1 China International Science and Technology Cooperation Base of Food Nutrition/Safety and Medicinal Chemistry, College of Bioengineering, Tianjin University of Science & Technology, 300457 Tianjin, China
2 Inner Mongolia Academy of Agricultural & Animal Husbandry Science, 010031 Hohhot, Inner Mongolia, China
3 School of life Sciences, Inner Mongolia University, 010070 Hohhot, Inner Mongolia, China
Academic Editor: Ananda Ayyappan Jaguva Vasudevan
Abstract
Fatty acids (FAs) are critical nutrients that regulate an organism’s health and
development in mammal. Long-chain fatty acids (LCFAs) can be divided into
saturated and unsaturated fatty acids, depending on whether the carbon chain
contains at least 1 double bond. The fatty acids that are required for humans and
animals are obtained primarily from dietary sources, and LCFAs are absorbed from
outside of cells in mammals. LCFAs enter cells through several mechanisms,
including passive diffusion and protein-mediated translocation across the plasma
membrane, the latter in which FA translocase (FAT/CD36), plasma membrane
FA-binding protein (FABPpm), FA transport
protein (FATP), and caveolin-1 are believed to have important functions. The
LCFAs that are taken up by cells bind to FA-binding proteins (FABPs) and are
transported to the specific organelles, where they are activated into acyl-CoA to
target specific metabolic pathways. LCFA-CoAs can be esterified to phospholipids,
triacylglycerol, cholesteryl ester, and other specialized lipids. Non-esterified
free fatty acids are preferentially stored as triacylglycerol molecules. The main
pathway by which fatty acids are catabolized is
Keywords
- fatty acid sensing
- lipid metabolism
- mTORC1
- homeostasis
In addition to serving as building blocks for lipid synthesis, fatty acids (FAs) are needed for membrane function, energy storage, and signaling. An adequate supplies of fatty acids is important for maintaining metabolism and energy homeostasis in cells. Animal cells obtain fatty acids primarily by extracellular uptake, de novo synthesis, and hydrolytic cleavage of ester bonds in triacylglycerol stored in tissues [1, 2, 3]. Moreover, essential fatty acids can be synthesized by rumen microorganisms in ruminant animals [4, 5, 6].
Fatty acids are classified by their carbon (C) chain length and degree of desaturation, each of which differs content in living cells or milk [7, 8]. Long-chain (LC) fatty acids refer to fatty acids with a chain length of 11/12–20 carbons and are precursors of various lipids that participate in various physiological processes, e.g., cellular metabolism, energy homeostasis, and cell proliferation [9, 10]. In general, Long-chain fatty acids (LCFAs) synthesis was found to vary, depending on the tissue type, the contribution of fatty acid synthesis de novo to the whole fatty acid pool is not dominant, and they are absorbed primarily from the outside of cells [11, 12]. The LCFAs that are taken up by cells are managed in many metabolic cascades, including their release from the inner leaflet of the plasma membrane, transport to specific organelles, and activation in cells. Mammalian cells possess the ability to properly sense both extracellular and intracellular nutrients for the maintenance of metabolic homeostasis, including lipid homoeostasis. However, the sensing mechanism by which fatty acids are taken up and used by human or animal cells is not fully understood.
Fatty acids are components of intracellular lipids, which store energy in the
form of triacylglycerol in mammalian cells. Fatty acids are classified by their
carbon (C) chain length. Short-chain (SC) FAs have a chain length of between 1
and 4 C atoms, comprising acetic (2:0),
propionic (3:0), and butyric (4:0) acids, and medium-chain (MC) FAs have lengths
of 6-10 Cs, including caproic (6:0), caprylic
(8:0), and capric (10:0) acids [13]. LCFAs have chain length of 11/12–20 Cs,
e.g., lauric (12:0), myristic (14:0), palmtic (16: 0), stearic (18:0), and
arachidic acid (20:0), of which C16/18 LCFAs are the most abundant FA species in
mammalian cells [13, 14]. Very long-chain fatty acids (VLCFAs) are defined as FAs
with C
The free fatty acids that are required for humans and animals are derived from endogenous synthesis, or come from exogenous sources. Long- and medium-chain fatty acids derived mainly from dietary triacylglycerol, and short-chain fatty acids (SCFAs), also called volatile FAs, produced by gut microbial fermentation—in particular, acetate, propionate and butyrate [18, 19]. Medium-chain fatty acids (MCFAs) are absorbed from dietary plant oils and milk directly into the portal blood, e.g., capric acid in coconut oil [13, 20]. In addition to being provided by the diet, SCFAs and MCFAs can be formed in mammalian and human tissues-primarily the liver, mammary gland, and adipose tissue [20]. LCFAs/LCPUFAs and VLCFAs, including SFAs, MUFAs, and PUFAs, are obtained mainly from the diet [13, 21], whereas sources of unsaturated fatty acids are vegetable oils, e.g., oleic acids in olive oil (Table 1, Ref. [13, 15, 19, 20, 21]) [13].
