1 Department of Pharmaceutical Sciences, Division of Pharmaceutical Technology, University of Basel, 4056 Basel, Basel-Stadt, Switzerland
2 Institute of Chemistry and Biotechnology, Zurich University of Applied Sciences (ZHAW), 8820 Wädenswil Zürich, Switzerland
3 Focal Area Infection Biology, Biozentrum, University of Basel, 4056 Basel, Basel-Stadt, Switzerland
Abstract
Introduction: Blood infections from multi-drug-resistant Salmonella pose a major health burden. This is especially true because Salmonella can survive and replicate intracellularly, and the development of new treatment strategies is dependent on expensive and time-consuming in vivo trials. The aim of this study was to develop a Salmonella-infection model that makes it possible to directly observe Salmonella infections of macrophages in vivo and to use this model to test the effect of antimicrobials against intra- and extracellular Salmonella in order to close the gap between in vitro and rodent-infection models. Methods: We established suitable Salmonella-infection conditions using genetically engineered zebrafish and Salmonella-expressing fluorescent proteins (green fluorescent protein (GFP) and/or mCherry). Results: We detected Salmonella inside and outside zebrafish larvae macrophages. Administration of the cell-impermeable antibiotic tobramycin removed Salmonella residing outside macrophages but did not affect Salmonella in macrophages, whereas ceftriaxone successfully cleared both types of Salmonella. Salmonella inside and outside macrophages experienced substantial DNA damage after administration of fluoroquinolones consistent with the excellent cell penetration of these antibiotics. Conclusions: The zebrafish-larvae model enables testing of antimicrobials for efficacy against extra- and intracellular Salmonella in a complex in vivo environment. This model thus might serve for antimicrobial lead optimization prior to using rodent models.
Graphical Abstract
Keywords
- Salmonella enterica
- infections
- macrophages
- zebrafish larvae
- antibiotic resistance
- drug screening
Salmonella is a major cause of systemic infections with a high fatality rate in low- and middle-income countries. As such, it is an significant health burden in those countries [1]. Salmonella can survive and replicate intracellularly in many cellular types, including macrophages [2]. This is particularly problematic because a number of regularly used antibiotics are unable to cross cell membranes and therefore cannot reach intracellular Salmonella [3, 4, 5]. After termination of the antibiotic intervention, these surviving intracellular pathogens can reemerge and lead to a reinfection of the organism [6].
Increasing resistance to currently used antibiotics [7, 8] and the low number of promising novel antibiotics pose a major threat to modern medicine and human health worldwide. This threat has the potential to become one of the most serious challenges in modern medical practice [9, 10, 11, 12, 13]. However, the development of anti-invective treatment regimens is expensive and the return of investment is relatively small, as efficient antibiotics rarely make it onto the market [14].
Due to poor biodistribution and pharmacokinetic-model properties, in vitro tests often do not mirror the efficiency of antibiotic compounds and formulations in vivo [15]. Thus, in addition to in vitro testing, novel antibiotics need to show activity in expensive and time-consuming in vivo trials. Several in vitro and in vivo models have been described, suitable to investigate Salmonella infections including underlying molecular mechanisms and pathogen-host interactions. The wide variety of described models includes in vitro models like enteroids and organoids, and in vivo models like C.elegans, insect larvae, zebrafish larvae, chicken embryos, rodents and calves [16, 17, 18, 19, 20].
