†These authors contributed equally.
Academic Editor: Ricardo Jorge Pinto Araujo
Background: RNA-binding proteins (RBPs), which form complexes or single/multiple RNA-binding domains, have a functional role in regulating and determining the function or stability of the bound RNAs in various cancers, including breast invasive carcinoma (BRCA). However, the biological functions and clinical implications of RBP-related long noncoding RNAs (lncRNAs) in BRCA remain largely unknown. Methods: Herein, we first identified and characterized RBP-related lncRNAs in BRCA. Then we built an RBP-related lncRNA signature (RBPLSig) and explored the clinical evaluation and prediction performance of the RBPLSig by bioinformatic analysis. In addition, to optimize treatment plans, prediction online tools was developed to predict the patient survival rate. Lastly, to verify the function of lncRNA WAC antisense RNA 1 (WAC-AS1), the experiments such as Quantitative real-time PCR (qRT-PCR), lncRNA knockdown, CCK-8, and terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining were performed. We also gained the potential mechanisms of the druggable compounds of the WAC-AS1 related RBP gene, putative NSUN6, using molecular docking. Results: The results showed that RBPLSig, as an independent prognostic factor for BRCA patients, was involved in numerous malignancy-associated immunoregulatory pathways. We found different immune statuses and responses to immunotherapy, chemotherapy, and targeted therapy between the high- and low-risk groups stratified by RBPLSig. Conclusions: Our data broaden the comprehensive understanding of the biological functions of RBP-related lncRNAs, and demonstrate a novel and independent RBPLSig to assess prognosis and the immune microenvironment, thus helping to guide treatment decisions for BRCA.
Breast carcinoma is a leading cause of death worldwide, accounting for 684,996 (6.9%) deaths globally in 2020 (Cancer Today-IARC, 2020), despite dramatic advances in diagnosis and treatment strategies [1, 2]. It is thus necessary to discover novel biomarkers for early diagnosis and therapeutic interventions. Long noncoding RNAs (lncRNAs) are a series of transcripts longer than 200 nucleotides that have limited protein coding capability [3, 4, 5]. Several studies have reported that the deregulation of lncRNAs, such as LncRNA RP11-400K9.4 [6], lncRNA DILA1 [7], and LncRNA OIP5-AS1 [8] affects the tumorigenesis and progression of breast invasive carcinoma (BRCA). However, the detailed mechanism underlying the regulation and alteration of these lncRNAs in BRCA over time remains unknown.
To date, more than 1500 RNA-binding proteins (RBPs) have been identified [9]. Studies have shown that several RBPs are abnormally expressed in tumors and regulate various biological functions, including coordinating RNA splicing, nuclear export, transcript stabilization, localization, and degradation of all types of RNAs [10]. Considering the important role of RBPs in transcriptional and post-transcriptional gene expression, it is not surprising that aberrantly regulated RBPs participate in the progression of human diseases, including cancers [11].
Recent studies have indicated that several cancer-associated lncRNAs act via transcriptional regulation and post-transcriptional regulation mediated by well-known RBPs [12]. For example, human antigen R (HuR)-regulated lncRNA NEAT1 is stabilized by HuR and functions as an oncogene in ovarian cancer [13]. LncRNA-HGBC is also stabilized by HuR and functions as an oncogene in gallbladder cancer [14]. HuR promotes the degradation of HOTAIR [15] by recruiting let7-Ago2 in breast cancer. Moreover, both the nuclear export and mitochondrial localization of lncRNA-RMRP are modulated by HuR and G-rich RNA sequence binding factor 1 (GRSF1) [16]. However, more direct evidence of the correlations and concordant links among RBPs, lncRNAs, and BRCA are needed to explore the potential of lncRNAs for future cancer diagnostic, therapeutic, or prognostic purposes. While immunotherapy is a promising remedial modality for BRCA treatment, the objective response rates to immune checkpoint inhibitor (ICI) blockade have remained low; notwithstanding, blocking ICIs is a widely used strategy in immunotherapy [17]. Therefore, it is critical to find a reliable biomarker that can be used to precisely screen BRCA patients for immunotherapy.
