- Academic Editor
†These authors contributed equally.
Background: Genetic mutations are quite common in non-small cell lung cancer (NSCLC), however, their prognostic value remains controversial. Methods: This study explored the mutational landscape of tumor samples from patients with advanced NSCLC by next-generation sequencing (NGS). A total of 101 NSCLC patients in stage III or IV receiving first-line treatment were included. Results: TP53 mutation was the most frequent genetic alteration in NSCLC tumors (68%), followed by EGFR (49%), CDKN2A (12%), LRP1B (9%), and FAT3 (9%) mutations. Among 85 patients with stage IV NSCLC, first-line targeted therapy remarkably prolonged progression-free survival (PFS) of patients compared with first-line chemotherapy (p = 0.0028). Among 65 patients with stage IV NSCLC whose tumors harbored EGFR, ALK, ROS, or BRAF mutations, first-line targeted therapy substantially prolonged the PFS of patients (p = 0.0027). In patients with TP53 mutations who received first-line targeted therapy or chemotherapy, missense mutation was the most common mutation type (36/78), and exon 5 represented the most common mutated site (16/78). Conclusions: TP53 mutation in exon 5 could independently predict poor PFS of patients with stage IV NSCLC after the first- line treatment. Moreover, mutations in TP53 exon 5 and LRP1B were associated with shorter PFS of such patients whether after first-line chemotherapy or targeted therapy, respectively. Thus, these patients should be given immunotherapy or immunochemotherapy.
Lung cancer is the most common malignancy worldwide, with non-small cell lung cancer (NSCLC) accounting for 85% of all cases [1, 2]. Despite the steady decline in mortality and rapid increase in survival in recent years, specifically for NSCLC, lung cancer remains the most deadly cancer in both men and women, with a 5-year overall survival of 21% for all stages combined [1].
Oncogenic driver mutations are responsible for the initiation and development of NSCLC. Clinical efficacy of targeting oncogenic driver genes such as epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), c-ros oncogene 1 (ROS1), and B-Raf proto-oncogene (BRAF) has been confirmed in NSCLC treatment [3, 4, 5, 6]. So far, several treatment guidelines have been proposed for NSCLC patients with targetable driver mutations [7, 8]. However, the prognostic value of those mutations remains controversial. Therefore, a better understanding of the mutational landscape and the prognostic value of the mutations in NSCLC may provide valuable information for risk stratification and refining of the treatment strategies.
Next-generation sequencing (NGS) has been widely used to detect germline and somatic mutations in human cancers, showing advantages compared with conventional genomic sequencing methods. Previous studies have shown that some somatic mutations are strongly associated with the survival of NSCLC patients, providing valuable information for clinical management of NSCLC patients. For example, it was found that the combination of 6 hotspot mutations of EGFR, PIK3CA, and tumor suppressor protein p53 (TP53) predicts poor survival of NSCLC patients [9]. The presence of specific DDR gene alterations was associated with worse prognosis of advanced NSCLC [10]. In addition, studies found that the tumor mutation burden, i.e., the approximate amount of gene mutation that occurs in the genome of a cancer cell, can predict the response of NSCLC to immunotherapeutic agents [11]. Therefore, using NGS to detect somatic mutations and identify prognostic indicators may provide important information to guide treatment selection and improve outcomes for NSCLC management.
TP53 is the most frequently mutated gene in human cancer, with a frequency of 50–60% [12]. Although many studies have shown that TP53 mutations are associated with worse prognoses in cancer carriers, the prognostic value of TP53 mutation in NSCLC remains somewhat controversial [13, 14] due to different mutated exons and different cancer stages [15, 16, 17]. TP53 contains 11 exons, and most TP53 mutations occur in exons 5–8 spanning 540 nucleotides that encode the DNA-binding domain of p53 protein [13]. Previously, all p53 mutants have been considered functionally equal; however, increasing evidence has revealed that mutations occurring in different positions of a single gene, e.g., TP53, could have a different effect on the prognosis of cancer [18, 19, 20], thus emphasizing the importance of stratifying NSCLC patients according to the classification of detailed gene mutations.