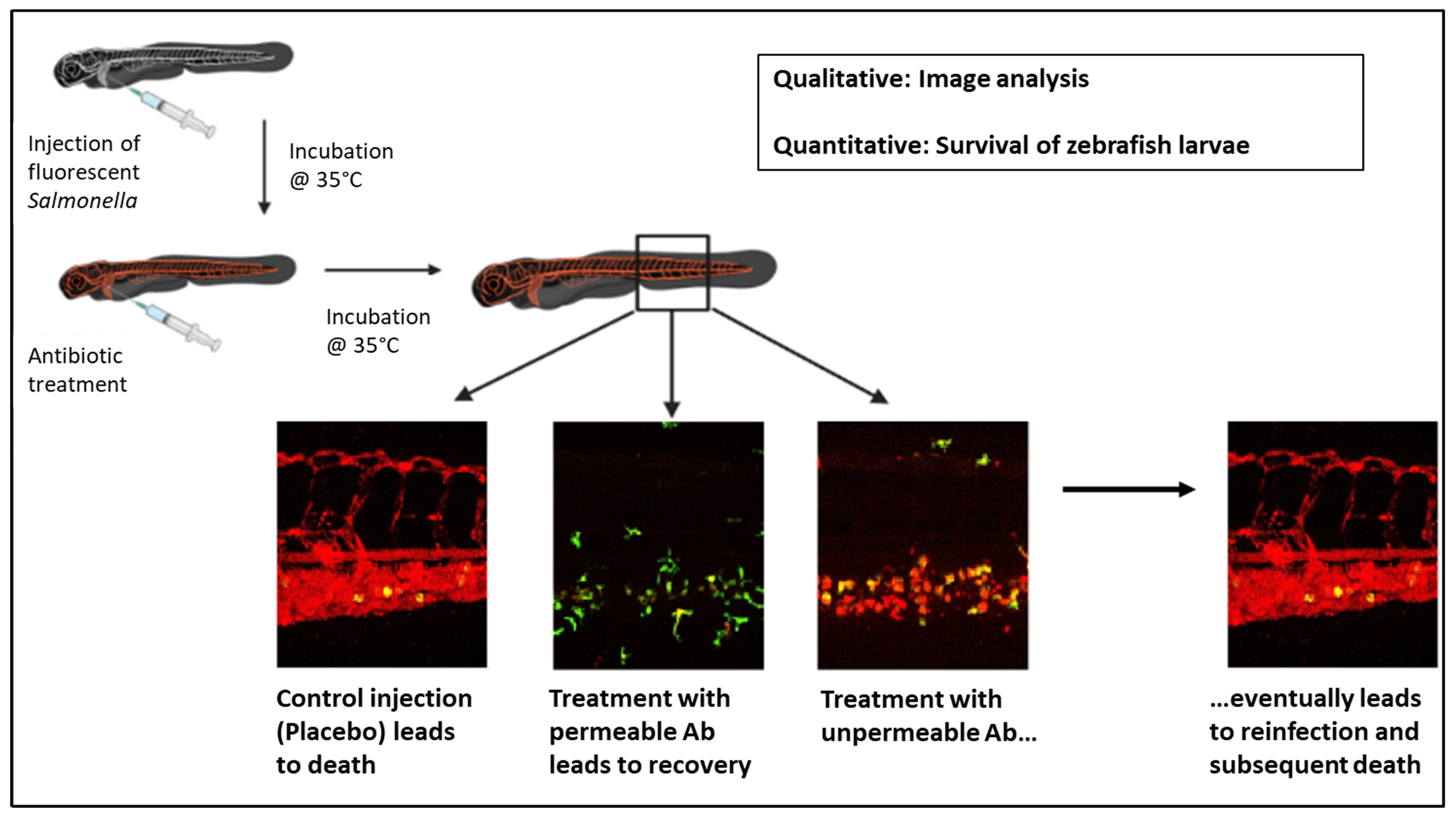
In this work, we describe a zebrafish-larvae-(ZFL)-based vertebrate model, optimized for the screening of antibiotics. We can distinguish between their intra- and extracellular efficiency against Salmonella. The proposed model is an alternative to commonly used rodent models, enabling researchers to reduce the costs for the development of novel anti-infectives or treatment regimens and to increase the throughput of compound screenings. The advantages of ZFL include the ease of nursing and drug injection, their transparency, which allows for fluorescence-based imaging, and a much higher throughput compared to other animal models due to their short reproductive cycle and high number of offspring [21]. In contrast to in vitro screening models, a ZFL-based model makes it possible to obtain pharmacological information, to identify the toxicity of compounds, and to observe the effects of compounds that only work in vivo (e.g., prodrugs, effects on pathogen–host interactions, etc.). Due to its unique properties, the ZFL animal model was already proposed as an early-stage screening tool for studying pharmacological aspects like the circulation behavior and the renal and macrophage clearance of a variety of drugs [22, 23, 24, 25]. Early macrophages in ZFL appear 22 hours post fertilization (hpf). Macrophages are capable of phagocytosing bacteria within the ZFL blood 30 hpf [26, 27]. ZFL have previously been used as infection models including for Salmonella [28, 29, 30] and in antibiotic activity testing against extracellular pathogens [29, 31, 32, 33]. Here, we established ZFL as a model for testing antimicrobials against intracellular Salmonella by infecting genetically engineered zebrafish selectively expressing KAEDE within macrophages [34] with Salmonella expressing fluorescent reporter proteins (GFP and/or mCherry).
