1 School of Medicine, Deakin University, 3216 Victoria, Australia
2 Institute of Mental and Physical Health and Clinical Translation, Deakin University, 3216 Victoria, Australia
Abstract
Background: Signal transducer and activator of transcription (STAT)
proteins play key roles in development, growth, and homeostasis. These roles have
principally been assigned to their “canonical” function as inducible
transcriptional activators acting downstream of cytokines and other factors.
However, variant “non-canonical” functions have also been identified. The
potential in vivo role for non-canonical STAT functions was investigated
in the zebrafish model. Methods: Two zebrafish Stat5.1 mutants were
generated using CRISPR/Cas9 that should impact canonical functionality: one with
a deleted transactivation domain (
Keywords
- cytokine
- JAK-STAT
- STAT5
- non-canonical
- immunity
- growth
Mammals possess seven signal transducer and activator of transcription (STAT) proteins: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6 [1]. The myriad roles of STATs across development, growth, and homeostasis have been largely attributed to their so-called “canonical” function as inducible transcription factors downstream of cytokines and other factors [2, 3]. In this modality, ligation of cytokine or other receptors triggers the tyrosine phosphorylation of inactive unphosphorylated STAT (uSTAT) monomers in the cytoplasm via associated kinases such as JAKs, allowing dimerization of phosphorylated STAT (pSTAT) molecules through reciprocal interactions between a phosphotyrosine-containing motif on one STAT protein with the SH2 domain on another. The subsequent nuclear translocation of these activated STAT dimers enables them to activate the transcription of target genes, impacting key cell processes, notably including differentiation, survival, proliferation, and activation [4, 5, 6]. However, STATs have also been demonstrated to regulate critical cell processes through alternative “non-canonical” functions [7].
Mammalian STAT5A and STAT5B proteins share more than 90% amino acid similarity and have both unique as well as overlapping functions [8]. Both STAT5A and STAT5B contribute to immune regulation, although STAT5B appears to play the dominant role, presumably via canonical signaling downstream of various interleukin (IL) receptors [9]. STAT5A is a master regulator of mammary gland development and lactogenesis attributed to a canonical signaling function downstream of prolactin [10], whereas STAT5B facilitates the regulation of growth and metabolism assigned to canonical growth hormone (GH) signaling [11, 12, 13]. Non-canonical functions of STAT5 proteins have also been reported. These include gene repression by pSTAT5B during oncogenesis [14], uSTAT5B participating in the regulation of megakaryocyte differentiation [15], and non-nuclear roles of STAT5A/B in maintaining the structural integrity of sub-cellular organelles such as the endoplasmic reticulum and Golgi body [16]. Various STAT5B mutations have been associated with disrupted growth and immunity but with differential clinical severity depending on the site of mutation, further suggesting the potential for non-canonical functions [12, 17, 18, 19].
Zebrafish possess two STAT5 proteins, Stat5.1 and Stat5.2 [20]. Stat5.1 displays the closest similarity with mammalian STAT5B and has been shown to contribute to lymphopoiesis throughout the life course [21], as well as the regulation of growth and sex-specific adiposity [21, 22]. However, these publications used loss-of-function (LOF) knockout (KO) mutant models that would abrogate both canonical and non-canonical functions. In this study, two Stat5.1 mutants were generated using CRISPR/Cas9-based mutagenesis to examine potential non-canonical or alternate functions. The impact of these Stat5.1 mutants on lymphopoiesis, growth, and adiposity was examined in direct comparison to a Stat5.1 KO mutant. The results provided evidence of predominantly canonical functions but also suggested that non-canonical and alternate Stat5.1 functions also exist.
Zebrafish were maintained using standard husbandry practices [23] following national guidelines for their care and use. All studies were approved by the Deakin University Animal Ethics Committee.
A single guide RNA (sgRNA) targeting the zebrafish stat5.1 gene was designed using ZiFiTargetor v4.2 [24] and CHOPCHOP v3 [25], which had a minimum of three mismatches with potential off-target sites, all on other chromosomes (Supplementary Table 1). The sgRNA template was cloned into pDR274 and sgRNA synthesized by in vitro transcription using a MEGAscript T7 Transcription Kit (#AM1354, Thermofisher Scientific, Australia), purified using a MegaClear Kit (#AM1908, Thermofisher Scientific, Australia), and quantified. One cell stage wild-type (WT) zebrafish embryos were injected with ~1 nL of 100 ng/µL sgRNA, 200 ng/µL TrueCut Cas9 protein v2 (#A36498, Thermofisher Scientific, Australia), and 0.4% (w/v) phenol red. Founder fish were raised to adulthood, out-crossed with WT fish, and screened for the presence of mutations. Confirmed heterozygous fish were further out-crossed to minimize potential off-target mutations, with heterozygous progeny in-crossed to obtain homozygous mutant lines.