In this study, we analyzed the effect of different firstline treatment strategies for advanced NSCLC patients. To factors that significantly associated with PFS of patients we carried out a detailed stratification of patients accoding to their TNM stages and genomic alterations. We then investigated the associations of high-frequency mutations with PFS of unresectable stage IV NSCLC patients receiving different first-line treatment regimens. Our results suggested that mutations in exon 5 of TP53 and low-density lipoprotein receptor-related protein 1B (LRP1B) were potential adverse prognostic indicators for stage IV NSCLC.
A total of 101 patients diagnosed with NSCLC at Shanxi Bethune Hospital (Taiyuan, Shanxi, China) were recruited for the present study. Formalin-fixed, paraffin-embedded tumor samples were collected between May 2013 and May 2021. Matched white blood cell samples were collected as reference controls for each tumor sample. Tumors were staged according to the 8th edition of the AJCC staging system [21]. Sixteen patients with stage III diseases received surgery and postoperative adjuvant chemotherapy, whereas 85 with unresectable stage IV diseases underwent first-line treatment according to their molecular signatures, which was in accordance with the Guidelines of the Chinese Society of Clinical Oncology (CSCO) Non-Small Cell Lung Cancer (http://meeting.csco.org.cn/MUser/CscoPeriodical/1/2?keyword=&leibie=&pyear=0&loc=E9317455E99FF2AE) [22]. Driver gene EGFR/ALK/ROS/BRAF mutations were detected in 65 patients with stage IV diseases. Of these patients, 44 received matched targeted-therapy, while the other 21 received chemotherapy (n = 34) or immunotherapy (n = 7) due to various causes.
This study was approved by the Ethics Committee of Shanxi Bethune Hospital (No. YXLL-2022-045) and conducted according to the Declaration of Helsinki. Informed consent was obtained from all patients.
DNA sequencing was performed as previously described [23]. Total genome DNA was isolated from paraffin-embedded tumor samples and matched white blood cells (germline mutations excluded) using a DNA extraction kit from Qiagen (Hilden, Germany) or Life Technologies (Carlsbad, CA, USA) according to the manufacturer’s instructions. The DNA concentration and purity were determined using NanoDrop 2000 (Agilent Technologies, Santa Clara, CA, USA) and Qubit 2.0. Genomic DNA was sonicated into 150–200 bp fragments using Covaris S220 focused ultrasonicator (Covaris, Woburn, MA, USA).
As previously described [23], a DNA library was constructed using a KAPA Hyper
Prep kit (Kapa Biosystems, Woburn, MA, USA) according to the manufacturer’s
instructions. A panel of 63 genes (Genetron Health, product catalog identifier:
fwa-p180-cancer) used in this study is summarized in Supplementary Table
1. The DNA library was enriched for regions of the custom-designed captured
probe manufactured by Agilent. A total of 750 ng DNA library was incubated with
two different hybridization reagents and blocking agents in a SureSelectXT target
enrichment system (Agilent Technologies). The enriched library was amplified
using P5/P7primer. The quality and quantity of the DNA library were assessed
using 2200 Bioanalyzer, Qbit3 (Thermo Fisher Scientific, Waltham, MA, USA), and a
qPCR NGS library quantification kit (Agilent Technologies). The library was
sequenced using a 150-bp paired-end strategy on the NovaSeq system (Illumina,
USA) with a depth of 3500
A total of 78 patients with stage IV NSCLC who received first-line targeted
therapy or chemotherapy were included in the analysis. Kaplan-Meier curve
analysis was used to assess the associations of variables of interest with the
PFS of patients. The patients with stage III diseases with follow-ups
Statistical analysis was performed using R (version 4.1.2, the R Foundation for
Statistical Computing, Vienna, Austria) and RStudio (version 1.4.1717, Posit
Community portal, Washington D.C., USA). Mutation interactions (co-mutations and
mutual exclusivity) were identified by performing Fisher’s exact test on pairs of
genes. The Cox proportional hazards model was used for multivariate survival
analysis. Variables with a p-value
A total of 101 patients, 43 females and 58 males (a mean age of 62 years) with stage III (n = 16) or unresectable stage IV (n = 85) NSCLC were included in this study. The clinicopathologic and molecular features of patients are summarized in Table 1. A total of 49 patients received first-line targeted therapy (e.g., gefitinib, osimertinib, and entrectinib), 45 patients received first-line chemotherapy, and 7 patients received first-line immunotherapy (e.g., pembrolizumab, sintilimab, and atezolizumab).
