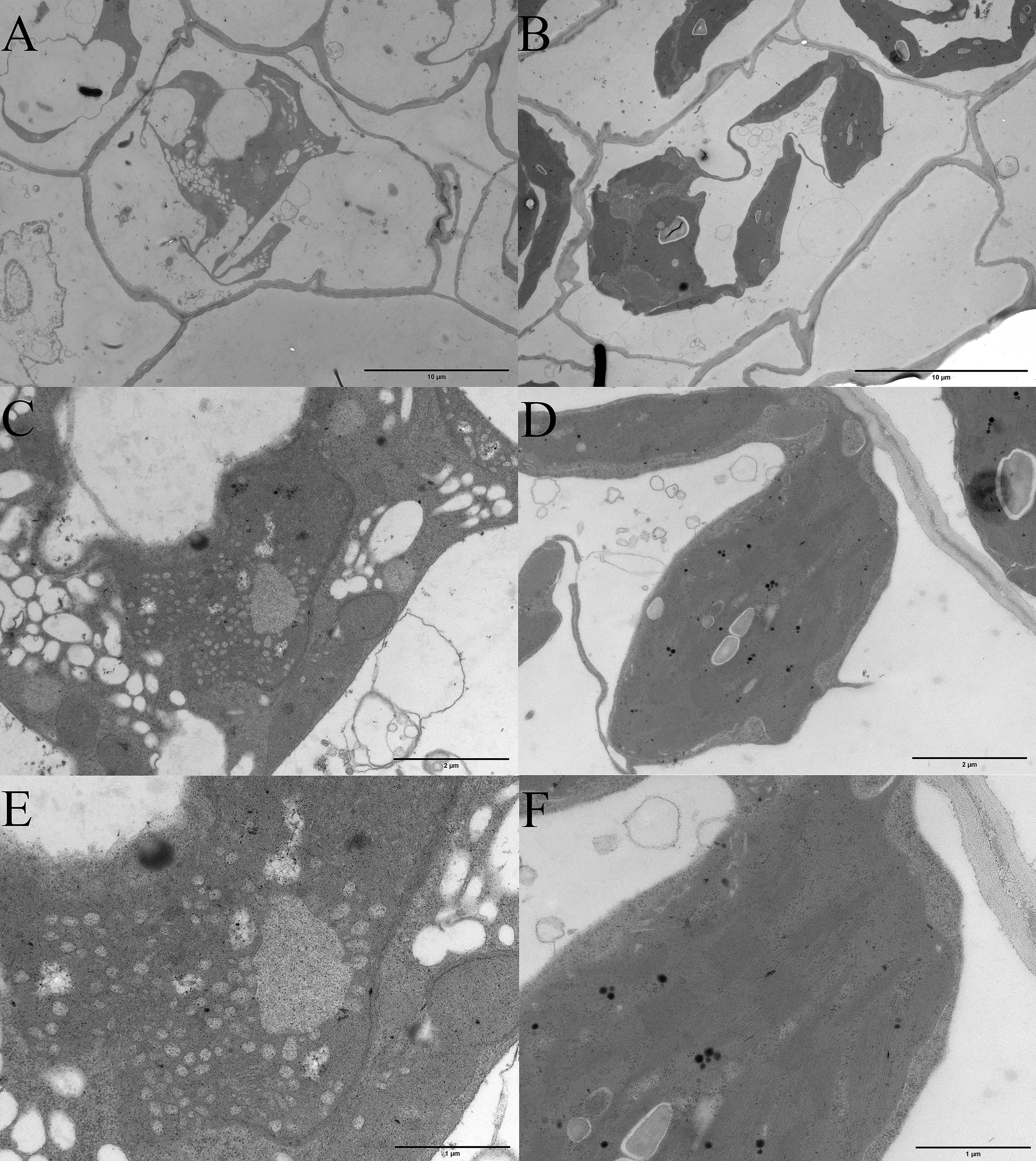 Fig. 2.
Fig. 2.Microscopic analysis of the plastids tissue between albino mutant and wild type seedlings of daylily. (A,C,E) Transmission electron microscopic examination of plastids of albino seedlings. (B,D,F) Transmission electron microscopic examination of plastids of wild-type. The bars in (A,B) represent 10 micrometers, (C,D) represent 2 micrometers, and (E,F) represent 1 micrometer.
To understand the mechanism of the decrease in chlorophyll content, the precursors of chlorophyll and carotenoids biosynthetic pathways were measured. Except PBG, the content of the precursors of chlorophyll biosynthesis significantly decreased in the leaves of albino mutants compared with that of wild-type plants (Supplementary Fig. 3).
Daylily ‘Black-eyed stella’ is a cross between two diploid parents ‘Stella de oro’ and ‘Little celena’ (https://www.daylilies.org/DaylilyDB/). Albino mutants were isolated from the self-pollinated offspring population of ‘Black eyed stella’ and one of its parents, ‘Stella de oro’. The F2 population of ‘Black-eyed stella’ was used for further analysis, and the segregation ratio was in accordance with the expected Mendelian ratio of 3:1 (Table 1). Furthermore, all cross-pollinated offspring between H. ‘black-eyed stella’ and H. Middebdorffii Trautv. & C. A. Mey displayed a wild-type phenotype. However, albino seedlings accounted for approximately a quarter of the F2 population.
| Generations | Total | Wildtype | Albino | Expected ratio | |
| F2 | 1505 | 1137 | 368 | 3:1 | 0.001 |
The surviving F2 plants should have consisted of two-thirds of heterozygous
plants and one-third of homozygous plants, because the albino mutant plants died
before flowering. The plants with self-pollination progeny that did not appear as
albino seedlings were designated as homozygous plants (Alb
RNA-seq was performed to compare gene expression patterns between the mutant and wild-type plants. Then, 41.96 G clean data were acquired. The effective data volume of each sample ranged from 6.74 G–7.17 G. The Q30 bases were distributed in 95.47–95.55%, and the average GC content was 46.59%. A total of 53479 unigenes were spliced and had a total length of 55899490 base pairs (bp) and an average length of 1045.26 bp (Supplementary Table 3). These results showed that the quality of the RNA-seq data was good enough for subsequent analysis. A total of 7952 DEGs, including 4069 up-regulated genes and 3883 down-regulated genes were identified between daylily albino mutant and wild-type plants.
GO function enrichment analysis showed that many DEGs were involved in biological processes related to cellular components and molecular function, such as cellular process, metabolic process, cell part, organelle, membrane, membrane part, membrane-enclosed lumen, binding, catalytic activity, and transporter activity (Supplementary Fig. 2). KEGG significant enrichment analysis revealed that the DEGs were involved in 125 metabolic pathways. The metabolic pathways with the most DEGs were carbohydrate, lipid, amino acid, and energy metabolism. The down-regulated DEGs were pyruvate, amino sugar, nucleotide, starch and sucrose metabolism (Supplementary Fig. 3, Supplementary Table 4).