| Fatty acids (FAs) classification | Dietary sources | In mammalian and human tissues | Reference |
| SCFAs: 1 and 4 C atoms | Produced chiefly by anaerobic fermentation and the metabolism of dietary fiber by gut rumen microbes. | Primarily the liver, mammary gland, and adipose tissue. | [19] |
| MCFAs: 6-10 C atoms | Absorbed from dietary plant oils and milk directly into the portal blood, e.g., coconut oil. | Primarily the liver, mammary gland, and adipose tissue. | [20] |
| LCFAs: 11/12-20 C atoms | Diet, e.g., olive oil. | [13, 21] | |
| VLCFAs: C |
Obtained mainly from the diet and through elongated and desaturated of endogenous FAs, e.g., peanut oil. | Skin, retina, meibomian gland, testis, and brain. | [15] |
| FAs, Fatty Acids; LCFAs, Long-chain fatty acids; MCFAs, Medium-chain fatty acids; SCFAs, Short-chain fatty acids; VLCFAs, Very long-chain fatty acids. | |||
VLCFAs are synthesized through elongation and desaturation of endogenous FAs,
such as palmitic acid (C16:0) and stearic acid (C18:0), in animals and humans
[15]. In the udder of dairy cows, FAs with C4:0-C14:0 and approximately 50% of
C16:0 are synthesized de novo from acetate and
In addition to external uptake and de novo synthesis, mammalian cells can obtain fatty acids through the hydrolytic cleavage of ester bonds in triacylglycerol from fat stores to maintain fatty acids homeostasis under fasting conditions [23, 24]. Dietary fat was broken down by lipases in small intestine to degrade triacylglycerol (TG) in human. The breakdown of TG generates fatty acids, glycerol and monoglycerides. In the process, three major lipases have been identified, including adipose triglyceride lipase (ATGL), hormonesensitive lipase (HSL) and monoglyceride lipase (MGL), which sequentially performs the TG hydrolysis generating diglycerides (DGs) and FAs, DGs hydrolysis generating monoglycerides (MGs) and MGs hydrolysis generates glycerol and the third FA [23]. In addition, the hydrolysis of cholesteryl esters also generates free fatty acids in human cells. Cholesterol can be derived from dietary sources in the intestine, and do novo synthesized in liver. Both endogenously synthesized and exogenously acquired cholesterol are processed into low-density lipoprotein chiolesterol (LDL-C) in blodstream, which can be taken up by peripheral cells. Excess cholesterol is esterified by acyl coenzyme A: cholesterol acyltransferase (ACAT) to cholesteryl esters for storage in lipid droplets in cells [24]. The cycle of esterification and hydrolysis of cholesterol esters is one of the important element for the lipid homoeostasis in cells. In spite of this, de novo synthesis of fatty acids constitutes a minor but necessary source (Fig. 1).
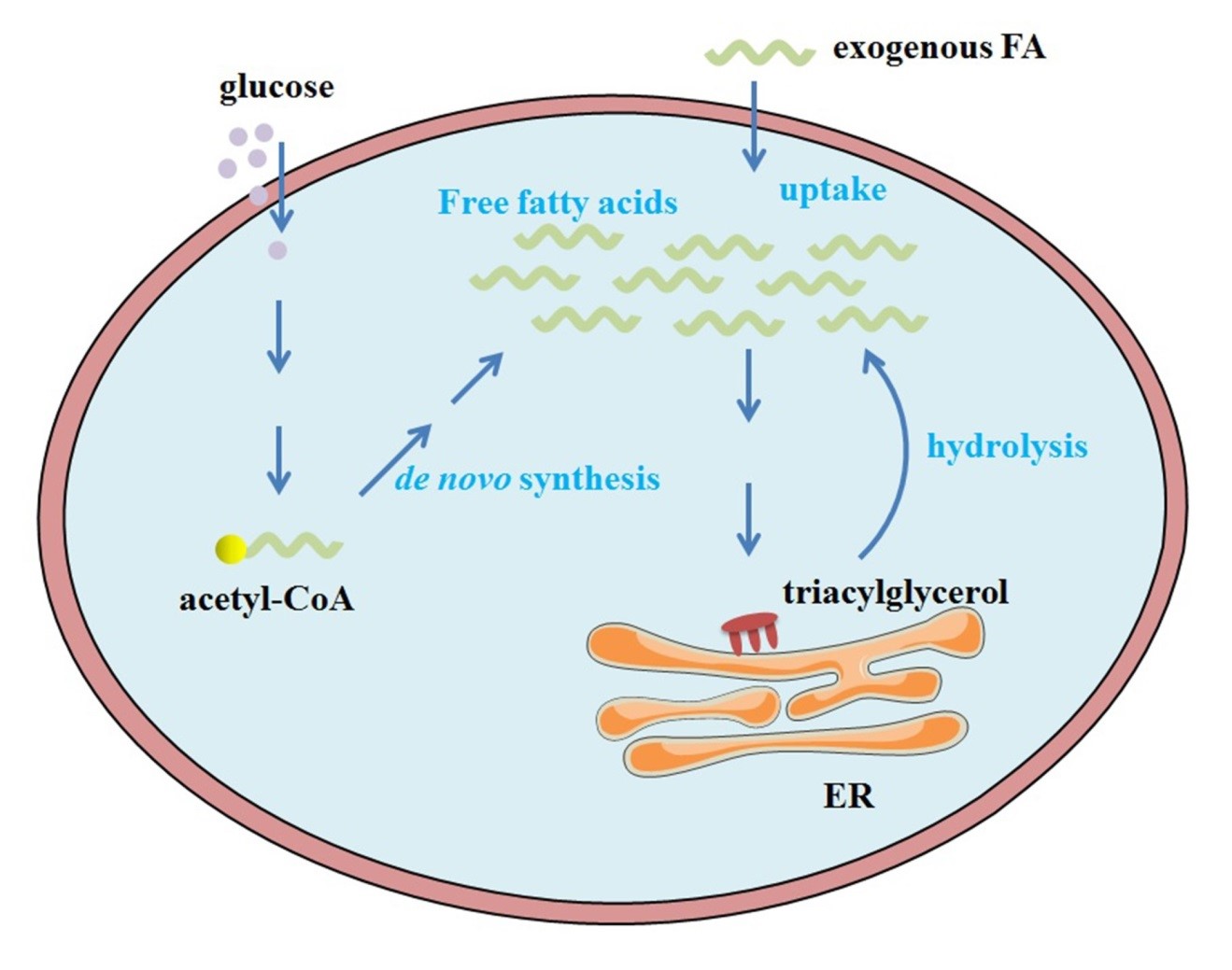 Fig. 1.
Fig. 1.The source of intracellular FAs. In mammalian cells, cellular FAs are generally absorbed from outside of cells; de novo synthesis of FAs is a minor but necessary pathway, and the catabolism of triacylglycerol to complement FAs occurs during nutrient deprivation. FAs, Fatty Acids; ER, endoplasmic reticulum.
LCFA uptake by cells is important in maintaining lipid homeostasis. Most LCFAs that circulate in body fluid exist in the form of free LCFA, complexes with albumin, and local lipoprotein lipase (LPL)-mediated LCFA release from membrane bound lipoprotein [25]. The cellular uptake of exogenous LCFAs for use in cells occurs through a cascade, comprising the dissociation of FAs from albumin-FA complexes and their binding to plasma membrane proteins, FA translocation across the plasma membrane, binding cytoplasmic FABP on the inner plasma membrane, and the activation of LCFAs into acyl-CoA, which is necessary for such metabolic processes as TG synthesis and oxidation [17].