| Characteristics | Value | |
| Total, n | 101 | |
| Sex n (%) | ||
| Female, n (%) | 43 (42.6) | |
| Male, n (%) | 58 (57.4) | |
| Age, median (range) | 62.00 (55.00, 68.00) | |
| Smoking | ||
| Non-smokers, n (%) | 64 (63.4) | |
| Smokers, n (%) | 37 (36.6) | |
| Primary site | ||
| Both | 2 (2.0) | |
| Left, n (%) | 37 (36.6) | |
| Right, n (%) | 62 (61.4) | |
| Pathology | ||
| Adenocarcinoma, n (%) | 87 (86.1) | |
| Squamous cell carcinoma, n (%) | 14 (13.9) | |
| Stage | ||
| IIIA, n (%) | 11 (10.9) | |
| IIIB, n (%) | 3 (3.0) | |
| IIIC, n (%) | 2 (2.0) | |
| IVA, n (%) | 72 (71.3) | |
| IVB, n (%) | 13 (12.9) | |
| Brain metastasis | ||
| No, n (%) | 80 (79.2) | |
| Yes, n (%) | 21 (20.8) | |
| Bone metastasis | ||
| No, n (%) | 52 (51.5) | |
| Yes, n (%) | 49 (48.5) | |
| Therapy | ||
| First-line therapy, n (%) | 46 (45.5) | |
| Second-line therapy, n (%) | 23 (22.8) | |
| Third-line therapy, n (%) | 32 (31.7) | |
| First-line therapy | ||
| Chemotherapy, n (%) | 45 (44.6) | |
| Immunotherapy, n (%) | 7 (6.9) | |
| Targeted therapy, n (%) | 49 (48.5) | |
| Second-line Therapy | ||
| Chemotherapy, n (%) | 14 (13.9) | |
| Immunotherapy, n (%) | 10 (9.9) | |
| None, n (%) | 46 (45.5) | |
| Targeted therapy, n (%) | 31 (30.7) | |
| Third-line Therapy | ||
| Chemotherapy, n (%) | 3 (3.0) | |
| Immunotherapy, n (%) | 4 (4.0) | |
| None, n (%) | 69 (68.3) | |
| Targeted therapy, n (%) | 25 (24.8) | |
To explore the degree of survival benefit of patients receiving first-line targeted therapy, we conducted Kaplan-Meier curve analysis in patients with stage IV NSCLC (n = 85) who were subsequently stratified into chemotherapy (n = 34), targeted therapy (n = 44), and immunotherapy (n = 7) groups. Kaplan-Meier curve analysis showed that first-line targeted therapy remarkably prolonged PFS of patients compared with first-line chemotherapy (522 days vs. 192 days, p = 0.0028). The first-line immunotherapy group was precluded from statistical analysis due to the small sample size (Fig. 1).