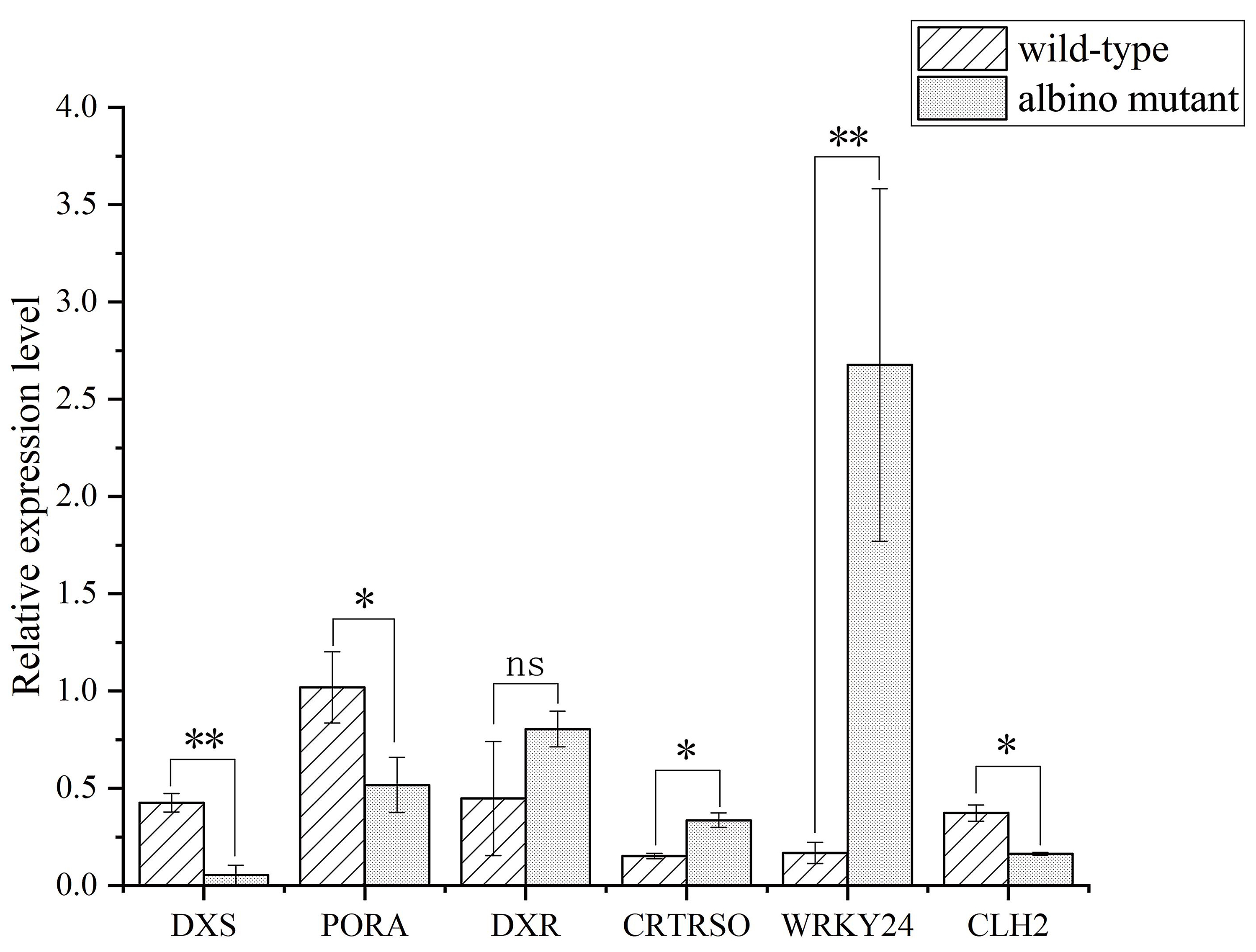
Six DEGs were randomly selected from genes encoding for terpene, carotenoid and chlorophyll metabolism and one transcription factor for quantitative real-time PCR (qPCR) validation, because all these genes were related to the albino trait (Fig. 3). The expression level of DEGs between qPCR and RNA-seq data was coincident (Supplementary Table 5). Therefore, the RNA-seq data described here can be considered reliable.
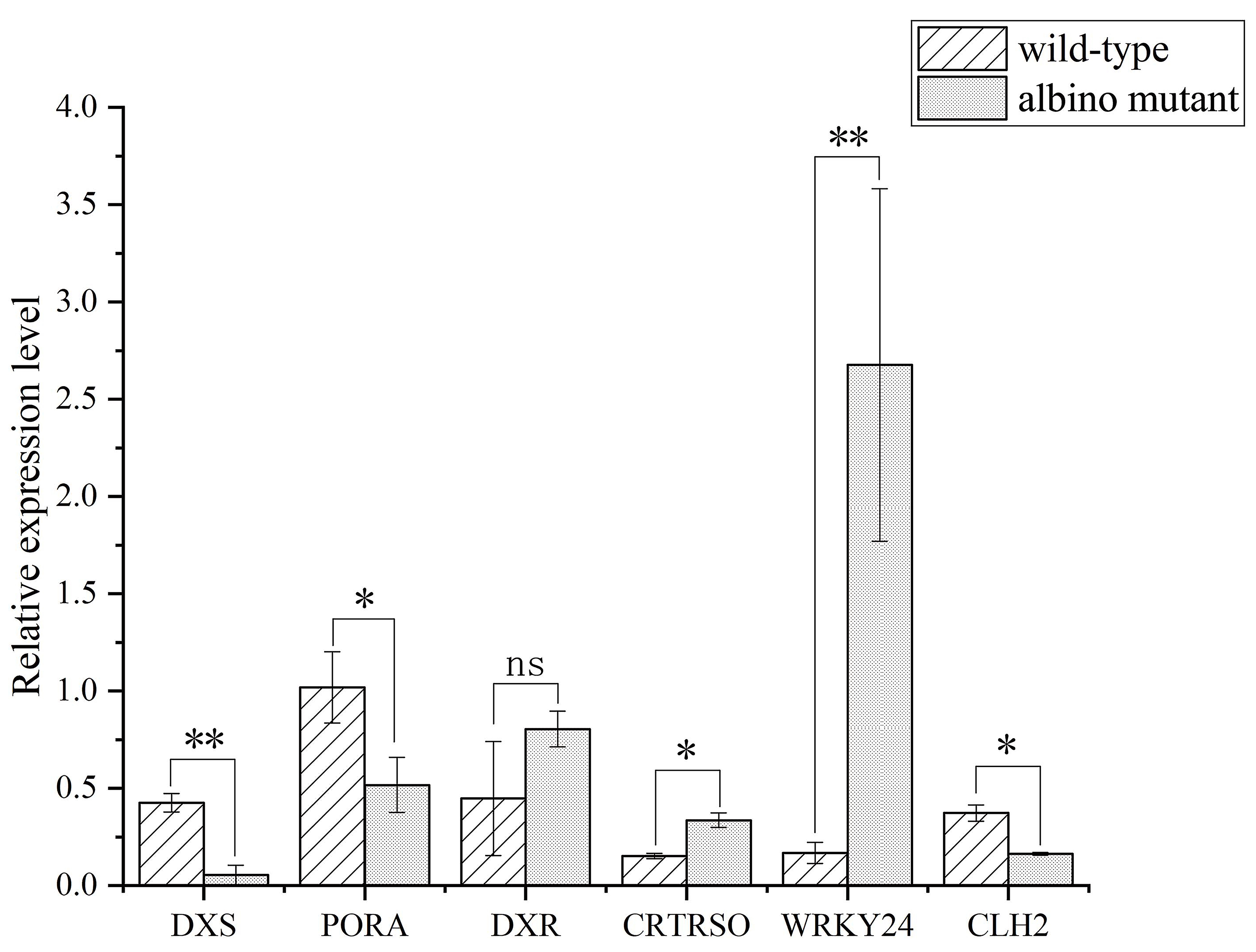 Fig. 3.