Genomic DNA from adult fin clips and whole embryos was isolated with
QuickExtract DNA Extraction Solution (#QE09050, Gene Target Solution, Australia)
following the manufacturer’s instructions. This was subjected to a polymerase
chain reaction (PCR) for high-resolution melt (HRM) analysis with the
stat5.1-specific primers 5
Embryos were collected and raised in egg water with 0.0003% (w/v)
phenylthiourea (PTU) for 5 days and then fixed in 4%
paraformaldehyde/phosphate-buffered saline (PFA/PBS) overnight at 4 °C,
followed by 100% methanol at –20 °C. Whole-mount in situ hybridization
(WISH) was performed on rehydrated embryos according to a published protocol
[26]. Embryos and juvenile fish were imaged with a MVX10 monozoom microscope
using a 1
Embryos and juvenile fish were imaged as described above and the length was
determined using ImageJ software. Adult fish were imaged using an iPhone 13 pro
max, wide camera-26 mm f1.5 12 MP, 3024
Total RNA was extracted from whole juvenile fish using TRIsure™
(#BIO-38033, Meridian Biosciences, OH, USA, and from the kidney and spleen of adult
male fish using an RNEasy Mini Kit (#74136, Qiagen, Australia) according to the
manufacturer’s instructions. RNA concentrations were determined using a NanoDrop
2000 Spectrophotometer (#ND-2000, Thermofisher Scientific, Australia), and cDNA
was synthesized from 500 ng RNA using a QuantiTect cDNA Synthesis Kit (#205311,
Qiagen, Australia). Samples were subjected to quantitative real-time
reverse-transcription PCR (qRT
A synthetic gene encoding wild-type Stat5.1 with a C-terminal Flag-tag was obtained from GeneArt (Thermofisher Scientific, Australia) and cloned into pBK-CMV vector, with KO, TAD, and TM mutants generated using a Q5 Site-directed Mutagenesis Kit (#E0554S, New England Biolabs, Australia). The resulting plasmids were transfected into HEK293 along with pBK-CMV-Jak3-A573V [30] using Lipofectamine 2000 Transfection Reagent (#11668019, Thermofisher Scientific, Australia) and Western blot performed as described [31] using mouse monoclonal antibodies for Flag M2 (#F1804, Sigma-Aldrich, Australia), pSTAT5A(Y694) (#ab30648, Abcam, Australia) or beta-actin (#A5441, Sigma-Aldrich, Australia), followed by rabbit anti-mouse Ig conjugated to horseradish peroxidase.
Statistical analysis was performed with GraphPad Prism v8.0.0 for Windows (GraphPad Software, San Diego, CA, USA) using One-way ANOVA with Tukey’s multiple comparison test.