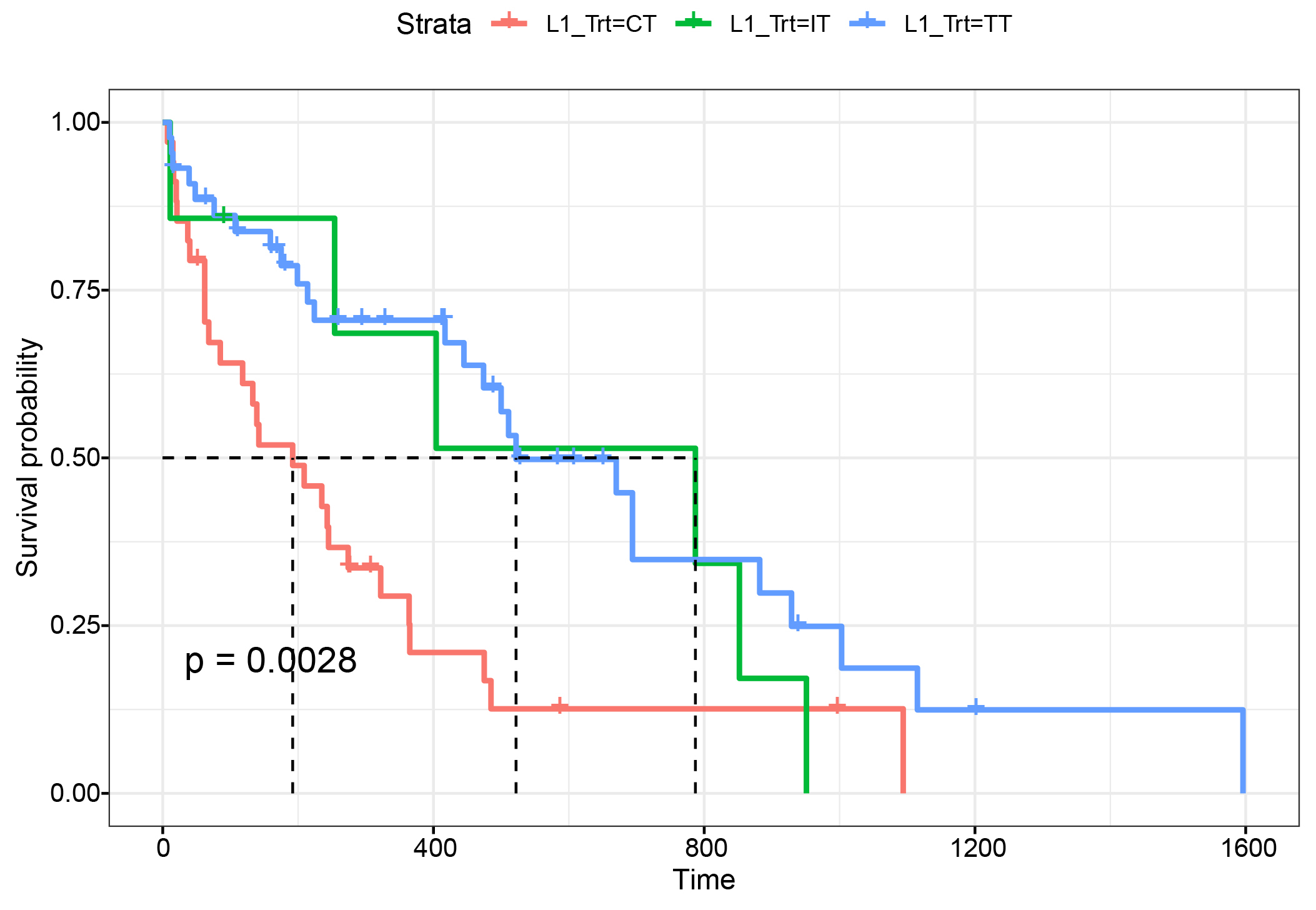 Fig. 1.
Fig. 1.Kaplan-Meier curves comparing PFS in stage IV NSCLC patients receiving different first-line therapies. A total of 85 patients with stage IV NSCLC were stratified into first-line chemotherapy (n = 34), targeted therapy (n = 44), and immunotherapy (n = 7) groups. Kaplan-Meier curves were generated to evaluate the association of first-line treatment with progression-free survival (PFS) of patients.
In order to identify somatic mutations associated with advanced NSCLC and the
survival of these patients, we performed NGS of tumor tissue and matched white
blood cell samples from 101 NSCLC patients in stage III or IV, finding that
97.03% (98/101) of tumor samples carried gene mutations, with TP53
mutation representing the most frequent alteration (68%, 69/101), followed by
EGFR (49%), CDKN2A (12%), LRP1B (9%), and
FAT3 (9%) mutations (Fig. 2). We also observed significant concurrent
mutations in 13 gene pairs, among which KRAS/EGFR were mutually
exclusive mutations (p
 Fig. 2.