Fig. 3.qPCR validation of DEGs obtained from RNA-seq data. DXS, PORA, DXR, CRTRSO, WRKY24 and
CLH2 stand for daylily 1-deoxy-D-xylulose-5-phosphate synthase,
protochlorophyllide oxidoreductase A, 1-deoxy-D-xylulose 5-phosphate
reductoisomerase, carotenoid isomerase, WRKY transcription factor 24 and
chlorophyllase 2, respectively. Mean values
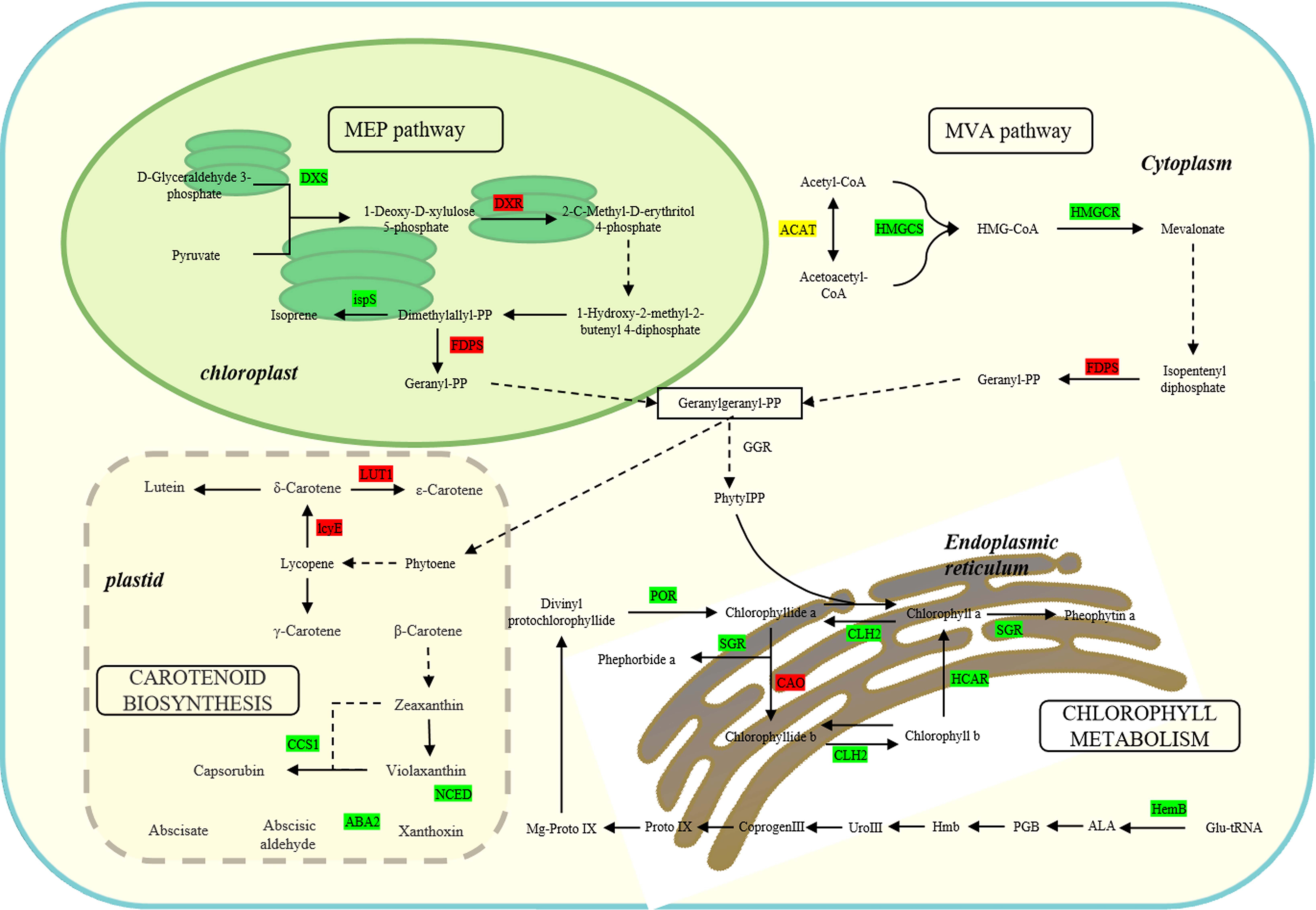
Given that the level of chlorophyll and carotenoid in albino mutants were much lower than those of wild-type plants (Supplementary Table 2), the DEGs related to chlorophyll biosynthesis were analysed with the RNA-seq data, showing that most of the DEGs related to chlorophyll biosynthesis were down regulated (Fig. 4, Ref. [61, 62, 63]). The fold change range was 0.20–0.49 (Supplementary Table 6). CAO, a gene necessary for transforming Chla to Chlb in chlorophyll oxygenase biosynthesis [64], was up-regulated (Fig. 4). This result might be responsible for the observed decrease in the Chlorophyll a/b ratio in the mutants.
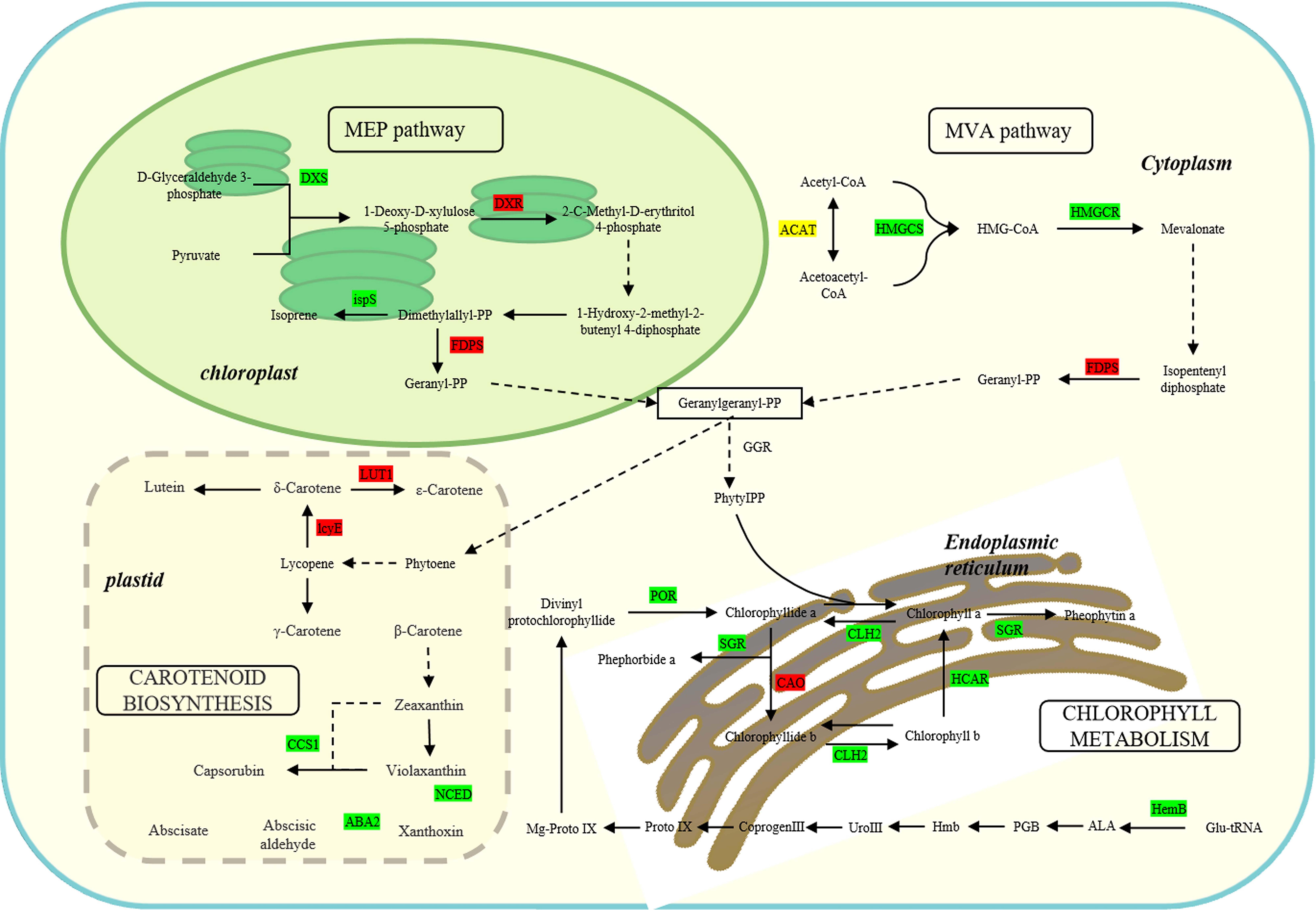 Fig. 4.