The aim of our study was to perform a comprehensive analysis of RNA-binding protein-related lncRNA in BRCA. We first explored the relationship between the expression of the RBP-related lncRNA signature (RBPLSig) and clinicopathological features. Then, for predicting the performance of the RBPLSig, a nomogram for BRCA and one nomogram for BRCA subtypes (luminal A and TNBC) were established, respectively. The latter is more appropriate for immunotherapy evaluation. In addition, to optimize treatment plans, prediction online tools was developed to predict the patient survival rate. Lastly, to verify the function of WAC-AS1, pathway enrichment, CCK-8, TUNEL, drug sensitivity, and molecular docking analyses were performed. These findings may increase our understanding of RBP-related lncRNAs in patients with BRCA.
RNA-sequencing data, updated clinical data, and sample information of breast invasive cancer (BRCA) cohort were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/) and 107 patients in GSE58812 for external validation was obtained from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) database. Patients who fulfilled the following selection criteria: (1) diagnosed with histologically confirmed BRCA; (2) available RNA-seq data; (3) available follow up were eligible for study enrollment. Finally, 1054 patients from TCGA-BRCA, 107 patients from GSE58812 were included for further analysis. For TCGA-BRCA data analysis, the established RNA matrix file was merged with Ref-Seq and used for all subsequent RNA-seq expression analyses. For the data from GEO datasets, the probe ID for each gene was converted to a gene symbols. All raw data were analysed using the ‘limma’ package in R (version 4.2.1, http://www.bioconductor.org/).
In the current study, we established 1542 encoding RNA-binding proteins (RBPs)
[9]. Correlations between lncRNAs and RBPs were calculated using Pearson’s
correlation coefficient using the limma R package [18]. A lncRNA with a
correlation coefficient
To better research and further explore the molecular mechanisms underlying the
prognostic difference between high- and low-risk groups the biological function
of GSEA was performed to by using the GSEA tools
(http://www.broadinstitute.org/gsea)
[19]. Significant predefined biological processes and pathways were enriched with
The univariate and multivariate Cox regression analyses were conducted with Cox proportional hazard regression models to analyze predictive independent factors. Then, a nomogram that included all independent factors predicting overal survival (OS) was developed using R version 4.0.3 (http://www.r-project.org/) with rms packages.
The immune cell infiltration level of each BRCA sample was quantified using
CIBERSORT (http://cibersort.stanford.edu/). The relationship between RBPLSig and
immune checkpoint inhibitor (ICI) gene expression was determined using the
ggstatsplot package and violin plot visualization. To clinically evaluate the
model for breast cancer treatment, we calculated the IC
Triple-negative breast cancer (TNBC) cell lines, MDA-MB-231 and HST578T cells,
were obtained American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in DMEM (Gibco, Waltham, MA, USA),
containing 4500 mg/L glucose and 10% fetal bovine serum in 5% CO
The Cell Counting Kit-8 (CCK-8) assay was used to examine the survival of TNBC cells following the protocol of the CCK-8 assay kit (Dojindo, Kyushu, Japan). Survival rates of the two TNBC cell lines after si-WAC-AS1 knockdown was measured at 450 nm [22]. Cellular apoptosis was quantified via terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining following the protocol of the One Step TUNEL Apoptosis Assay Kit (Keygen, Nanjing, China) [22]. The treated TUNEL-positive TNBC cells were counted under a fluorescence microscope (Olympus, Tokyo, Japan).
We used the cell miner interface (https://discover.nci.nih.gov/cellminer/), containing 727 FDA-approved drugs and clinical trial drugs along with their corresponding protein targets in 60 cancer cell lines, to download the DTP NCI-60 database and RNA-seq data. Then, Spearman or Pearson correlation were used to analyze the association between RBP gene (NSUN6) expression and drug treatment response in five BRCA cell lines.
The 2D structure of the drug molecules was searched using PubChem (https://pubchem.ncbi.nlm.nih.gov/), and a mol2 file was converted using Chem3D (PerkinElmer, Waltham, MA, USA). Receptor proteins were searched from the Protein Data Bank (http://www.rcsb.org/). Dehydration and ligand elimination were performed using PyMOL software (PyMOL2.3, https://pymol.org). The docking site was set in a cubic box in the center of the initial ligand, and a grid map of each atom type in the box was computed. The AutoDockTools 1.5.6 software (https://autodocksuite.scripps.edu/adt/) was used to simulate the molecular docking of potential targets and components. The best scoring conformer of each compound was analyzed and visualized in AutoDockTools-1.5.6 and PYMOL. Concurrently, we used Gromos96 (G43A1, http://gromos.net/) for dynamic simulation, which was performed for 10 ns in combination with the Extended Simple Point Charge (SPCE) water model [23]. System stability was evaluated by the root-mean-square deviation (RMSD) method.