The first step is the release of FAs from albumin-FA complexes for presentation to the cell surface [25]—a process that remains poorly understood. The dissociation of FAs from albumin-FA complexes is believed to be facilitated by membrane-associated proteins, including FA translocase (FAT/CD36), plasma membrane FA-binding protein (FABPpm), and FA transport protein (FATP) [26]. FAT/CD36, FABPpm, FATP, and caveolin-1 are thought to mediate the translocation of LCFAs across the plasma membrane.
In 1993, a cDNA clone from a rat adipocyte cDNA library was isolated by screening and implicated in the transport of LCFAs. It was found to be homologous to human CD36 and was termed FAT/CD36 [27]. CD36 is a multifunctional membrane protein, and its relative molecular weight depends on its post-translational modification [28]. Glycosylated CD36 increases the uptake of LCFAs [29]. Its structure is divided into 5 regions: carboxy-terminal (COOH-terminal) and amino-terminal cytoplasmic domains (NH2-terminal), 2 transmembrane regions, and an extracellular domain. The COOH-terminus contains 2 palmitoylation sites and 2 ubiquitination sites, and the NH2-terminus contains only 2 palmitoylation sites—palmitoylated CD36 is located in the lipid rafts of the cell membrane where it mediates adsorption and transport of fatty acids [29, 30].
The extracellular domain is a large, highly glycosylated hydrophobic neck ring and contains 3 pairs of disulfide bonds, 10 glycosylation sites, and 2 phosphorylation sites. These modified sites can interact with a variety of extracellular substances, such as oxidized low-density lipoprotein (ox-LDL) and LCFAs [30, 31]. In dairy cows and goats, CD36 is expressed by the mammary glands and respond to LCFAs to improve milk lipid synthesis [32, 33]. Palmitic acid upregulates CD36 and promotes its translocation from the cytoplasm to plasma membrane in mouse podocytes [34]. CD36 appears to be the most important translocator of FAs, based on current evidence.
In 1985, FABPpm was isolated and identified by the Berk group from rat liver plasma membranes and jejunal microvillous membranes [35], adipocytes [36], and cardiac myocytes [37]. Subsequently, the protein was determined to be identical to the mitochondrial isoenzyme glutamic-oxaloacetic transaminase (mGOT)/mitochondrial aspartate aminotransferase (mAspAT) [35]. FABPpm is anchored to the outer leaflet of the plasma membrane, with its hydrophobic tail binding to FAs with high affinity, their facilitating dissociation from albumin [17]. The overexpression of FABPpm in vitro or in vivo increases the rate of LCFA transport and metabolism [38, 39, 40]. The mechanism by which FABPpm transports LCFAs is not fully understood.
The FATP family comprises 6 highly homologous FA transport proteins in human/mouse/rat [41], also known as solute carrier protein family 27A (SLC27A) [42, 43]. FATP1, originally called FATP, the first of these proteins, was identified by Schaffer and Lodish in 1994 with an expression cloning strategy and a cDNA library from 3T3-L1 adipocytes. FATP1 is localized to the plasma membrane and augments the uptake of LCFAs when expressed in cultured cells [44]. FATP1 has a distinct membrane topology, and its N-terminus lies outside of the plasma membrane. Amino acids 1-190 contains at least 1 transmembrane domain, the fragment from 190–257 contains the AMP-binding motif, and the C-terminus faces the cytosolic space [45, 46]. FATP1-6 are expressed in a variety of tissues and, as membrane proteins, are associated with the import of FAs [41, 47, 48].
In addition to promoting cellular FA uptake, FATP has acyl-coA synthase activity that is central in downstream metabolic pathways, and overexpression of FATP1 increased acyl-CoA synthetase activity and fatty acid uptake in 3T3-L1 adipocytes [17, 49]. A constitutive interaction between FATP1 and A ligases/acyl-CoA synthetase 1 (ACSL1) contributes to the efficient cellular uptake of LCFAs in adipocytes [48, 50]. FATP1 and FATP4 localize to the endoplasmic reticulum to facilitate uptake and utilization of LCFAs by catalyzing the esterification of FAs with CoA [48, 49, 51].
Caveolae, specialized rafts, are 50- to 100-nm flask-shaped invaginations of the cell surface plasma membrane that are found in many cell types; transcytosis is one of the first functions of caveolae that transport macromolecules into the cell [52, 53]. Caveolae are associated with signal transduction and endocytosis of pathogens [54]. Pohl et al. [55] found a significant function for caveolae-mediated uptake and intracellular trafficking of LCFAs in HepG2 cells.
The biogenesis and function of caveolae depend on caveolins, of which there are 3 in mammals: Cav-1, -2, and -3 [53]. Caveolin-1 is the principal marker and major structural protein of caveolae, and its loss results in a complete lack of caveolae from the plasma membrane and influences FA uptake by regulating the availability of FAT/CD36 at the surface [56, 57]. Cav-1 can bind FAs directly [58], and the transport of FAs across the plasma membrane is modulated by caveolin-1 [59, 60]. Further, caveolin-1 accumulates LCFAs on the inner leaflet and presents them to cytoplasmic FABPs for further shuttling of LCFAs to various organelles [61].