Fig. 4.DEGs in MEP, MVA, and metabolism of carotenoids and chlorophylls. Green represents down-regulated genes, red represents up-regulated genes. The MEP, MVA, and metabolism of carotenoids and chlorophyll map are summarized according to previous reports [61, 62, 63]. DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; ISPS, isoprene synthase; FDPS, farnesyl diphosphate synthase; ACAT, acetyl-CoA C-acetyltransferase; HMGCS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase; GCR, unclear receptor subfamily 3, group C, member 1; LUT1, Cytochrome P450 superfamily protein; ICYE, lycopene epsilon cyclase; CCS1, copper chaperone CCS1; NCED, putative 9-cis-epoxycarotenoid dioxygenase 3; ABA2, NAD(P)-binding Rossmann-fold superfamily protein; POR, cytochrome p450 oxidoreductase; SGR, AP2-domain transcription factor SGR; CLH2, chlorophyllase 2; HCAR, coenzyme F420 hydrogenase family / dehydrogenase, beta subunit family; CAO, chloroplast signal recognition particle component; HemB, porphobilinogen synthase; PGB, porphobilinogen; Hmb, hydroxymethylbiliin; UroⅢ, uroporphyrinogen Ⅲ; CoprogenⅢ, Coproporphyrin Ⅲ; Proto IX, protoporphyrin Ⅸ; Mg-Proto IX, Mg-Protoporphyrin IX.
The direct precursor of carotenoids biosynthesis is geranylgeranyl pyrophosphate
(GGPP), which is also a precursor of chlorophyll biosynthesis [24]. Therefore,
the DEGs of terpene metabolism were analysed in detail. The results indicated
that HfDXS, which encodes a key enzyme in the MEP pathway, was
significantly negatively regulated (Figs. 3,4). The expression level of
HfDXS in albino mutant plants was down-regulated 3.5 times compared with
that in the wild-type control (Supplementary Table 5). Regarding the
mevalonate pathway (MVA) pathway for terpene metabolism, the genes encoding
acetyl-CoA C-acetyltransferase and hydroxymethylglutaryl-CoA synthase were
down-regulated (Fig. 4). These DEGs act upstream of the metabolic pathways of MEP
and MVA. Therefore, changes in these DEGs likely substantially contributed to
decreases in chlorophyll and carotenoid content in the leaves of the mutant
plants. Regarding other genes relevant to carotenoid biosynthesis, two
up-regulated and three down-regulated DEGs were found (Table 2). The up-regulated
DEGs were genes involved in the biosynthesis of
| Gene id | Gene description | Fold Change | Regulation | Pathway |
| TRINITY_DN24954_c0_g1_i7_1 | lcyE, lycopene epsilon-cyclase | 2.40 | Up | Carotenoids metabolism |
| TRINITY_DN6095_c0_g1_i1_2 | LUT1, carotenoid epsilon hydroxylase | 8.62 | Up | Carotenoids metabolism |