Zebrafish Stat5.1 is highly homologous to human STAT5B, both at the level of
protein domains as well as individual amino acid residues [20], including the
region spanning the SH2 domain, the tyrosine motif (TM) phosphorylated in
response to the external stimuli [32], and the transactivation domain (TAD) (Fig. 1A). The region adjacent to the TM was targeted using CRISPR-Cas9 genome editing
[33, 34] with a specific sgRNA (Fig. 1B). Two different mutant alleles were
generated, which were out-crossed twice and bred to homozygosity and sequenced
(Fig. 1C): mdu033, a 4 bp deletion causing a frameshift that results in
the complete deletion of the TAD (
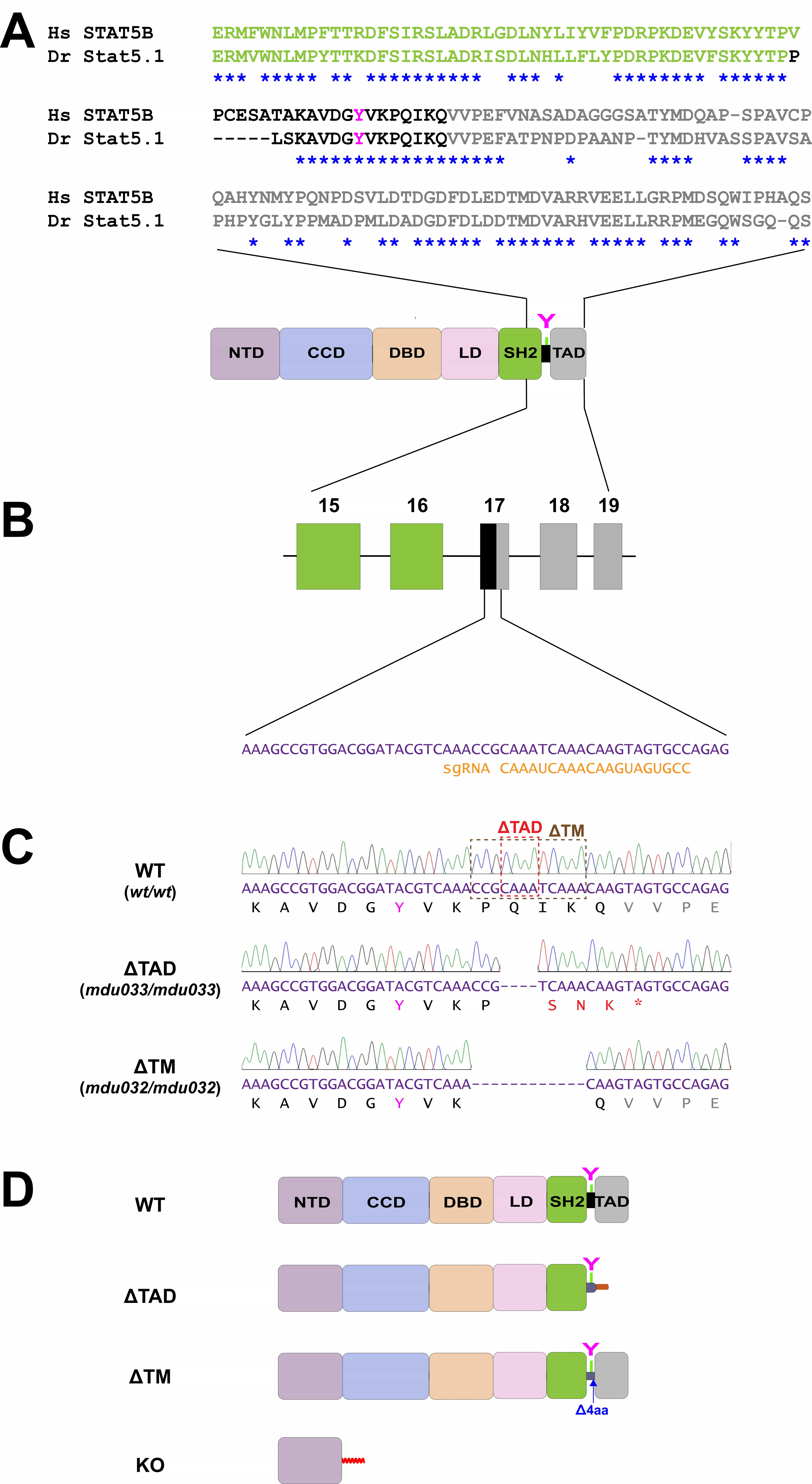 Fig. 1.
Fig. 1.Generation of zebrafish Stat5.1 mutants using CRISPR/Cas9. (A)
Schematic diagram of STAT5B/Stat5.1 and its constituent N-terminal domain (NTD),
coiled-coil domain (CCD), DNA-binding domain (DBD), linker domain (LD),
Src-homology 2 domain (SH2), tyrosine residue (Y), and transactivation domain
(TAD). An amino acid alignment for the indicated region of Homo sapiens
(Hs) STAT5B and Danio rerio (Dr) Stat5.1 is shown above, with identical
residues indicated by asterisks. (B) Part of the zebrafish stat5.1 gene.
Exons are shown as boxes in color matching the corresponding domain, along with
the sequence targeted by the sgRNA. (C) Sequence traces, corresponding
nucleotides, and encoded amino acids for homozygous wild-type (WT)
(wt/wt),
Mammalian STAT5 proteins play important roles in the development of lymphoid cells, particularly STAT5B [35]. We have recently shown that zebrafish Stat5.1 KO mutants exhibited severe disruption in T lymphopoiesis throughout the life course, as well as, other lymphoid defects [21]. Thus, we compared the impact of the other Stat5.1 mutants on this aspect of biology.