Flip-flop. In addition to the 4 proteins above that help transport LCFAs across the plasma membrane, passive diffusion moves LCFAs from one half of the bilayer to the other—through the so-called “flip-flop” mechanism of diffusion (Table 2, Ref. [32, 33, 38, 39, 44, 53, 55, 62, 63]) [62, 63]. In early research on FA transport, passive diffusion was suggested and proven in a protein-free model of the phospholipid bilayer, and it was believed that this mechanism was efficient for shuttling FAs through simple membrane models [64]. However, this mode of LCFA transport across cell membranes remained controversial until now, and the focus has shifted to transport efficiency [65].
| Name | Function | Molecular mass | Mechanism | Reference |
| FAT/CD36 | Protein mediated facilitated diffusion | 78–88 kDa | CD36 is expressed by the mammary glands and respond to LCFAs to improve milk lipid synthesis. | [32, 33] |
| FABPpm | Protein mediated facilitated diffusion | 40–43 kDa | The overexpression of FABPpm increases the rate of LCFA transport and metabolism. | [38, 39] |
| FATP | Protein mediated facilitated diffusion | 63 kDa | FATP1 augments the uptake of LCFAs when expressed in cultured cells. | [44] |
| Caveolin-1 | Protein mediated facilitated diffusion | 17–24 kDa | Function for caveolae-mediated uptake and intracellular trafficcking of LCFAs. | [53, 55] |
| Flippases, floppases and scramblases | Passive diffusion | Passive diffusion moves LCFAs from one half of the bilayer to the other—through the so-called “flip-flop” mechanism of diffusion. | [62, 63] | |
| FAT/CD36, FA translocase; FABPpm, plasma membrane FA-binding protein; FATP, FA transport protein; LCFA, Long-chain fatty acid; flippases, P4-ATPases; Floppases, ATP-binding cassette (ABC) transporters, ATP-Binding Cassette A1 (ABCA1); Scramblases, the TMEM16 family (anoctamins). | ||||
The free fatty acid transport model through the cell membrane is divided into 3 steps: adsorption of the FA to the membrane, translocation across the membrane (“flip-flop”), and subsequent desorption of the fatty acid into the cytosol [63, 66]. Several groups have suggested that the “flip-flop” step is not limiting—that desorption is the rate-limiting step in the phospholipid bilayer [67, 68]—and that FAs can diffuse freely by flip-flopping, even in the biological membrane [69, 70]. In contrast, other studies have suggested that the flip-flop of LCFAs is prohibitively slow and that transbilayer flip-flopping is rate-limiting in lipid bilayer membranes—that flip-flop across the lipid phase alone is unable to support the metabolic requirements of cells [66].
Although the debate regarding whether this process is fast or slow continues,
many proteins that are associated with flip-flop have been identified [71, 72],
falling into 3 broad categories: flippases, floppases, and scramblases; the first
2 groups are ATP-dependent, whereas scramblases facilitate the bidirectional
movement of lipids in an ATP-independent manner [73]. Type 4 P-type ATPases
(P4-ATPases) are flippases that mediate out-to-in lipid movement through the
plasma membrane, from the exoplasmic to cytosolic side [74]. Floppases are
transmembrane ATP-binding cassette (ABC) transporters that use the hydrolysis of
ATP to facilitate the in-to-out movement of various substrates across the cell
membrane [75]. ATP-Binding Cassette A1 (ABCA1) flops cholesterol from the inner
to the outer leaflet of the plasma membrane [76]. The TMEM16 family of proteins,
also known as anoctamins, contains lipid scramblases that are activated by
increases in intracellular Ca
The lipid flip-flop model (passive diffusion) has been challenged by the discovery of several transporters that mediate LCFA translocation across the plasma membrane [27, 36, 44], generating alternative hypotheses, such as passive diffusion and protein-mediated FA transport through membranes (Fig. 2). It is likely that membrane proteins are important in the transmembrane transport of FAs across the plasma membrane.
 Fig. 2.
Fig. 2.Cellular uptake of LCFAs. Non-esterified LCFAs in blood exist as complexes with serum albumin. The cellular uptake of exogenous LCFAs requires the dissociation of FAs from albumin-FA complexes. Free LCFAs enter cells through passive diffusion and protein-mediated translocation across the plasma membrane. The free FA transport model alone, through a “flip-flop” mechanism across the lipid phase, is unable to support metabolic requirements, and the membrane proteins FAT/CD36, FABPpm, FATP, and caveolin-1 play key roles in LCFA translocation across the plasma membrane. The proteins associated with “flip-flop” fall into three broad categories: flippases, floppases, and scramblases, the first 2 of which are ATP-dependent, whereas scramblases facilitate the bidirectional movement of lipids in an ATP-independent manner. Membrane proteins play key roles in the transmembrane transport of LCFAs across the plasma membrane. ABC-transporters, ATP-binding cassette transporters; ACSL, A ligases/acyl-CoA synthetase; ER, endoplasmic reticulum; FAT/CD36, FA translocase; FABPpm, plasma membrane FA-binding protein; FATP, FA transport protein; LCFA, Long-chain fatty acid; P4-ATPases, Type 4 P-type ATPases; SLC27A, solute carrier protein family 27A.
LCFAs are released from the inner leaflet of the plasma membrane into the cytoplasm after translocation across the plasma membrane by passive diffusion or protein-mediated diffusion [17]. The released FAs bind to cytoplasmic FA-binding proteins (FABPs), which are LCFA carriers with high cytosolic concentration [17, 78] to transport LCFAs to sites of metabolic conversion (e.g., oxidation, esterification) or subcellular targets [79, 80]. Cytoplasmic FABPs comprise a family of proteins that bind LCFAs with high affinity [81, 82], and at least 9 FABPs have been identified in human [82, 83]. FABPs have tissue-specific expression patterns and abound in tissues with active FA metabolism [84, 85].