Statistical analyses were conducted using the R software V4.2.1
(http://www.bioconductor.org/) and SPSS V18.0 (IBM Corp., Armonk, NY, USA). The
Kaplan–Meier method was used for survival analysis. The statistical analysis of
clinical information was performed using a chi-square test or Fisher’s exact
test. The statistical significance of the difference between two groups was
assessed using a Student’s t-test, and p
We first screened 1542 RBP-related encoding genes (mRNAs) and identified the
available expression data of 1550 total lncRNAs as RBP-related lncRNAs in the
TCGA-BRCA set (
After multivariate Cox proportional hazards regression analysis, the 20 lncRNAs
were used to establish a prognostic RBP-related lncRNA signature (RBPLSig). The
estimated RBPLSig was calculated as follows: = (–0.258
 Fig. 1.
Fig. 1.The prognostic value of the RBPLSig. (A) Heatmap demonstrating the expression levels of 20 prognostic lncRNAs between the high- and low-risk groups. (B,C) Hierarchical clustering analysis of survival status of patients with increased risk score. (D) Kaplan–Meier analysis of the prognostic model in TCGA-BRCA.
Univariate and multivariate Cox regression analyses were performed to determine
whether the RBPLSig is an independent prognostic factor for BRCA. The hazard
ratio (HR) of the risk score was 1.104 (95% confidence interval [CI]
1.081–1.128, p
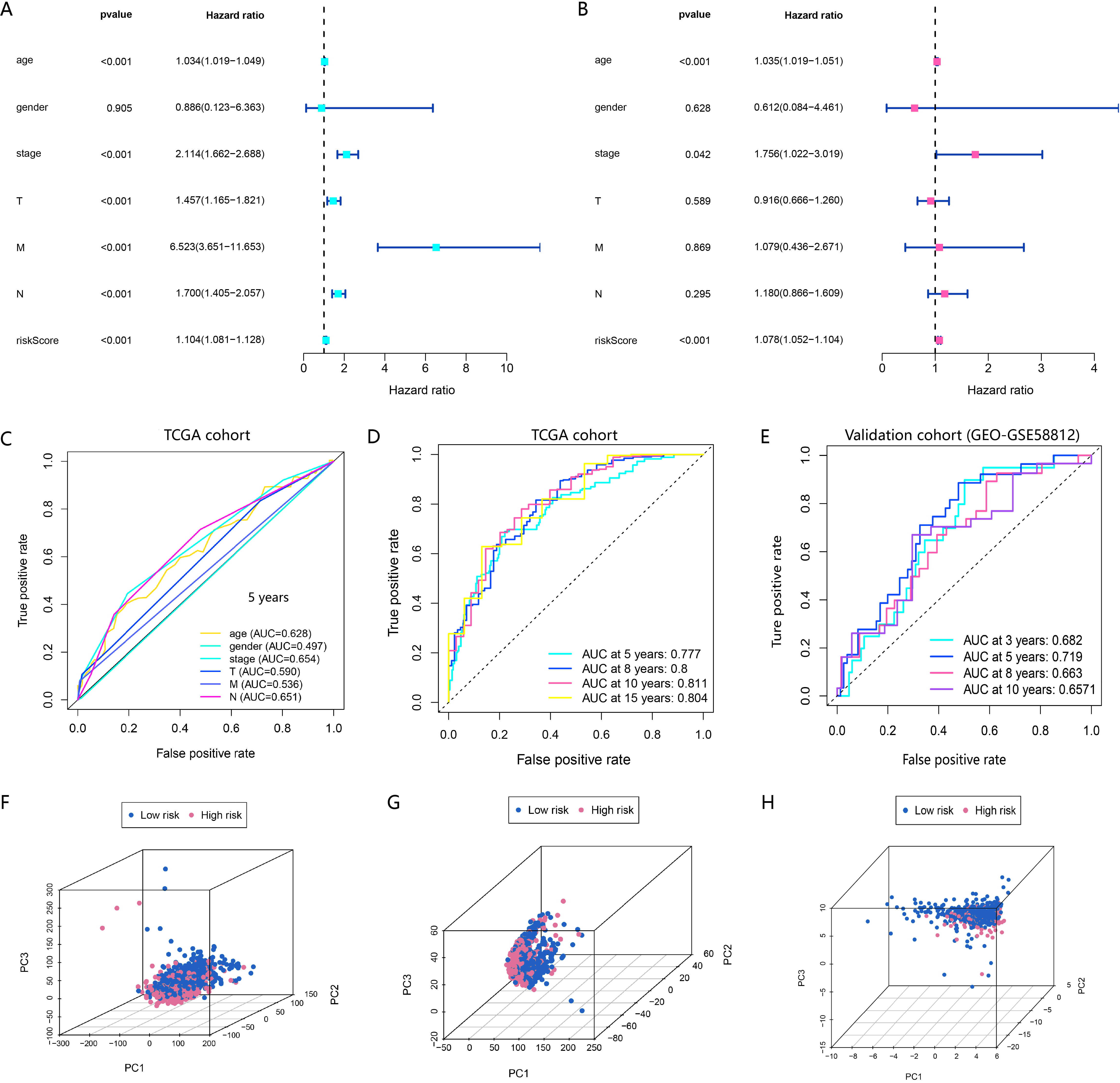 Fig. 2.
Fig. 2.The RBPLSig is an independent prognostic factor for OS prediction. (A,B) Univariate and multivariate Cox regression analyses to screen OS-related factors. (C) Receiver operating characteristic (ROC) curve analysis of clinicopathological features. (D) Time-dependent ROC curves of OS at 5-, 8-,10- and 15-years in TCGA-BRCA cohort. Their AUC values are all greater than 0.75. The closer the AUC is to 1.0, the higher the authenticity of the detection method. (E) Time-dependent ROC curves of OS at 3-, 5-, 8- and 10-years in the validation cohort. Their AUC values are all greater than 0.65. (F–H) Principal component analysis (PCA) of the high- and low-risk groups based on the whole-genome (F), RBP-related lncRNAs (G), and RBPLSig expression profiles (H).
To further assess whether the 20 RBP-related lncRNAs participated in the
development of BRCA, the relationship between the risk score and
clinicopathological factors was analyzed. As shown in Fig. 3A–E, an association
was found between the risk score and clinicopathological factors (p
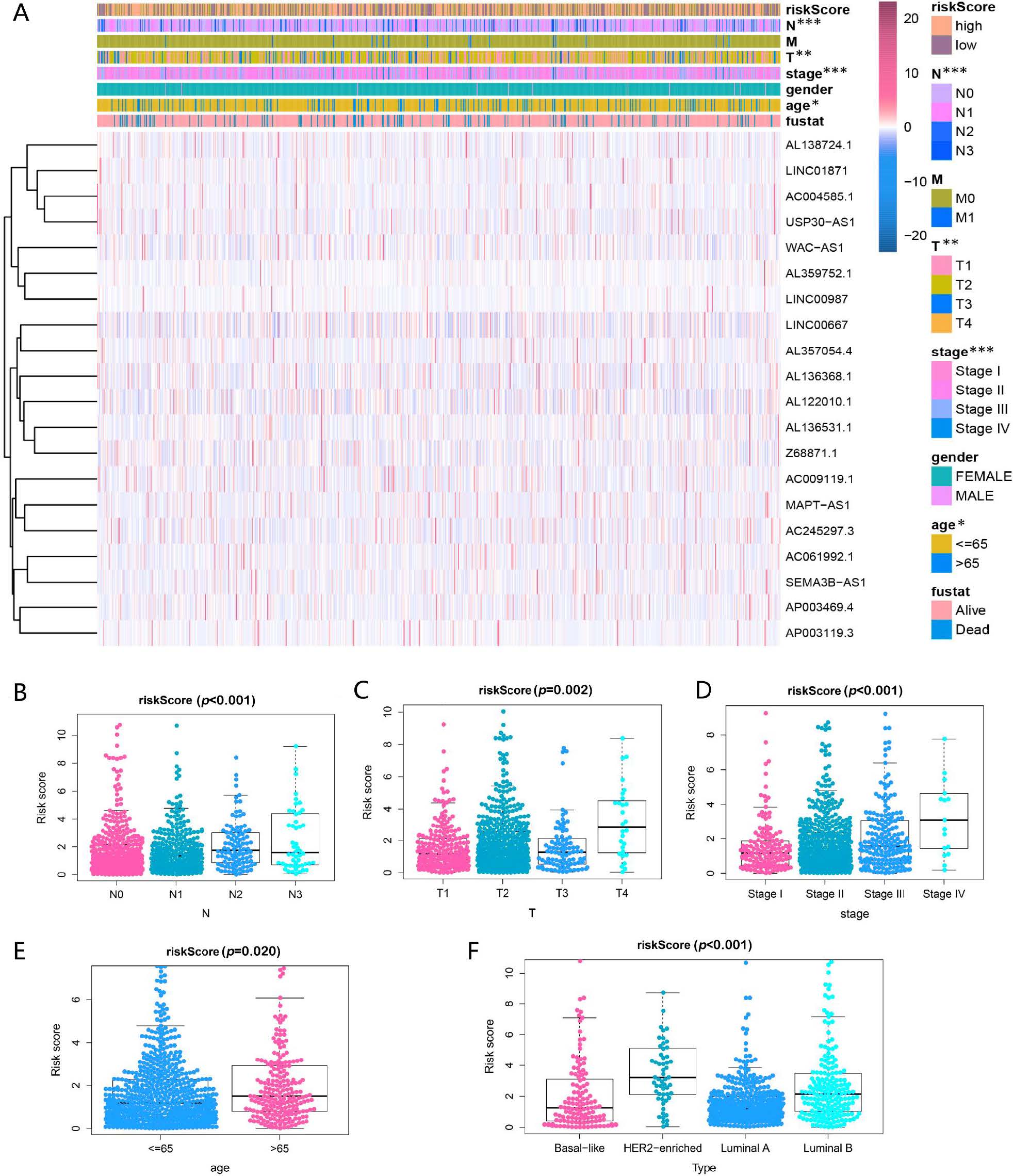 Fig. 3.
Fig. 3.Prognostic risk score associated with the clinical