| TRINITY_DN18331_c0_g1_i1_2 | CCS1, capsanthin/capsorubin synthase | 0.43 | Down | Carotenoids metabolism |
| TRINITY_DN16727_c0_g1_i1_3 | NECD, 9-cis-epoxycarotenoid dioxygenase | 0.19 | Down | Carotenoids metabolism |
| TRINITY_DN17994_c0_g1_i1_1 | ABA2, xanthoxin dehydrogenase | 0.38 | Down | Carotenoids metabolism |
| TRINITY_DN16388_c0_g1_i1_1 | atoB, acetyl-CoA C-acetyltransferase | 2.00 | Up | Terpenoid biosynthesis |
| TRINITY_DN24924_c0_g1_i1_3 | atoB, acetyl-CoA C-acetyltransferase | 0.23 | Down | Terpenoid biosynthesis |
| TRINITY_DN24275_c0_g1_i1_3 | E2.3.3.10, hydroxymethylglutaryl-CoA synthase | 0.31 | Down | Terpenoid biosynthesis |
| TRINITY_DN24659_c0_g1_i9_3 | HMGCR, hydroxymethylglutaryl-CoA reductase (NADPH) | 0.46 | Down | Terpenoid biosynthesis |
| TRINITY_DN24770_c0_g1_i2_3 | dxs, 1-deoxy-D-xylulose-5-phosphate synthase | 0.29 | Down | Terpenoid biosynthesis |
| TRINITY_DN22835_c2_g3_i1_1 | dxr, 1-deoxy-D-xylulose-5-phosphate reductoisomerase | 4.06 | Up | Terpenoid biosynthesis |
| TRINITY_DN17066_c0_g1_i1_2 | ispS, isoprene synthase | 0.14 | Down | Terpenoid biosynthesis |
| TRINITY_DN17405_c0_g1_i2_2 | ispS, isoprene synthase | 0.07 | Down | Terpenoid biosynthesis |
| TRINITY_DN25913_c0_g2_i19_2 | GPS, geranyl diphosphate synthase | 2.62 | Up | Terpenoid biosynthesis |
| TRINITY_DN27270_c0_g1_i15_1 | GPS, geranyl diphosphate synthase | 2.12 | Up | Terpenoid biosynthesis |
| TRINITY_DN23792_c1_g1_i3_3 | hemB, porphobilinogen synthase | 0.49 | Down | Chlorophyll metabolism |
| TRINITY_DN24157_c0_g1_i6_2 | por, protochlorophyllide reductase | 0.42 | Down | Chlorophyll metabolism |
| TRINITY_DN24157_c0_g2_i1_2 | por, protochlorophyllide reductase | 0.23 | Down | Chlorophyll metabolism |
| TRINITY_DN25035_c0_g3_i3_3 | por, protochlorophyllide reductase | 0.34 | Down | Chlorophyll metabolism |
| TRINITY_DN17815_c1_g1_i1_3 | SGR, magnesium dechelatase | 0.21 | Down | Chlorophyll metabolism |
| TRINITY_DN17815_c1_g2_i1_3 | SGR, magnesium dechelatase | 0.27 | Down | Chlorophyll metabolism |
| TRINITY_DN24503_c1_g4_i2_1 | CAO, chlorophyllide a oxygenase | 2.41 | Up | Chlorophyll metabolism |
| TRINITY_DN17158_c0_g1_i1_3 | E3.1.1.14, chlorophyllase | 0.47 | Down | Chlorophyll metabolism |
| TRINITY_DN26828_c0_g4_i1_3 | HCAR, 7-hydroxymethyl chlorophyll a reductase | 0.20 | Down | Chlorophyll metabolism |
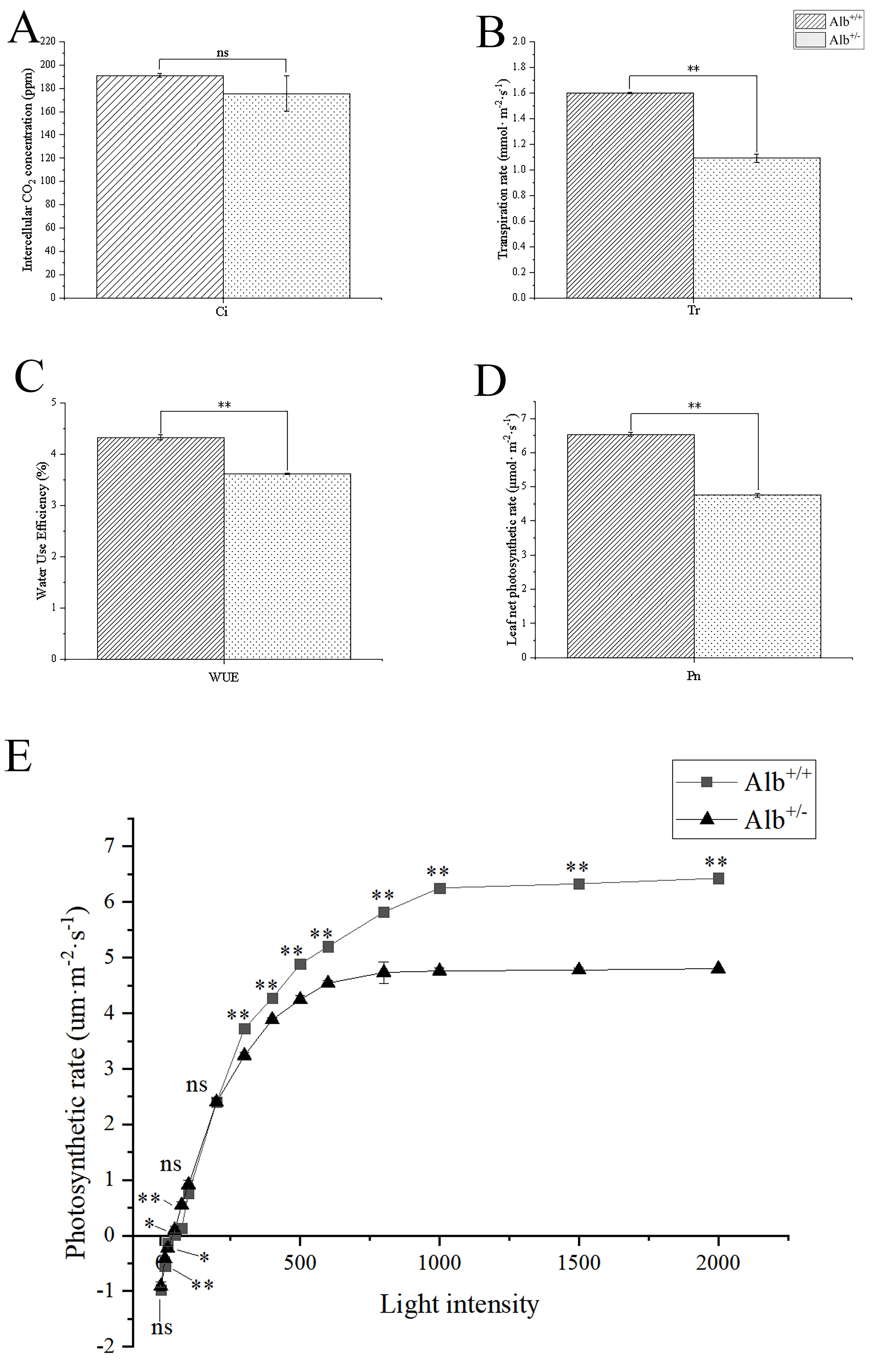
The photosynthetic pigment contents of Alb
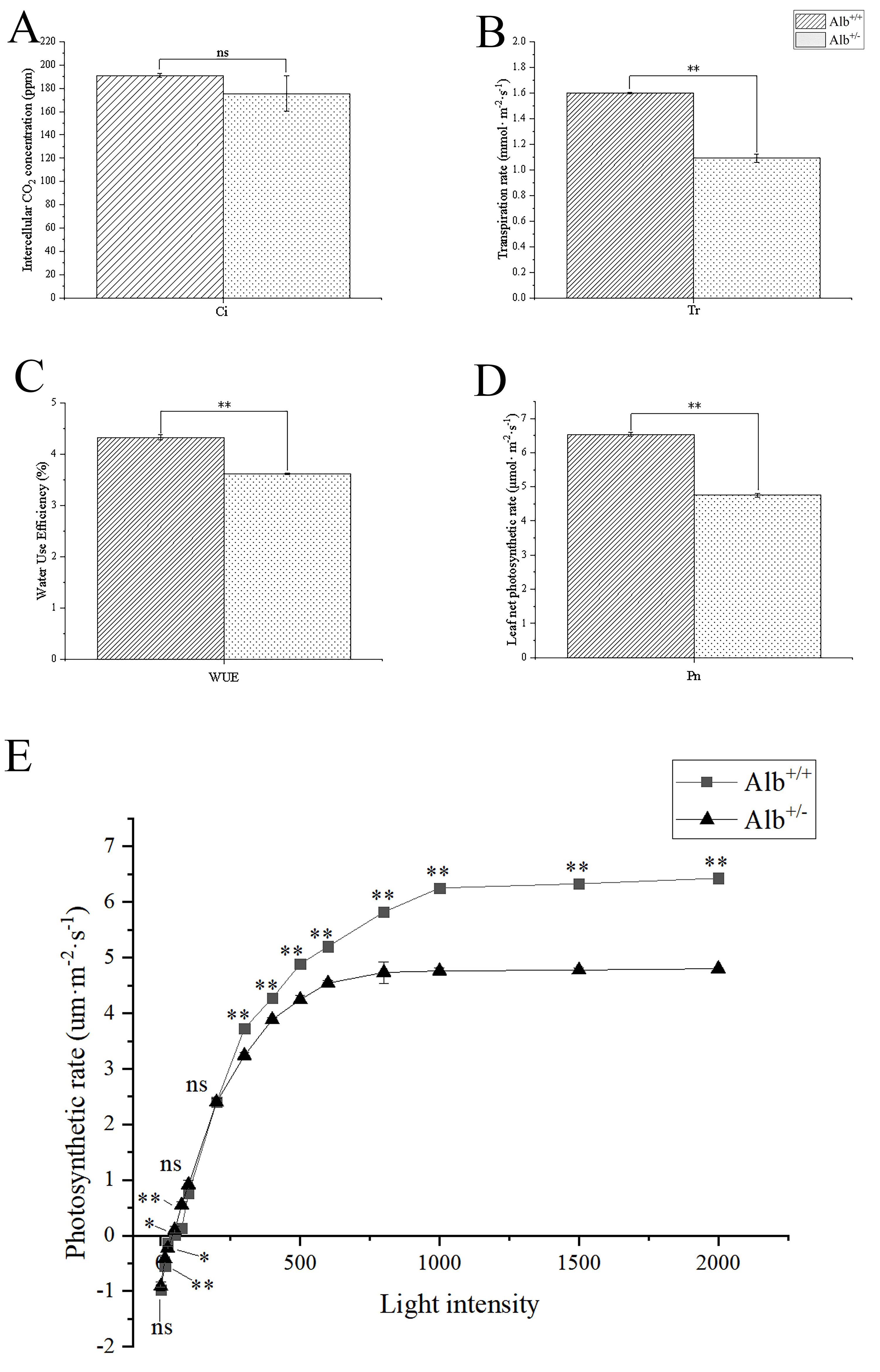 Fig. 5.