The primary function of FABP family members is LCFA intracellular transport;
they might also promote LCFA desorption from the cytoplasmic face of the plasma
membrane [81]. Further, desorbed LCFAs are bound by FABP and transported to
various organelles for metabolism, such as mitochondria for
In this review, we discuss the main metabolic pathways of palmitic acid (PA,
C16:0) and stearic acid (SA, C18:0) in cells as examples of LCFAs metabolism
(Fig. 3). Cytoplasmic non-esterified free palmitic acid or stearic acid is
activated to palmityl-CoA or stearoyl-CoA by
ACSLs [92, 93] and then used in various metabolic pathways. Metabolically,
intracellular PA and SA primarily undergo esterification,
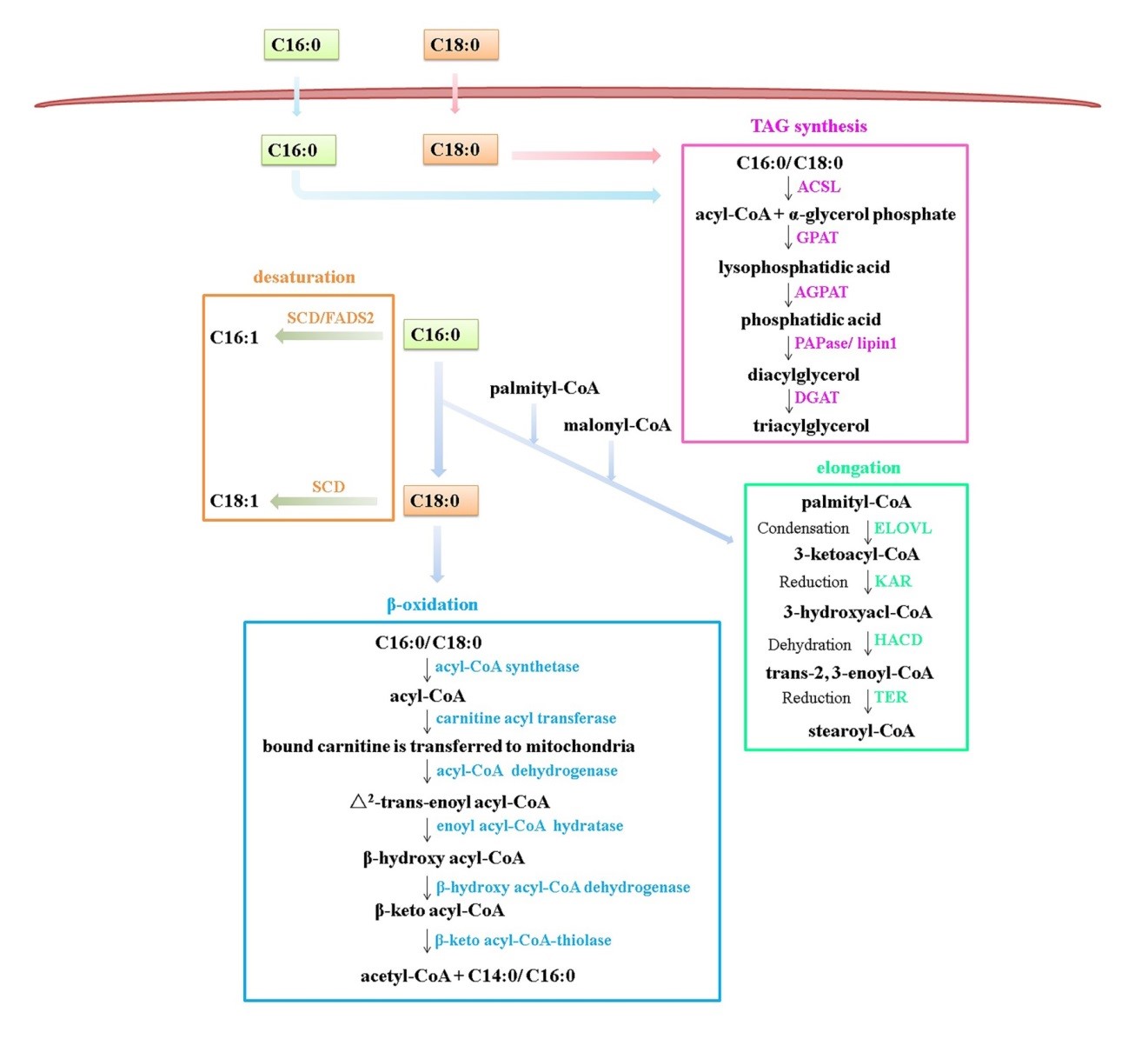 Fig. 3.
Fig. 3.Intracellular LCFA metabolic pathways. Cytoplasmic
non-esterified free palmitic acid (PA, C16:0) or stearic acid (SA, C18:0) must be
activated to acyl-CoA by their acyl-CoA synthetase to enable them to target
specific metabolic pathways—primarily esterification,
Esterification is the chief means by which FAs are used as cellular energy stores. After entering cells, non-esterified free LCFAs are activated by their acyl-CoA synthetase and targeted to phospholipids, triacylglycerol, or cholesteryl esters by FABP. NEFAs are preferentially stored as triacylglycerol via the sequential activities of glycerol-3-phosphate acyltransferase (GPAT), 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), PAPase/Lipin 1, and diacylglycerol acyltransferase (DGAT) [94, 95]. The triacylglycerol structure and interesterification of palmitic and stearic acids likely affect the physical characteristics of fat [96, 97] and their oxidative stability [98]. Recent reports revealed the crystal structure of the lipin/Pah phosphatidic acid phosphatase, suggesting its mechanisms [99], wherein the middle lipin domain adopts a novel membrane-binding dimeric protein fold, with which the functions of lipin oligomerization can be determine in the regulation of phospholipid and triacylglycerol synthesis [100]. In addition, LCFAs stored in phospholipids are presented in the plasma membrane. The arachidonic acid (AA), a polyunsaturated 20 carbon fatty acid, is released when the membrane phospholipids were hydrolyzed by phospholipase A2 (PLA2). AA can be subsequently metabolized by enzymes to generate prostaglandins (PGs) and other productions of the eicosanoid, which are involved in the regulation of many biological processes [101].
Desaturation or elongation converts LCFAs into other types of FAs. The
desaturation of LCFAs is ubiquitous—e.g., the saturated stearic acid (C18:0) is
converted to monounsaturated oleic acid (C18:1n-9) by stearoyl CoA desaturase
(SCD) [108]. SCD has 2 isoforms (SCD1 and SCD5) in humans and catalyzes
LCFAs can be used to synthesize membrane or other phospholipids, for which LCFAs are often elongated [111, 112]. In metabolic experiments with [1-14C]-labeled myristic acid (C14:0) and palmitic acid (16:0), myristic acid was strongly elongated to radiolabeled palmitic acid, and palmitic acid was lengthened to stearic acid [113]. Palmitic acid (16:0), which is synthesized de novo and taken up from the diet, can be elongated into stearic acid (C18:0) and further to VLCFAs. Formation of VLCFAs is performed mainly in the endoplasmic reticulum by membrane-bound enzymes [114], and elongation occurs by cycling through a 4-step process (condensation, reduction, dehydration, and reduction), corresponding to 4 enzymes: elongases (elongation of very long chain FAs, ELOVL), 3-ketoacyl-CoA reductase (KAR), 3-hydroxyacyl-CoA dehydratase (HACD), and trans-2, 3-enoyl-CoA reductase (TER) (Table 3, Ref. [94, 95, 102, 108, 109, 111, 112] ).