characteristics of patients with BRCA. (A) Heatmap and clinicopathologic
features of high- and low-risk groups. *p
GSEA was performed to further examine RBPLSig-associated signaling pathways in
BRCA. We observed that transforming growth factor
 Fig. 4.
Fig. 4.Bioinformatics analysis between the two groups stratified by
RBPLSig. (A) GSEA for biological pathways and processes correlated with immune
score values in the cohort from TCGA. (B) Association of 20 lncRNAs expression
with different immune infiltrate subtypes tested with ANOVA (analysis of
variance) in TCGA-BRCA. ns, not significant; *p
To evaluate the correlation between the risk score and tumor immune cell
infiltration, the CIBERSORT algorithm was used to investigate the scale of 22
immune cell types. Patients with high-risk scores had higher macrophage M0 and
macrophage M2 infiltration levels than those with low-risk scores (p
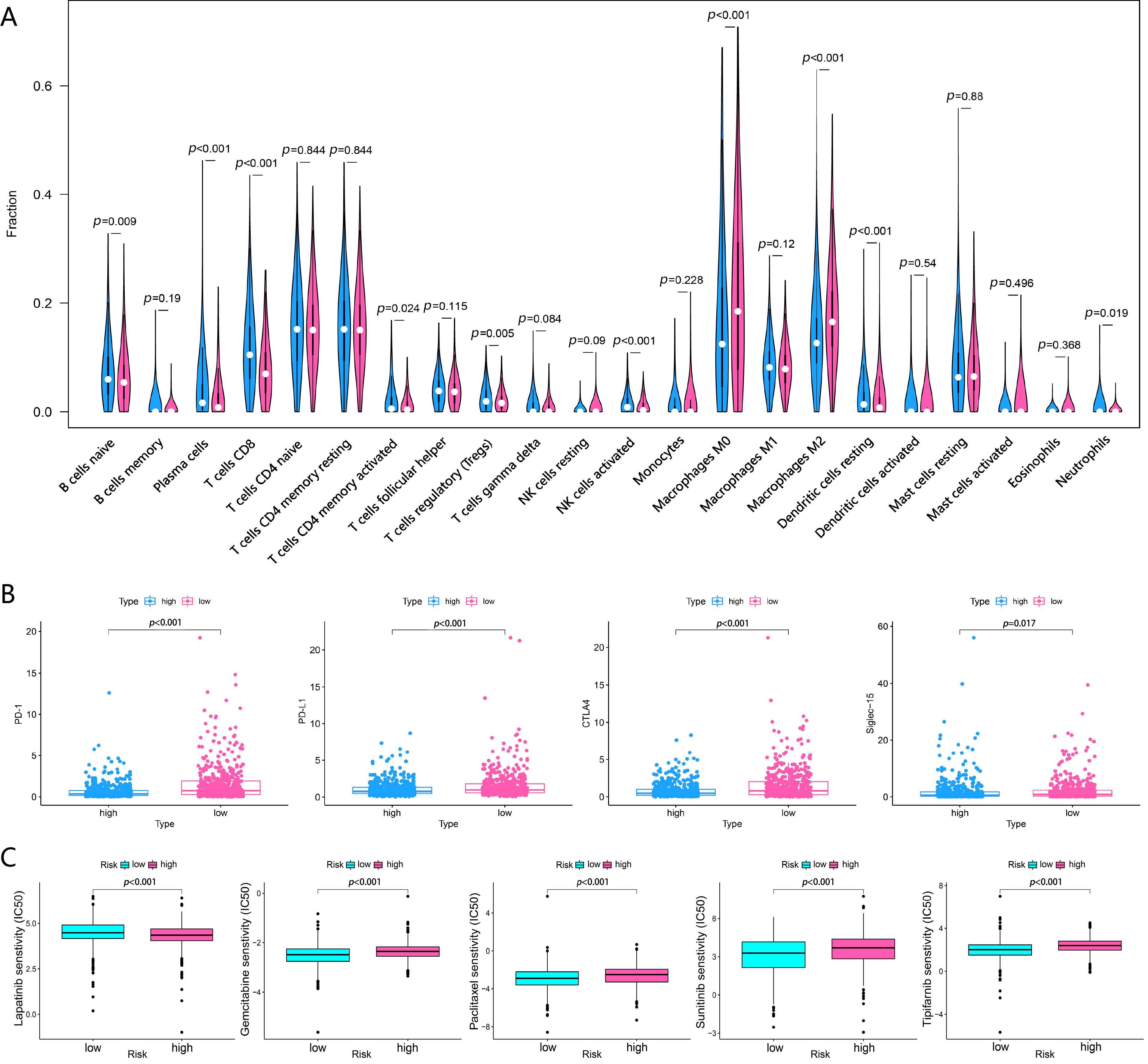 Fig. 5.
Fig. 5.Different responses to immunotherapy, chemotherapy and targeted
therapy between the two groups stratified by RBPLSig. (A) Violin plot of 22
immune-related terms were incorporated to assess the abundance of immune cells
between the two groups stratified by RBPLSig, in which red represents the
high-risk samples, and blue the low-risk samples. (B) The expression of