Embryonic lymphopoiesis was investigated using WISH on 5 dpf embryos using
specific T cell markers: rag1 for early T cells [36] and lck
for mature T cells [37, 38]. KO embryos showed a significant reduction in the
area of rag1 staining in comparison to WT, as described [21]. Both
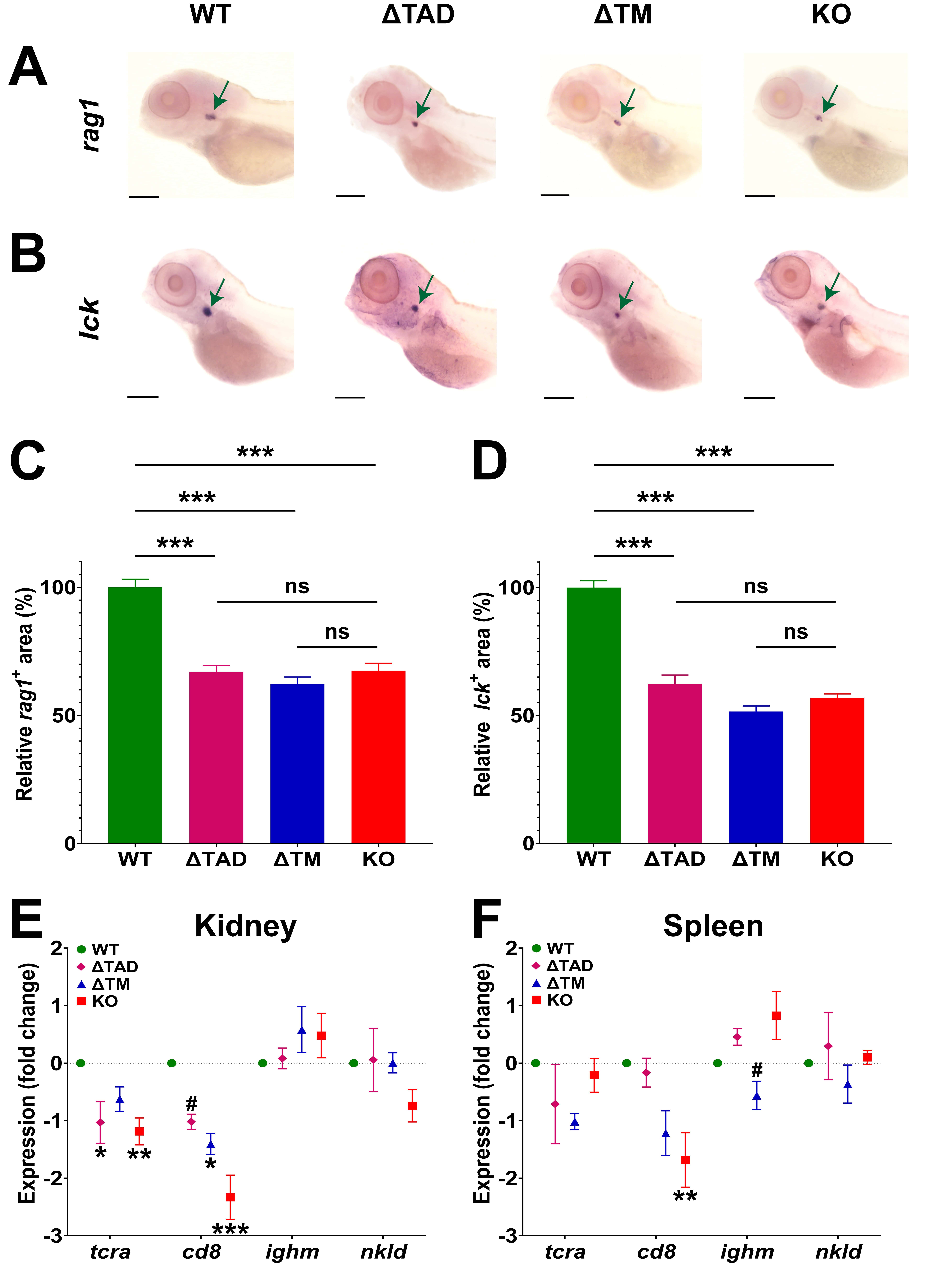 Fig. 2.