Fig. 2.Mutation landscape of patients with stage III or IV non-small cell lung cancer (NSCLC) (n = 101). Each horizontal line represents a single gene, and each vertical line represents different samples. Different colors indicate the type of mutations. Representative clinical features of the patients, such as tumor stage, metastasis status, and sex, are shown at the bottom of the plot. N, no; Y, yes; F, female; M, male; A, adenocarcinoma; S, squamous cell carcinoma; CT, chemotherapy; IT, immunotherapy; TT, targeted therapy.
In order to personalize stratification patients more accurately, we further performed survival analysis of stage III and IV NSCLC patients, respectively. The results showed that only stage IV patients statistically significantly benefit from molecular typing in our cohort. Analysis of the 65 stage IV patients whose tumors harbored driver gene EGFR/ALK/ROS/BRAF alterations showed that compared with non-targeted therapy (n = 21), matched targeted therapy (n = 44) significantly prolonged PFS of patients harboring these mutations (500 days vs. 139 days, p = 0.0027; Fig. 3). These data suggest that first-line targeted therapy improves the prognosis of NSCLC patients harboring druggable mutations, highlighting the importance of identifying genetic mutations by NGS in direct therapeutic decision-making.
 Fig. 3.
Fig. 3.Comparison of PFS of stage IV NSCLC patients receiving matched targeted therapy and non-targeted therapy. A total of 65 patients with EGFR/ALK/ROS/BRAF mutations in the tumor were analyzed, including 44 patients receiving matched targeted therapy and 21 receiving non-targeted therapy. Kaplan-Meier curves were generated to evaluate the association of first-line treatment with the PFS of patients.
Previous studies have shown that different positions of TP53 mutation have a differential effect on the prognosis of cancer [18, 19, 20]. According to those findings we grouped patients more detailed based on their somatic TP53 mutation subtypes. As shown in Fig. 4A, among 101 NSCLC patients, the frequency of TP53 mutation was 68% (69/101). We conducted a more thorough analysis of TP53 mutations in 78 patients receiving first-line chemotherapy (n = 34) or targeted therapy (n = 44). The first-line immunotherapy group was excluded from statistical analysis due to the small sample size (n = 7). The most common mutation type was a missense mutation (46%, 36/78), followed by nonsense mutation (14%, 11/78) and multiple mutations (4%, 3/78) (Fig. 4B). The most common mutated site was exon 5 (20%, 16/78), followed by exon 7 (14%, 11/78), exon 8 (11%, 9/78), and exon 6 (10%, 8/78) (Fig. 4C). Mutations in exons 5–8 accounted for 65% of all TP53 mutations.
 Fig. 4.
Fig. 4.TP53 mutation profiles of NSCLC patients. (A) Frequency of TP53 mutation in 101 patients with NSCLC. (B) Composition of mutation types in 69 NSCLC patients with TP53 mutation. (C) Distribution of TP53 mutation subtypes in 101 patients with NSCLC.
Then, the prognostic value of popular mutations were further evaluated in this
study. We carried out univariate cox regression analysis to test the association
between gene altaeration and PFS. Those mutations significant associated with PFS
identified in the univariate analysis were then selected to conduct multivariate
Cox regression analysis, which showed that mutations in TP53 exon 5
(p = 0.033, hazard ratio (HR) = 2.3 (95% CI [1.069–5.1])), EP300 (p = 0.005, HR = 23.8 (95% CI [2.654–212.5])), and PTPN11 (p
 Fig. 5.
Fig. 5.Multivariate Cox regression analysis of the association between gene mutation and PFS after the first-line treatment. Multivariate Cox regression analysis was performed for 78 patients with stage IV NSCLC, including 34 patients receiving first-line chemotherapy and 44 patients receiving first-line targeted therapy. A forest plot showed that TP53 exon 5, EP300, and PTPN11 could independently predict poor PFS after the first-line treatment. * indicated statistically significant; ** and *** indicated very significant.