Peptone (No. 8952.2), yeast extract (No. 2904.4), and agar (No. 5210.2) were obtained from Carl Roth GmbH, Karlsruhe, Germany. Methanol (Art. No. 34860), triethylamine (No. 471283), triton X-100 (No. 648466), N-phenylthiourea (No. P7629), tricaine (ethyl-3-aminobenzoate methane sulfonate) (No. E10521), ceftriaxone (No. C5793), tobramycin (No. T4014), ciprofloxacin (No. 17850), moxifloxacin (No. SML1581), and gemifloxacin (No SML1625). were purchased from Merck & Cie, Buchs, Switzerland. Ethanol (No. 24562) was obtained from Honeywell, Riedel-de-Haën, Seelze, Germany. Sodium chloride (No. A2942) was obtained from AppliChem GmbH, Darmstadt, Germany. LB Lennox medium: peptone (20 g), yeast extract (10 g), and sodium chloride (10 g) were dissolved in 2 liters of water, and the pH was adjusted to 7. Agar plates: microbiological grade agar (15 g) was dissolved in 1 liter of water and autoclaved for 20 min at 15 psi liquid cycle; 50 μg/mL kanamycin was added, and the medium was distributed to 10 cm petri plates (25 mL per plate) and allowed to cool down to 25 °C. An attenuated Salmonella SDB15 SL1344
Bacteria from a glycol stock kept at –80 °C were streaked on LB agar containing 50 μg/mL kanamycin at 37 °C o/n. A single colony of bacteria was taken the next day, inoculated in 3 mL of LB medium with 50 μg/mL kanamycin, and incubated overnight at 37 °C and 250 rpm. Then 0.5 mL of the culture was added to 50 mL fresh LB medium with 50 μg/mL kanamycin and incubated at 37 °C and 200 rpm. The optical density at 600 nm was measured, and samples were taken from the culture every 15 min for 3 h. The samples were each diluted in 1:10 steps in D-PBS until a final dilution of 10
Bacteria were prepared for injections with an overnight culture followed by inoculation of a 50 mL culture as described above. Then 1 mL samples were taken in 30 min steps until the OD
Zebrafish (Danio rerio) larvae (kindly provided by Prof. Dr. M. Affolter and Dr. H. Belting, University of Basel, Switzerland) were obtained from adult Tg(mpeg1:Gal4;UAS:Kaede) fish [34, 43] with macrophages expressing EGFP or wild-type (AB/TU) fish [44] and were kept at 28 °C in a zebrafish-culture medium supplemented with 30
The ZFL were mechanically dechorionized 2 days post fertilization (dpf) using two jeweler’s forceps (Dumont No. 5, L 4 ¼ in., Inox alloy), anaesthetized with 0.01% tricaine, embedded in 0.3% agarose containing tricaine and PTU, and injected with the indicated amounts of the selected Salmonella strain into the duct of Cuvier (if not stated differently) using a micromanipulator (Wagner Instrumentenbau KG, Schöffengrund, Germany), a pneumatic PicoPump PV830 (WPI, Sarasota, Florida, USA), and a Leica S8APO microscope (Leica, Wetzlar, Germany). From then on, the ZFL were kept at 35 °C until the end of the experiment unless stated differently. At the indicated time points, the infected ZFL were injected with the given amounts of antibiotics.
At the indicated time points, the fish were imaged using either a confocal-laser scanning microscope (either an SP5-II-MATRIX, Leica, Wetzlar, Germany, equipped with a 25x HCX IRAPO L (NA 0.95) objective or an Olympus FV 1000 inverted microscope, Olympus Ltd, Tokyo, Japan, with a 20x UPLSAPO (NA 0.75) objective) and, where indicated, the number of surviving fish was counted. Visual inspection to discriminate between living and dead larvae was done with a Leica S8APO microscope (Leica, Wetzlar, Germany). The presence of a heartbeat was chosen as the survival criterion. Only living larvae (with an observable heartbeat) were used for confocal imaging. The image analysis was carried out using Fiji ImageJ v. 1.52n (U.S. National Institutes of Health, Bethesda, Maryland, USA) and OMERO.web v. 5.9.1 (https://www.openmicroscopy.org/). A minimum of 3 ZFL were imaged in all settings. The area for the quantification of the fluorescent area fraction was chosen based on a phase-contrast image of the tail region of one ZFL. Subsequent measurements in the same experiment were done based on the same region. The statistical analysis and data plotting was carried out using OriginPro 2018 (64-bit) SR1 b9.5.1.195 (Academic) Software (OriginLab Corporation, Northampton, MA, USA). A minimum of 10 ZFL per group were used for survival analysis.