Fig. 2.Analysis of lymphopoiesis in Stat5.1 mutants. (A–D) Expression
analysis of lymphoid markers using Whole-mount in situ hybridization (WISH) in 5
dpf WT (stat5.1
Adult lymphopoiesis was examined in the kidney marrow, a primary lymphoid organ
that plays an equivalent role to mammalian bone marrow [39, 40], and the spleen,
a secondary lymphoid organ [41]. Expression of four different lymphocyte-specific
markers was analyzed: tcra (T cells) [42], cd8 (cytotoxic T
cells) [43], ighm (B cells) [44], and nkld (NK cells) [45]. In
the kidney, expression of tcra and cd8 were both significantly
reduced in the KO compared to the WT, as described [21]. Expression of
cd8 was also significantly reduced in the
Mammalian STAT5B/Stat5b plays a crucial role in the regulation of growth and
adiposity [46]. Moreover, zebrafish Stat5.1 KO mutants have also been found to
exhibit reduced growth but increased adiposity along with dysregulation of growth
and lipid metabolism genes [21, 22]. During all developmental stages and into
adulthood for both sexes, the
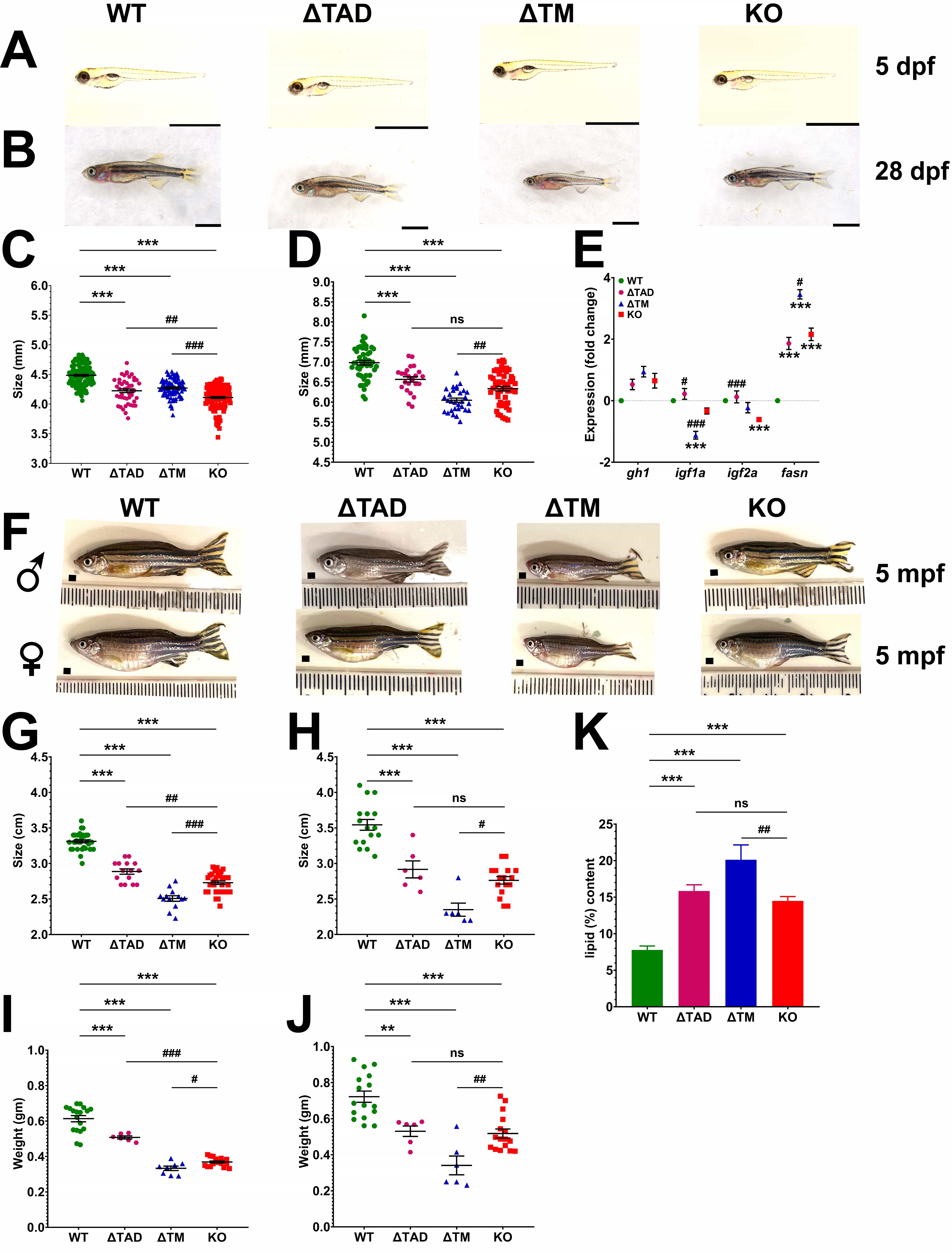 Fig. 3.