Fig. 5.Comparison of the net photosynthetic capacity, and light
response curve between Alb
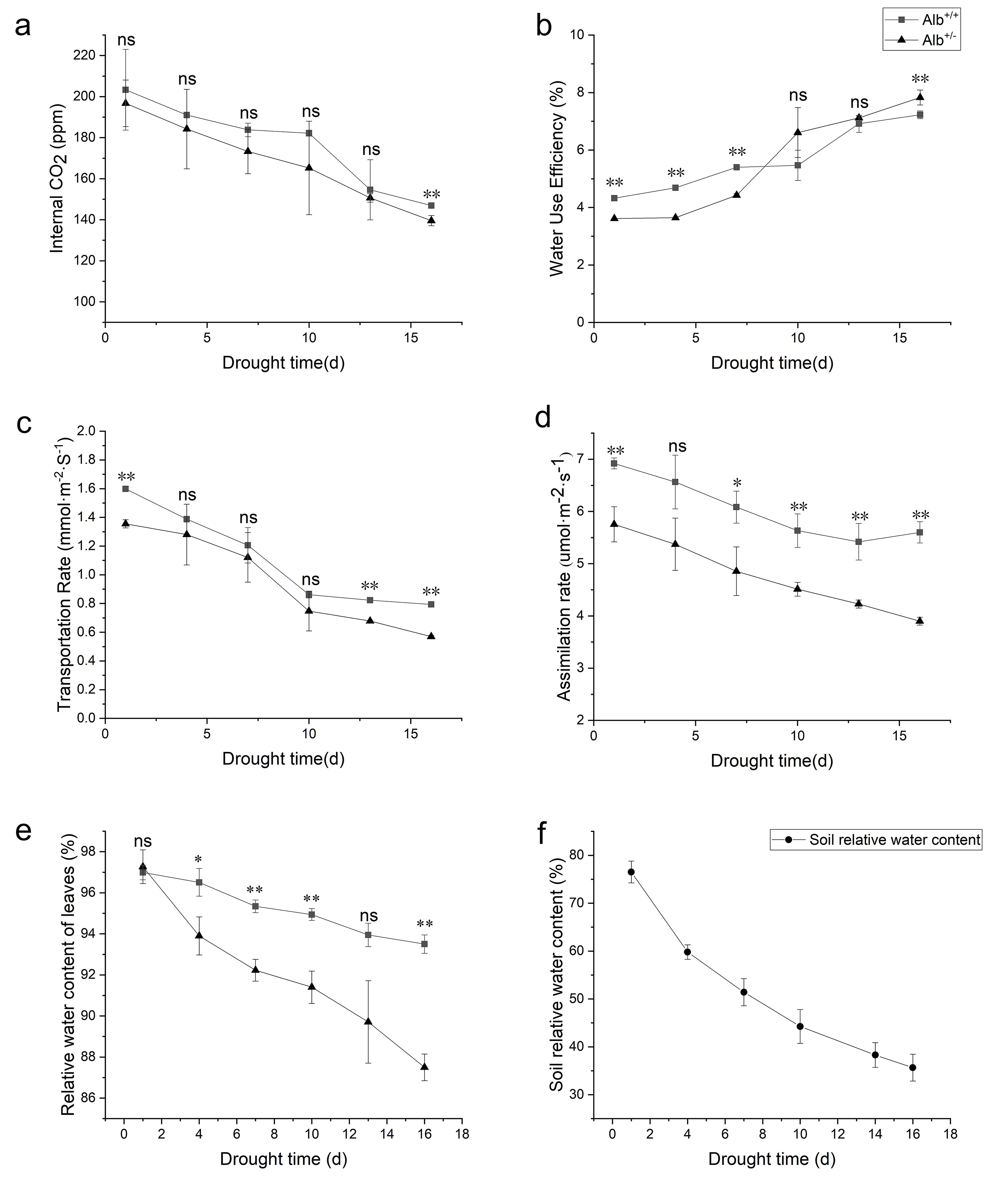
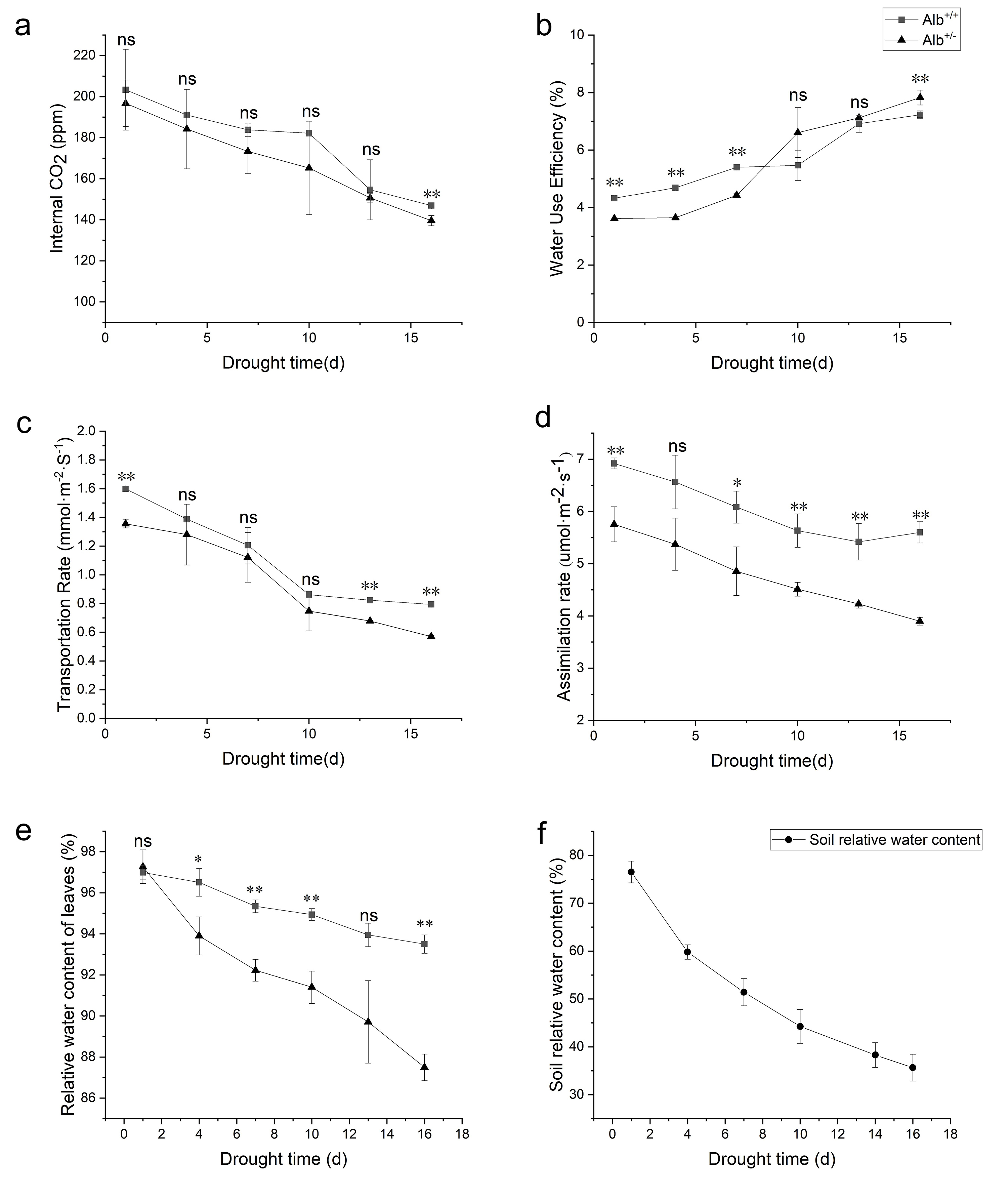 Fig. 6.