We further performed survival analysis on the 78 patients. As shown in Fig. 6A, patients with TP53 mutation in exon 5 (n = 13) had shorter PFS than those with wild-type TP53 or non-exon 5 mutations (n = 65) after first-line treatment (p = 0.0218, HR = 2.22). Among 34 patients receiving first-line chemotherapy, patients with TP53 mutation in exon 5 (n = 7) had shorter PFS after first-line treatment than those with wild-type TP53 or non-exon 5 mutations (n = 27) (p = 0.00616, HR = 3.37; Fig. 6B), which indicated that TP53 mutation in exon 5 was an adverse indicator for the prognosis of advanced NSCLC after first-line treatment or first-line chemotherapy. This further suggests that NSCLC patients with TP53 exon 5 mutations are insensitive to first-line chemotherapy and could hardly obtain survival benefits from first-line chemotherapy.
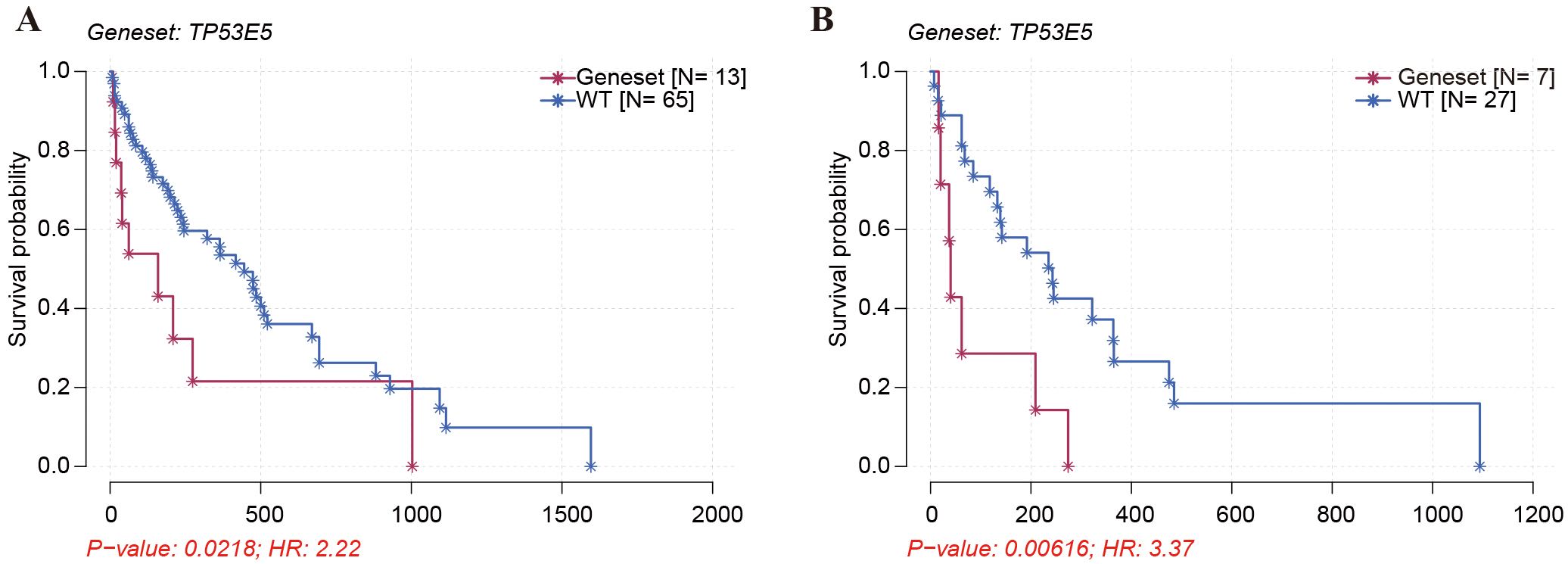 Fig. 6.
Fig. 6.The association of TP53 exon 5 mutation with the PFS of stage IV NSCLC patients. (A) Kaplan-Meier survival analysis was carried out to assess the association of TP53 exon 5 mutation with the PFS of stage IV NSCLC patients receiving first-line treatment. (B) Kaplan-Meier survival analysis was conducted to assess the association of TP53 exon 5 mutation with the PFS of NSCLC patients after first-line chemotherapy.