| Metabolic pathways | Function | Products | Reference |
| Esterification | The chief means by which FAs are used to form triacylglycerol and complex lipids. | phospholipid, triacylglycerol, or cholesteryl ester | [94, 95] |
| The main pathway of LCFA catabolism, which occurs in the mitochondria and peroxisome. | acetyl-CoA | [102] | |
| Desaturation | Desaturation or elongation converts LCFAs into other types of FAs. | C16:1, C18:1 | [108, 109] |
| Elongation | LCFAs can be used to synthesize membrane or other phospholipids, for which LCFAs are often elongated. | stearoyl-CoA | [111, 112] |
| FAs, Fatty acids; FADS2, Fatty acid desaturase 2; LCFAs, Long-chain fatty acids; SCD, stearoyl-CoA desaturase. | |||
VLCFA elongation has been reviewed extensively [114]. The condensation that is catalyzed by elongase (ELOVL) is the rate-limiting step in the sequential VLCFA elongation cycle, and a recent study reported the first crystal structure of a membrane-bound FA elongase-condensing enzyme, revealing a new reaction mechanism: FA elongation by ELOVL depends on a histidine nucleophile [115, 116]. These novel crystal structures of lipid metabolism enzymes provide important insights into the reaction mechanism of LCFAs.
Mammalian cells must adapt their metabolism to maintain their energy homoeostasis and respond to nutrient availability during their proliferation. Thus, the ability to properly sense both ingested and circulating nutrients is crucial for the maintenance of metabolic homeostasis [117]. Cellular nutrient sensing mechanisms engage anabolism and storage when food abundance, and scarcity triggers homeostatic mechanisms, such as uptake from outside the cell or the mobilization of internal stores [118].
Mammalian target of rapamycin (mTOR), which is now referred to as mechanistic target of rapamycin, has been implicated as a sensor of nutrient sufficiency in cells and is activated by essential amino acids, glucose, and phosphatidic acid (PA) [119, 120, 121]. mTOR is a kind of Ser/Thr kinase in mammalian cells, and it combines with other proteins to form two mTOR complexes, mTORC1 and mTORC2. mTORC1 integrates input signals from nutrients, growth factors, energy, oxygen and environmental stress to control cellular growth and metabolism homeostasis [122]. Nutrients, including amino acids, glucose and nucleotide, drive the recruitment of mTORC1 to the lysosomal surface via the Rag GTPases and the sensors of several types of amino acid, glucose and purines in cells mediated nutrient signals to excite mTORC1 signaling are reported in recent years [122, 123]. mTORC1 is also involved in lipid metabolism and several types of fatty acid signals can excite mTORC1 [124, 125, 126, 127]. However, the sensing mechanisms of fatty acid in cells and how mTORC1 are regulated by fatty acid signals remain unclear. Therefore, understanding the cellular mechanism behind the fatty acid sensing and homeostasis is important for cell growth and metabolism.
The fluctuation of internal fatty acid levels and intracellular and extracellular LCFA sensing mechanisms exist in mammals. 2 types of pathway may exist in mammalian cells, they are the direct binding of the LCFA molecule to the sensor protein, or indirect mechanism relying on the detection of metabolites of LCFA [118]. Extracellular and intracellular LCFAs may sense by sensors which located on cytoplasmic membrane and in cytoplasm. Increasing evidence in human or rodent models indicates that G protein-coupled receptors (GPRs), as free fatty acid receptors (FFARs), can sense the level of extracellular LCFA and affect the biological characteristics of cells. The free fatty acid receptors include FFAR1 (GPR40), FFAR2 (GPR43), FFAR3 (GPR41) and FFAR4 (GPR120) which are seven transmembrane-spanning proteins show different expression patterns on different cells, and GPR40 (FFAR1) and GPR120 (FFAR4) are involved in sensing medium- and long-chain fatty acids while GPR43 (FFAR2) and GPR41 (FFAR3) are activated by SCFA [17, 128]. In addition to GPR40 and GPR120, other GPR-independent mechanisms have been suggested in mediating LCFA sensing, e.g., CD36 [117, 118, 129]. CD36, as mentioned earlier, is a multifunctional membrane protein, and is considered as the most important translocator of LCFAs. Importantly, CD36 has been reported to be a LCFA receptor and involved in LCFA sensing [130, 131, 132, 133].
GPRs couple to diverse intracellular downstream G proteins and then activate downstream signaling pathways. In general, GPR40 or GPR120 are activated by the extracellular LCFAs, and then transduce signals downstream cAMP and the phospholipase C (PLC) signaling cascade. PLC cleaves the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2) into the second messengers diacylgycerol (DAG) and inositol 1,4,5-trisphosphate (IP3), leading to calcium release, protein kinase C (PKC) activation, and the phosphatidylinositol 3-kinase (PI3K)/Akt (protein kinase B) signaling pathway [134, 135]. BSA-conjugated palmitic acid increases Akt/mTORC1 pathway via GPR40, and the mechanism by which palmitic acid regulate mTORC1 activity is probably its translocation onto lysosomal membranes [136, 137]. Moreover, oleic acid activates Akt/mTORC1 and ERK/mTORC1 pathways via GPR40 or GPR120 [138], and inhibits AMPK signaling which is a negative regulator of mTORC1 [139, 140, 141, 142]. There are few reports on stearic acid and mTORC1 activity, it can upregulate mTOR expression [143].