To evaluate the appropriate number of colony-forming units (CFU) that need to be injected into the ZFL for drug screening, we injected either 30,000, 20,000, 10,000, 7500, 5000, 3000, or 1500 CFU Salmonella into the ZFL and kept them at 28 °C. At each inoculum size, the Salmonellae were easily detectable within the whole ZFL using confocal imaging (Supplementary Figs. 1,2). Some colocalization of the green (KAEDE of the macrophages) and the red (mCherry of Salmonella) fluorescence could be observed 2 h post injection of 3000 and 1500 CFU, indicating a fraction of intracellular Salmonella, while other Salmonella seemed to remain in the vasculature. After injections lower than 5000 CFU, the red mCherry signal of Salmonella largely vanished from ZFL circulation within 24 h. Survival was clearly observable to be dependent on the dose: doses higher than 5000 CFU led to a rapid death of the fish within 24 h.
We tested infections at 35 °C, which lies within the optimal temperature range for Salmonella growth (35–37 °C) [45]. While 35 °C is still well tolerated by ZFL, higher temperatures (nearer to 37 °C) lead to an unsatisfactory survival rate [46]. Keeping the model as close as possible to human body temperature is important since the efficacy of some antibiotics is highly temperature dependent. For example, the EC
We determined the impact of ceftriaxone and tobramycin treatment on ZFL infected with 300 CFU of Salmonella. Differentiating between intra- and extracellular efficiency is a major strength of the proposed model. Ceftriaxone and tobramycin were chosen to validate the model because of their known distinct efficiencies in killing intracellular Salmonella [48, 49]. Ceftriaxone is used to treat salmonellosis in humans [50].
The ED
 Fig. 1.
Fig. 1.Infection-model validation: Treatment of systemic Salmonella infection with ceftriaxone or tobramycin. (A) Experimental procedure. ZFL expressing KAEDE (green) under macrophage-specific promoter mpeg, were injected intra venously (i.v.) with mCherry-expressing Salmonella (300 CFU) and incubated at 35 °C. Then 1 h after Salmonella injection, the fish were injected i.v. with either PBS (control), ceftriaxone, or tobramycin (600 pg per antibiotic per fish). Confocal images were taken 7 h and 1 day after Salmonella injection, and survival studies were carried out until 2 days post Salmonella injection. (B) Kaplan–Meier curve of survival studies 0–48 h after Salmonella injection. Survival probability including 95% confidence intervals (dotted lines, n
We also tested a model using ZFE embryos infected with DNA-damage reporter Salmonella, in which genotoxic stress induces red and green fluorescence. These reporter Salmonella carry the following two promoter fusions responding to DNA damage on a pSC101 backbone: The Pcad promoter of colicin D fused to gfp-ova encoding an unstable variant of the green fluorescent protein and the PrecA promoter fused to mCherry [38, 39, 40, 41, 42]. PrecA has moderate activity even in absence of genotoxic stress resulting in baseline red fluorescence of the reporter strain, with an increasing mCherry expression and red fluorescence after genotoxic stress. A detectable GFP signal is expected to only appear after induction of a DNA damage response. Infection of ZFE with these bacteria without any additional treatment yielded detectable mCherry but no GFPsignals, as expected (Fig. 2).
 Fig. 2.
Fig. 2.The effect of gyrase inhibitors on SOS-DNA damage-reporter Salmonella in ZFL blood vessels. (A) ZFL were injected i.v. with SOS-DNA damage-reporter Salmonella (300 CFU) and incubated at 35 °C. Reporter Salmonella constitutively express mCherry with an increased mCherry expression after DNA damage. gfp is exclusively expressed after DNA damage. Ninety min after Salmonella injection, the fish received either no treatment (control), or an i.v. injection of either ciprofloxacin, moxifloxacin, or gemifloxacin (600 pg of antibiotic per fish). Confocal images were taken 90 min after the antibiotic injection. (B) Confocal images of GFP and mCherry fluorescence. Green: GFP (cda promoter), red: mCherry (recA promoter). (C) Percentages of areas with GFP or mCherry fluorescence, respectively, 90 min after antibiotic injection. Bars: average; horizontal line: median; whiskers: SE; n
To validate the capability of this model to assess the in vivo efficiency of antibiotic compounds targeting bacterial DNA in- and outside host Macrophages, we tested the effect of 3 distinct fluroquinolones (which inhibit DNA gyrase and cause DNA damage), known to be effective against Salmonella, namely ciprofloxacin, moxifloxacin and gemifloxacin [53, 54, 55].