The association of LRP1B mutation with PFS of patients was also assessed since LRP1B was one of the most frequently altered genes in the cohort of this study. As shown in Fig. 7A, similar to TP53 mutation, LRP1B mutation was significantly associated with poor PFS of NSCLC patients (n = 78) after first-line treatment (p = 0.0212, HR = 2.9). Furthermore, of the 44 patients receiving first-line targeted therapy, patients harboring LRP1B (n = 2) had significantly shorter PFS than those with wildtype LRP1B (n = 42) (p = 0.0414, HR = 4.21) (Fig. 7B). These data suggests that patients with LRP1B mutation are insensitive to first-line targeted therapy and could hardly achieve a survival benefit from first-line targeted therapy.
 Fig. 7.
Fig. 7.Kaplan–Meier curve showing the difference in PFS among patients harboring LRP1B mutation or not. (A) Kaplan-Meier survival analysis was carried out to assess the association of LRP1B mutation with the PFS of patients with stage IV NSCLC, including 5 patients with and 73 patients without LRP1B mutation. (B) Kaplan-Meier survival analysis was conducted to assess the association of LRP1B mutation with the PFS of NSCLC patients after first-line targeted therapy, including 2 patients with and 42 patients without LRP1B mutation.
In the present study, we sought to identify somatic mutations associated with PFS in patients with advanced NSCLC after first-line treatment. Analysis of tumor tissue samples from 101 NSCLC patients in stages III and IV using NGS revealed that TP53 was the most frequently mutated gene, with exon 5 representing the most common mutated site. In patients with stage IV NSCLC, TP53 mutation in exon 5 was an independent predictor of poor PFS after the first-line treatment. Furthermore, mutations in TP53 exon 5 and LRP1B were associated with worse PFS after first-line chemotherapy and targeted therapy, respectively. These results suggested that TP53 exon 5 and LRP1B mutations are negative prognostic indicators for advanced NSCLC.
The NGS data may guide the treatment options to improve clinical outcomes. Cao et al. [24] have demonstrated that after identifying driver mutations by NGS, patients receiving matched targeted therapy achieve significantly longer PFS than those receiving non-targeted therapy. A recent meta-analysis, which included 11 studies and 2874 participants, has shown that ALK inhibitor targeted therapy causes a significant increase in PFS of patients with ALK-rearranged NSCLC compared with chemotherapy [25]. Similarly, in the present study, the first-line targeted therapy remarkably prolonged PFS of patients compared with first-line chemotherapy, regardless of the mutation status. In patients who harbored driver gene EGFR/ALK/ROS/BRAF mutations matched targeted therapy substantially prolonged the PFS of patients compared with non-targeted therapy. Thus, NGS data can indeed inform therapy decisions in patients with advanced NSCLC.
TP53 is the most frequently altered gene in NSCLC, occurring in 30–65% of cases [15, 26, 27]. The prevalence rate of TP53 mutation in this study was 68%. About 80–90% of TP53 mutations encode missense proteins with a reduced capacity to bind to a specific DNA sequence that regulates gene transcription [12, 28]. This study found that missense mutation was the most common type of TP53 mutation, found in 46% of patients. TP53 mutations frequently occur in the ‘hot-spot’ region of exons 5–8 encoding the DNA-binding domain [20, 29, 30, 31]. In the present study, mutations in exons 5–8 accounted for 65% of all TP53 mutations, and exon 5 was the most common mutated location. Vega et al. [32] explored the clinical significance of TP53 exon 5 mutation, demonstrating that patients with squamous cell lung tumors carrying TP53 mutations in exon 5 exhibit worse prognoses than those carrying mutations in other locations. Nonetheless, since then, many studies have focused on the prognostic value of TP53 exon 8 mutations, revealing that TP53 mutations affecting exon 8 but not exon 5 indicate poor prognosis in NSCLC patients [20, 26, 33]. In the current study, multivariate Cox regression analysis showed that TP53 exon 5 mutation was an independent predictor for worse PFS in patients with stage IV NSCLC after the first-line treatment. Moreover, TP53 mutation in exon 5 was correlated with poor PFS after first-line treatment regardless of the treatment option, suggesting that mutation in exon 5 might identify a subgroup of NSCLC patients with unfavorable prognoses.