LCFA, e.g., palmitic acid, can be used to synthesize phosphatidic acid (PA) or be degraded to produce acetyl-CoA after LCFAs are transported inside cells. Phosphatidic acid interacts with the FK506-binding protein–12-rapamycin-binding (FRB) domain of mTOR to activate mTORC1 [120, 144, 145], and lower total cytosolic acetyl-CoA levels led to decreased raptor acetylation and reduced lysosomal localization of mTOR, resulting in impaired activation of mTORC1 [126], indicating changes in the level of LCFA metabolites can be sensed by mTORC1. However, whether there is a mechanism for mTOR to distinguish phosphatidic acid species with LCFAs remains to be determined (Table 4, Ref. [18, 134, 138, 141, 144]).
| Protein name | FA type | Signaling pathway | mTORC1 signaling | Reference |
|---|---|---|---|---|
| GPR40 | MCFA and LCFA | (a) cAMP/PKA/ERK/mTORC1 | GPR40 mediates extracellular LCFA signals to excite mTORC1 signaling | [18, 134] |
| (b) PLC/DAG/PKC/ERK/mTORC1 | ||||
| GPR120 | MCFA and LCFA | (a) PLC/IP3/AMPK/mTORC1 | GPR120 mediates extracellular LCFA signals to excite mTORC1 signaling | [138, 141] |
| (b) PI3K/Akt/mTORC1 | ||||
| CD36 | LCFA | (a) PA/ mTORC1 | Intracellular LCFAs translocated by CD36 are further metabolized to produce PA or acetyl-CoA, which regulates mTORC1 activity | [144] |
| AMPK, AMP-activated protein kinase; Akt, Protein kinase B; cAMP, Cyclic adenosine monophosphate; DAG, Diacylgycerol; ERK, Extracellular signal-regulated kinase; GPR40, G protein-coupled receptor 40; GPR120, G protein-coupled receptor 120; IP3,Inositol 1,4,5-trisphosphate; LCFA, Long-chain fatty acid; MCFA, Medium-chain fatty acid; mTORC1, Mammalian target of rapamycin complex 1; PKA, Protein kinase A system; PLC, Phospholipase C; PKC, Protein kinase C; PI3K, Phosphatidylinositol 3-kinase; PA, Phosphatidic acid. | ||||
The downstream signaling pathways mediated by GPR40 or GPR120, the second messenger cAMP can activate downstream effector PKA and regulate ERK sequentially [138, 146, 147], and another downstream pathway PLC/DAG/IP3 in which DAG can excite PKC and ERK/mTORC1 [147, 148], and IP3 leads to calcium release activating AMPK which is an upstream negative regulator of mTORC1 [139, 140, 142, 149], and the third downstream pathway, PI3K/Akt pathway, can activate mTORC1 signaling [136, 138, 140, 147]. Different G proteins mediate LCFAs to regulate mTORC1 signaling. Furthermore, although whether CD36 can act as a LCFAs receptor to activate mTORC1 remains determined, and it was definite that intracellular LCFAs translocated by CD36 are further metabolized to produce phosphatidic acid (PA) or acetyl-CoA, which regulates mTORC1 activity (Fig. 4). Whether mTORC1 senses intracellular LCFA levels or existing other sensors of LCFAs remains to be determined.
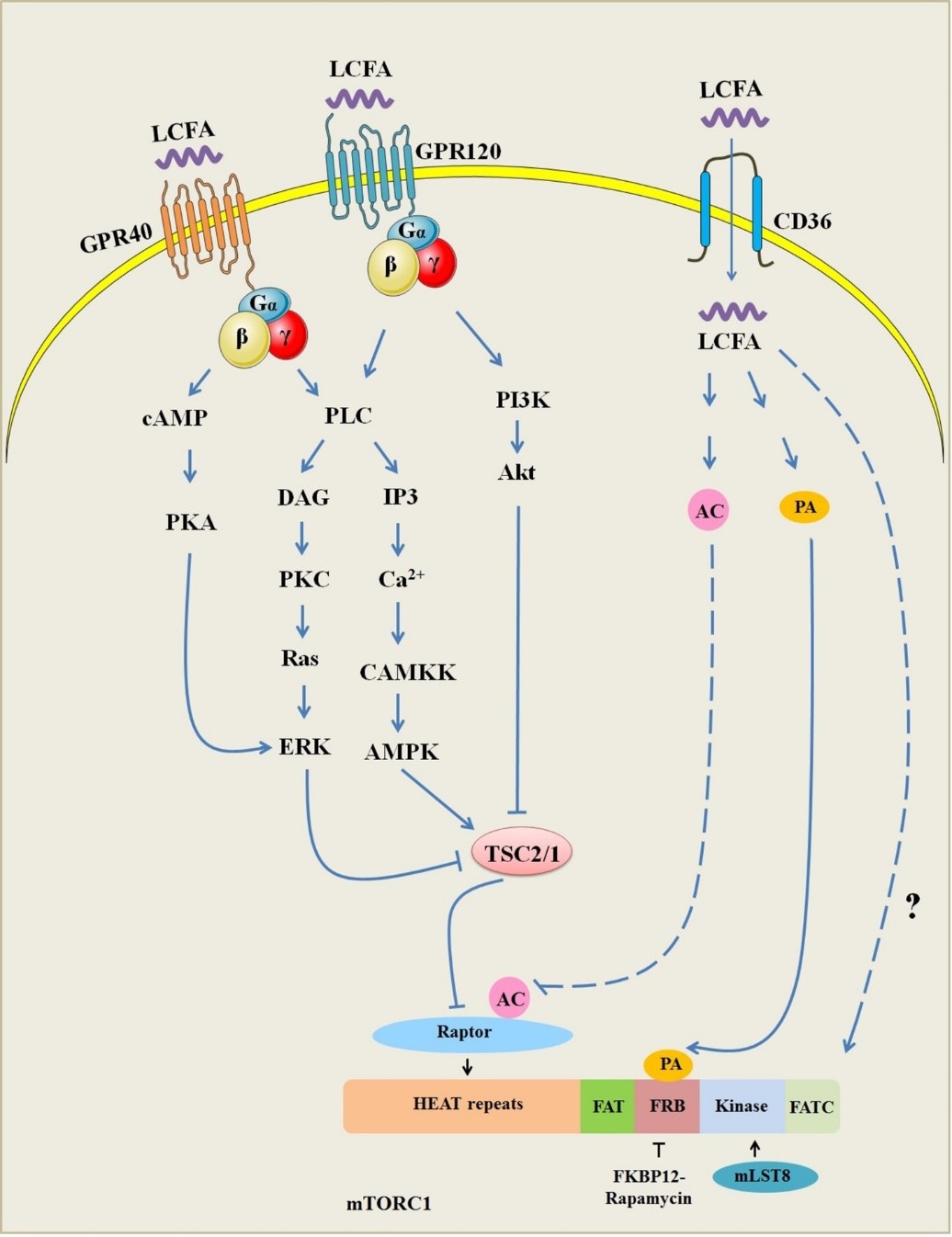 Fig. 4.