Fig. 3.Analysis of growth and adiposity in Stat5.1
mutants. (A–B, F) Images of representative WT (stat5.1
To investigate the molecular underpinnings of the growth deficiency and enhanced
adiposity, 28 dpf juvenile fish were analyzed for the expression of genes
specific for growth, gh1 (growth hormone 1), igf1a and
igf2a (insulin-like growth factors) [47], and lipid metabolism, fasn
(fatty acid synthases) [48]. As described [21], KO juveniles showed reduced
igf2a and elevated fasn expression, but no significant changes
in gh1 or igf1a compared to the WT (Fig. 3E). Expression of
gh1, igf1a and igf2a in the
The critical function of STAT proteins as inducible transcription factors downstream of cytokines is well documented [2, 49]. However, there is growing evidence that in addition to such canonical roles, STATs also participate in non-canonical functions [7]. However, the in vivo impact of these non-canonical functions remains poorly understood since most studies are based on KO/LOF mutants that abrogate both canonical and non-canonical functions. Zebrafish Stat5.1 is structurally and functionally conserved with STAT5B, and ablation of either impacted growth, adiposity, and lymphopoiesis [11, 21, 22]. This study sought to investigate potential non-canonical functions of zebrafish Stat5.1.
Two Stat5.1 mutants were successfully generated using CRISPR-Cas9 genome
editing. The first mutant,
As we have recently described, the Stat5.1 KO mutant exhibited disrupted growth
at all developmental stages compared to WT fish with enhanced adiposity observed
in adult females along with reduced igf2a and increased fasn [21]. This finding suggests that altered metabolism was responsible, consistent
with studies on an alternate Stat5.1 KO mutant [56]. The Stat5.1
The Stat5.1 KO mutants further displayed decreased embryonic and adult T
lymphopoiesis, but the B and NK cells of these mutants were largely unaffected,
as described [21]. The Stat5.1
The mechanism by which non-canonical Stat5.1 functions regulate aspects of adult
lymphopoiesis remains to be determined, although precedents exist for
non-canonical STAT5 functions impacting mammalian immune cell development. For
example, human STAT5B mutations that delete most of the protein—and
so likely LOF—are associated with severe T and NK lymphopenia [12, 17], but
shorter truncations that would still ablate canonical functions result in milder
effects, such as reduced CD8
The distinct phenotypes of the
This study explored the relative involvement of canonical versus non-canonical and other functions for zebrafish Stat5.1 in vivo. The results revealed strong impacts of Stat5.1 mutations on embryonic lymphopoiesis, growth, and adiposity that largely appear to be due to loss of canonical functions. However, there was also evidence of potential non-canonical Stat5.1 functions regulating adult T lymphopoiesis and growth, with growth and adiposity also impacted by alternate functions.
All data generated or analyzed during this study are included in this article, or available upon request.
ACW and CL designed the research study. NA and CL performed the research. NA, ACW and CL analyzed the data. NA and ACW wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
This study was approved by the Deakin University Animal Ethics Committee under projects 23-2019, 24-2019, 25-2019, 14-2022, 15-2022, and 16-2022.
The authors would like to thank the Deakin University Animal House staff for superb aquarium management and Somayyeh Heidary for helpful advice.
The Research was supported by Deakin University Postgraduate Research Scholarship (DUPRS) to NA.
The authors declare no conflict of interest. Given his role as Guest Editor, ACW had no involvement in the peer-review of this article and has no access to information regarding its peer-review. Full responsibility for the editorial process for this article was delegated to Amedeo Amedei.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.