Fig. 6.Photosynthetic capacity and relative water content of
Alb
As photosynthetic efficiency affects the drought tolerance of plants [51], RWC
of Alb
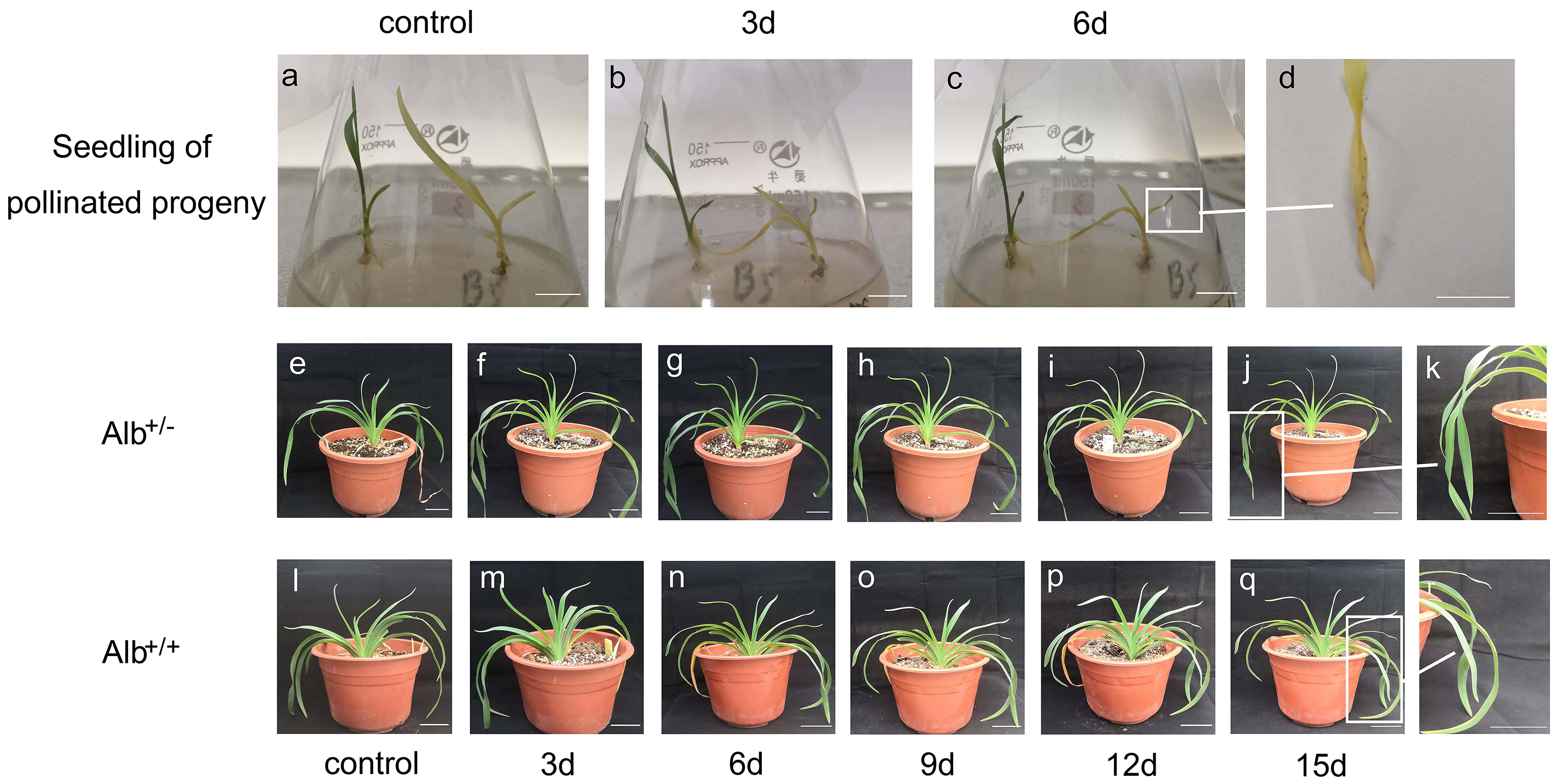
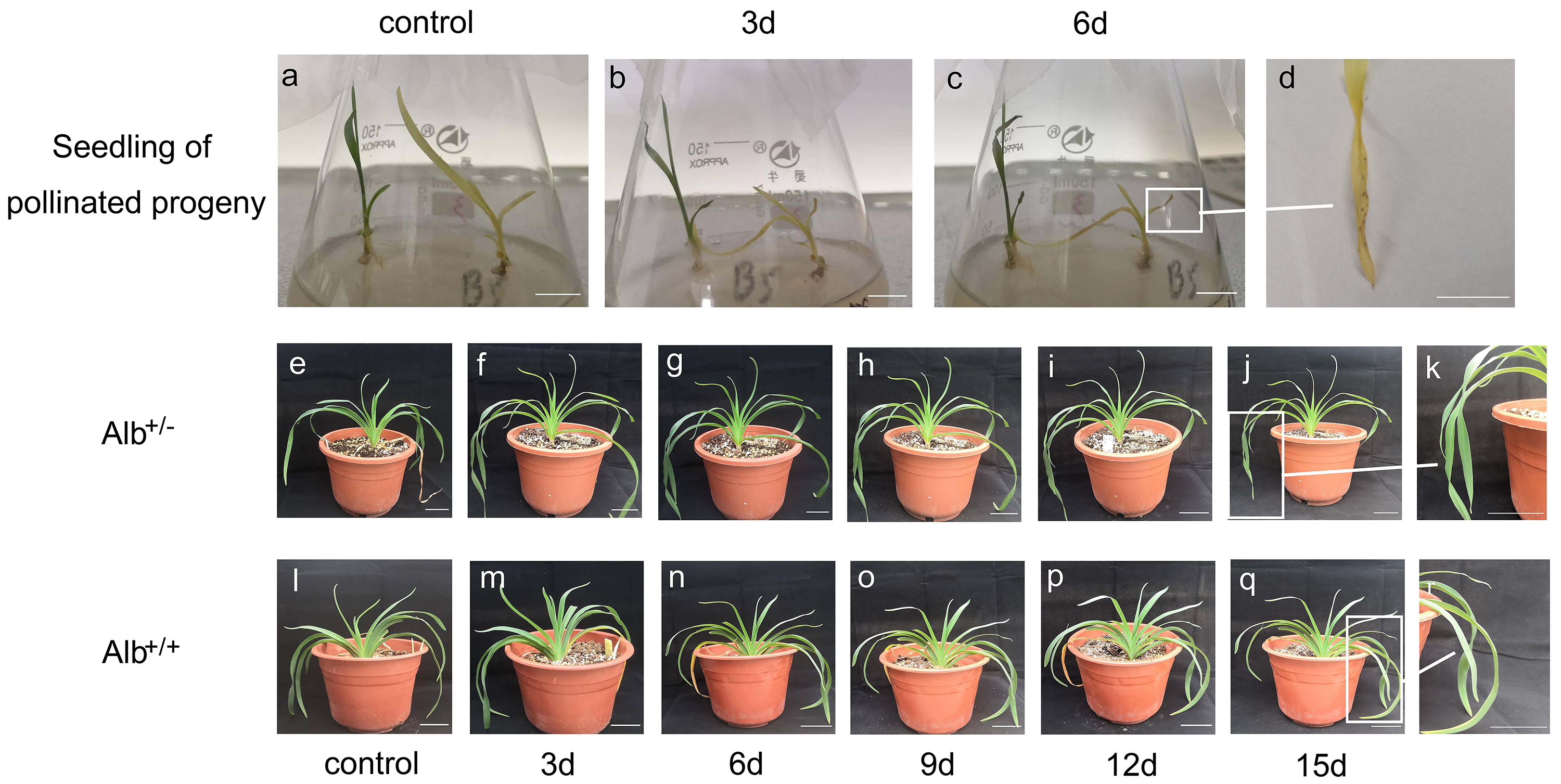 Fig. 7.