With comprehensive genomic profiling, concurrent genetic alterations have been
discovered in human cancers. The presence of TP53 co-mutations can
identify high-risk ALK
LRP1B encodes low-density lipoprotein receptor-related protein 1b, which is frequently mutated in NSCLC and acts as a putative tumor suppressor [38, 39]. In this study, we observed LRP1B mutation in 9% of patients with advanced NSCLC, which was remarkably lower than that observed in other NSCLC cohorts, possibly because the samples being investigated here only include stage III&IV NSCLC patients [40, 41]. Accumulating evidence has suggested that LRP1B alterations are correlated with high herterogeineity of tumors which may lead to poor prognosis [40]. Multiple studies have shown that LRP1B mutation improved outcomes of immunotherapy in patients with different types of cancer, including NSCLC [40, 42, 43, 44, 45, 46, 47]. One possible mechanism is that NSCLC patients with somatic mutation of LRP1B usually have higer tumor mutation burden which postitively associated with immunotherapy [40]. We found that patients with LRP1B mutation had significantly shorter PFS compared to those with wild-type LRP1B after first-line targeted therapy, which suggests that NSCLC patients with LRP1B mutation may benefit from immunotherapy rather than targeted therapy.
The present study has some limitations that need to be addressed in the future. First, 7 patients who received first-line immunotherapy were excluded from the correlation analysis between TP53 mutation and PFS due to the small sample size. To investigate the prognostic values of immunotherapy-related genes such as LRP1B, larger sample size is needed. Second, the predictive potential of TP53 exon 5 mutation in the prognosis of NSCLC patients should be tested in large validation cohorts. Long-term follow-up data are also needed to investigate whether TP53 exon 5 mutations correlate with the overall survival of the patients. Third, although many patients have received second and third-line therapies, numerous confounding factors such as loss of follow-up prevented us from data collection and analysis.
We used NGS to characterize the mutational landscape in tumor samples from patients with stages III or IV NSCLC. Mutations in TP53 exon 5 and LRP1B resulted in being negative prognostic factors for patients with stage IV NSCLC after first-line chemotherapy and targeted therapy, respectively. According to these findings, immunotherapy or immunochemotherapy are more recommended treatment stratigies for advanced NSCLC patients with somatic mutations in TP53 exon 5 and LRP1B. These findings reinforce the importance of genetic testing to guide first-line therapy for NSCLC management.
ALK, anaplastic lymphoma kinase; BRAF, B-Raf proto-oncogene; EGFR, epidermal growth factor receptor; LRP1B, low-density lipoprotein receptor-related protein 1B; NGS, next-generation sequencing; NSCLC, non-small cell lung cancer; PFS, progression-free survival; ROS1, c-ros oncogene 1; TP53, tumor suppressor protein p53.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
XR, JZ and FL contributed to the conception, design, Supervision, and guidance of the study; HF and HX, performed the data collection and analyses and wrote the manuscript; GD, CY and LL assisted in project design, guided the statistical analysis, and reviewed the manuscript; ZJ assisted in bioinformatic analysis; XY and HG contributed to data collection and collation; XS helped perform the analysis with constructive discussions. XR, JZ and FL critically reviewed the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
This study was approved by the Ethics Committee of Shanxi Bethune Hospital (No. YXLL-2022-045) and conducted according to the Declaration of Helsinki. As this was a retrospective study, the requirement for informed consent was waived.
This work was supported by Cancer Biobank of Tianjin Medical University Cancer Institute and Hospital.
This work was supported by grants from the Health Commission of Shanxi Provincial [grant number 2019014] and the CAPTRA-Lung Scientific research fund of Beijing Cancer Prevention and treatment Society [grant number CAPTRALung2022010].
LL and ZJ are both Medical Advisors of Genetron Health Inc and they declare no conflicts of interest. Other authors declare no potential conflicts of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