Fig. 4.LCFAs sensing by mTORC1. The downstream signaling pathways of GPR40 and GPR120, including cAMP, PLC/DAG/IP3, and PI3K/Akt. cAMP activates downstream effector PKA and regulate ERK sequentially; PLC/DAG/IP3 excites PKC and ERK/mTORC1; IP3 leads to calcium release activating AMPK/mTORC1; PI3K/Akt activates mTORC1 signaling. Phosphatidic acid (PA) or acetyl-CoA (AC) regulates mTORC1 activity. Whether mTORC1 can directly sense intracellular LCFA levels remains to be determined. AC, acetyl-CoA; AMPK, AMP-activated protein kinase; Akt, Protein kinase B; cAMP, Cyclic adenosine monophosphate; DAG, Diacylgycerol; ERK, Extracellular signal-regulated kinase; GPR40, G protein-coupled receptor 40; GPR120, G protein-coupled receptor 120; IP3, Inositol 1,4,5-trisphosphate; LCFA, Long-chain fatty acid; mTORC1, Mammalian target of rapamycin complex 1; PKA, Protein kinase A system; PLC, Phospholipase C; PKC, Protein kinase C; PI3K, Phosphatidylinositol 3-kinase; PA, Phosphatidic acid.
In this review, we have provided the most up-to-date information available on the absorption of LCFAs, their translocation across the plasma membrane, and their metabolic pathways as well as their sensing mechanisms in cells. We have not covered all possible metabolic pathways and regulatory mechanisms of LCFA, such as the regulation of the transcriptome by fatty acids, or whether mTOR to distinguish phosphatidic acid species with different types LCFAs in cells. However, based on the literature reviewed, we can make some concluding comments and propose some forward-looking predictions for each main topic covered in this review.
FAs are important biocompounds that participate in complex metabolic pathways to ensure human health and development. FAs are obtained by mammalian cells through external uptake (primarily), de novo synthesis, hydrolysis of triacylglycerols. LCFAs and VLCFAs are absorbed mainly from outside of cells in mammals. LCFAs enter cells through passive diffusion and protein-mediated FA translocation across the plasma membrane, in the latter of which FAT/CD36, FABPpm, FATP, and caveolin-1 are believed to be critical. The LCFAs that are absorbed by cells bind to FABPs, are transported to metabolic organelles, and converted into acyl-CoA to target specific metabolic pathways. LCFA-CoA is esterified to phospholipids, triacylglycerols, and cholesteryl esters or other specific lipids.
NEFAs are preferentially stored as triacylglycerol, and the triacylglycerol
structure and interesterification of LCFAs likely affect the physical
characteristics of fat and oxidative stability. The mechanisms by which
triacylglycerol synthase function will be illustrated by the crystal structure of
these synthases.
FA elongation occurs by cycling through a 4-step process, comprising condensation, reduction, dehydration, and reduction. The first crystal structure of a membrane-bound FA elongase-condensing enzyme has revealed a new reaction mechanism. Understanding the mechanism of the cellular uptake and metabolism of FAs is important for human nutrition and metabolism, and the novel crystal structures of lipid metabolism enzymes provide insights into the reaction mechanism of LCFAs.
GPR40 or GPR120 mediates extracellular LCFA signals to excite mTORC1 signaling, and intracellular LCFA’s sensor remains to be determined. However, mTORC1 activation can be regulated by phosphatidic acid (PA) and acetyl-CoA which are the metabolites of LCFAs. CD36 is a potential receptor of LCFAs to mediate extracellular LCFA signals.
The metabolism of LCFAs is closely related to the occurrence of human obesity, nonalcoholic fatty liver disease (NAFLD), cardiovascular disease, and hyperlipidemia. The metabolism of LCFAs is diverse and is related to genes, living environment and dietary nutrition. Thus, the metabolism of different LCFAs, and the regulatory mechanism in human health and diseases should be considered comprehensively.
ABC-transporters, ATP-binding cassette transporters; ACSL, A ligases/acyl-CoA synthetases; AMPK, AMP-activated protein kinase; Akt, Protein kinase B; cAMP, Cyclic adenosine monophosphate; DG/DAG, Diacylgycerol; PKC, Protein kinase C; ERK, Extracellular signal-regulated kinase; ER, endoplasmic reticulum; FAs, Fatty Acids; FAT/CD36, FA translocase; FABPpm, plasma membrane FA-binding protein; FATP, FA transport protein; FATP1 originally called FATP; FADS2, Fatty acid desaturase 2; GPR40, G protein-coupled receptor 40; GPR120, G protein-coupled receptor 120; IP3, Inositol 1,4,5-trisphosphate; LCFAs, Long-chain fatty acids; mTORC1, Mammalian target of rapamycin complex 1; MCFAs, Medium-chain fatty acids; PA, Phosphatidic acid; PKA, Protein kinase A system; PLC, Phospholipase C; PI3K, Phosphatidylinositol 3-kinase; SCFAs, Short-chain fatty acids; SLC27A, Solute carrier protein family 27A; SCD, Stearoyl-CoA desaturase; TG/TAG, Triacylglycerol; VLCFAs, Very long-chain fatty acids.
PY—writing - reviewing and editing, supervision. QH—writing - original draft reviewing and editing. YC—visualization, investigation. ZW—conceptualization, writing - reviewing and editing. HH—writing - reviewing and editing. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Not applicable.
Not applicable.
This work was funded by the Inner Mongolia Agriculture and Animal Husbandry Innovation Fund (2019CXJJM11 and 2022CXJJM09), by Inner Mongolia Autonomous Region Science and Technology Program Project (2019GG354).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.