Fig. 7.Sensitivity of albino mutant plants of daylily to drought
conditions. Seedlings of self-pollinated progeny of daylily cultivar ‘black-eyed
stella’ were transplanted to a medium containing 10% PEG2000 after culturing on
a PEG free medium for 2 weeks. (A) Seedlings before PEG-treatment. (B,C)
Seedlings being treated for 3 and 6 days, respectively. (D) The albino leaves
showing brown speckles after PEG-treatment. (E–R) One-year-old plants, grown in
pot with soil, were treated with drought. (E,L) Potted Alb
This study reports the isolation of a natural albino daylily mutant
(Alb
The analysis of RNA-seq and quantitative PCR indicated that expression of many
genes was altered in mutant plants compared with wild-type plants (Figs. 3,4,5, Supplementary Fig. 1; Table 2). To further understand the
RNA-seq data, the top ten up-regulated and down-regulated DEGs have been
analysed. Regarding the top 10 down-regulated genes, there were 4 genes encoding
photosystem II Psb, 1 gene encoded for a heat shock protein, 2 genes encoded for
cytochrome, 1 gene encoded for isoprene synthase, 1 gene encoded chalcone
synthase, and 1 gene encoded for NAD (P) H-quinone oxidoreducase
(Supplementary Table 6). Psb plays a role in protecting the PS
II system [68]. So far no reports of downregulation of psb genes that
affect leaf colour have been published. Regarding heat shock proteins, abnormal
expression of genes encoding heat shock proteins was shown to cause abnormal
chloroplast development, which in turn affects the colour of leaves [69, 70]. The
expression of a gene encoding cytochrome was showed altered in albino mutants,
but its gene function is related to plant growth and development, as well as
responding to biotic and abiotic stresses, and does not appear to be a key gene
that causes leaf colour mutations in plants [71, 72]. Isoprene synthase regulates
an important reaction within the MEP pathway [63, 64, 65]. However, no reports of
downregulation of isoprene synthase-encoded genes to affect leaf colour have been
published so far. Overexpression of the chalcone synthase-encoded gene increases
the tolerance of plant leaves to strong light due to its high level of
anthocyanin synthesis [73]. The top 10 genes upregulated were genes encoded
beta-glucosidase, polygalacturonase, allene oxide synthase, autophagy-related
protein, probable linoleate 9S-lipoxygenase, heat shock protein,
ubiquitin-ribosomal protein, NADPH-dependent aldo-keto reductase, acetyl-CoA
acyltransferase and DNA-directed RNA polymerase subunit beta
(Supplementary Table 6). These genes are related to the activity of
The results showed changes in the precursor contents of chlorophyll biosynthesis
and a decrease in the Chl a/b ratio in mutant plants compared with wild-type
plants. The possible reason was that Chla may not be as stable as Chlb. The
decrease in the Chl a/b ratio can be attributed to the increased expression of
CAO (Fig. 4), which encodes the synthesis of chlorophyll oxygenase.
Changes in the content of chlorophyll biosynthesis precursors between albino and
wild-type seedlings showed that urogen III synthase is the key enzyme for the
decreasing chlorophyll content (Supplementary Fig. 2). However,
the expression level of this gene revealed from the RNA-seq data did not change
significantly. Changes in urogen III synthase activity may be a side-effect of a
reduction in the expression level of HfDXS in the albino seedlings.
Moreover, the chloroplast development of the mutant was impeded, indicating that
the mutated gene of daylily affected not only photosynthetic pigment
biosynthesis, but also chloroplast development. The results were consistent with
the DXS loss-of-function mutants of Arabidopsis and tomato [9, 10, 88], supporting the notion that the albino phenotype described here might be
influenced by the down-regulation of HfDXS. The higher expression level
of HfWRKY24 in both the Alb
The present results also showed that the light saturation point and the net
photosynthesis rate of the Alb
Our results showed that the RWC of the Alb
The first natural albino mutant of daylily was isolated in this study. According
to Mendelian proportions of the phenotypes in the progeny of heterozygous plants,
the albino phenotype is caused by a single recessive mutation. The homozygous
mutant plants suffered the pronounced downregulation of HfDXS1, causing
a dramatic decline in chlorophyll and carotenoid levels and were unable to live
in the soil for longer than 15 days. Heterozygous Alb
ALA, 5-aminolevulinic acid; Car, carotenoids; ALB3, 63 kDa inner membrane family
protein; CAO, chlorophyllide a oxygenase; Coprogen Ⅲ, coproporphyrinogen III;
Chla, chlorophyll a; Chlb, chlorophyll b; Ci, intercellular CO
Sequence data from this article can be found in the NCBI database (https://www.ncbi.nlm.nih.gov) with the following accession numbers: Hf EF-1a (MT096368), Hf UBQ (MT096370), HfDXS (OP913381), HfPORA (OP913382), HfDXR (OP913380), HfCLH2 (OP913378), HfCRTRSO (OP913379), HfWRKY24 (OP913383).
Conceptualisation: SD, MR and DN. Methodology: DN, QQ and KD. Resources: DN and ZZ. Funding acquisition: DN and MR. Investigation: SD and MF. Data curation: SD, ZZ and DN. Formal analysis: SD. Visualisation: SD. Validation: TĆ, MR, ZZ and DN. Writing - original draft preparation: SD. Writing - review and editing: MF, QQ, ZZ, KD, TĆ, MR and DN. Supervision: TĆ, MR and DN. Project administration: DN. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
The plants used in this study were Hemerocallis Middebdorffii Trautv. & C. A. Mey and Hemerocallis ‘black-eyed stella’ and ‘stella de oro’. Hemerocallis ‘black-eyed stella’ and ‘stella de oro’ were popular. These two varieties were purchased from the local flower market. And their information could be obtained by the American Daylily Association (https://www.daylilies.org/DaylilyDB/). Hemerocallis Middebdorffii Trautv was a wild-type specie, which was collected from Northeast China.
We would like to thank Ms. Jiaying Zhang (Shanghai Academy of Agricultural Sciences) for her assistance in photosynthesis measurements.
This research was funded by Shanghai Municipal Commission of Science and Technology, capacity building project for local universities (23010504800), China Education Association for International Exchange (2022144), and the Ministry of Science, Technological Development and Innovation of Republic of Serbia, contract no. 451-03-47/2023-01/200007.
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/j.fbl2902060.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.