After administering all three of the tested fluoroquinolones, GFP and intensified mCherry fluorescence appeared, indicating that the antibiotics had reached their Salmonella targets (Fig. 2). To evaluate the effectiveness of gyrase inhibitors on intracellular Salmonella, extracellular Salmonella were eradicated by co-administering tobramycin. The remaining intracellular Salmonella still reported substantial DNA damage consistent with excellent cell penetration of the fluoroquinolones (Fig. 3).
 Fig. 3.
Fig. 3.The effect of gyrase inhibitors on SOS-DNA damage-reporter Salmonella within Salmonella containing vacuoles (SCVs). ZFL were injected i.v. with SOS-DNA damage-reporter Salmonella (300 CFU) and incubated at 35 °C. Reporter Salmonellae constantively express mCherry, with an increased mCherry expression after DNA damage. gfp is exclusively expressed after DNA damage. (A) Sites of imaging. Imaging of (B): A2 and of (D): A1. (B) GFP and mCherry fluorescence (DNA damage response within SCVs) 4 h after moxifloxacin injection. Ninety min after Salmonella injection, fish were injected i.v. with either 600 pg tobramycin only or 600 pg tobramycin and 600 pg moxifloxacin. Confocal images were taken 4 h after antibiotic injection. (C) Percentages of areas with GFP or mCherry expression, respectively, 4 h after antibiotic injection. (D) GFP and mCherry expression (DNA damage response within SCVs) 1 day after gyrase inhibitor treatment. 90 min after Salmonella injection, fish were injected i.v. with 600 pg tobramycin to eradicate free Salmonella outside of SCVs. Ninety min after tobramycin injection, ZFL received either no second treatment (control), or a second injection of either ciprofloxacin, moxifloxacin, or gemifloxacin (600 pg per antibiotic per fish). Confocal images were taken 1 day post Salmonella injection. (E) Percentages of total areas with GFP signal 1 day after gyrase inhibitor treatment. (B) and (D): Green: GFP (cda promoter), red: mCherry (recA promoter). (C) and (E): Bars: average; horizontal line: median; whiskers: SE; n
Testing antimicrobials in vivo in rodents is resource consuming and raises ethical concerns. Efficacy data are usually end-point measurements only. ZFL are easy to generate in large numbers, and their transparency enables real-time monitoring of infections and treatment responses using fluorescence microscopy. We established suitable infection conditions and localized Salmonella inside macrophages using fluorescent zebrafish lines and fluorescent Salmonella strains. Using these methods, we could demonstrate differential access of tobramycin and ceftriaxone to intra- versus extracellular Salmonella. Using DNA damage-reporter, we could even directly monitor the action of fluoroquinolones on extra- and intracellular Salmonella. The results from this ZFL model were entirely consistent with the well-characterized permeability properties of the three antibiotics and their suitability for treating systemic Salmonella infections in human patients [56, 57, 58, 59]. Even the effective doses of tobramycin were comparable to recommendations for extracellular infections in humans. Our ZFL model thus appeared to be suitable as an informative and predictive in vivo model for antimicrobial-activity testing against intra- and extracellular pathogens in vivo, closing the gap between in vitro assays and rodent models.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
PH, JB, DB, and JH designed the research study. PH, JB, RP, CLA, and SS performed the research. JF, BC, and DB provided help and advice on bacteria cultivation and the design of the infection models. PH, JB, and RP analyzed the data. PH, DB, and JH wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity.
All fish were kept in accordance with Swiss animal-welfare regulations [22, 23].
We thank the group of Prof. Markus Affolter, Biozentrum, University of Basel, and particularly the group member Dr. Heinz-Georg Belting for providing us with zebrafish larvae and their valuable inputs. Illustrations were created with https://www.biorender.com/.
This research was funded by a grant (SCAHT-GL 21-09) from the Swiss Centre for Applied Human Toxicology – SCAHT and by the Swiss Nanoscience Institute (SNI-P1801).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.