PD-1 (p
Expression of the ICI (immune checkpoint inhibitor) genes and/or their ligands
have attracted widespread attention as BRCA biomarkers for immune checkpoint
blockade therapy. It is critically important to effectively screen BRCA patients
who might benefit most from different ICI immunotherapies. To further investigate
the association between risk score and ICIs, cytotoxic T-lymphocyte-associated
antigen 4 (CTLA-4), sialic acid binding Ig-like lectin 15
(Siglec-15), programmed cell death-1 (PD-1), and programmed cell
death-ligand 1 (PD-L1) were selected to compare the expression patterns
between the two groups stratified by the RBPLSig. The result indicates that
patients in the low-risk group tended to have higher PD1,
PD-L1, and CTLA-4 expression levels than those in the high-risk
group, whereas patients in the high-risk group tended to have higher
Siglec-15 expression levels (Fig. 5B, p
| Cancer types | Over survival | High risk | Low risk | AUC | ||
| (p-value) | (n) | (n) | 1-year | 3-year | 5-year | |
| BLCA | 202 | 202 | 0.648 | 0.687 | 0.714 | |
| CESC | 145 | 146 | 0.7 | 0.737 | 0.726 | |
| COAD | 182 | 182 | 0.718 | 0.74 | 0.743 | |
| KIRC | 263 | 263 | 0.713 | 0.704 | 0.739 | |
| KIRP | 143 | 143 | 0.868 | 0.824 | 0.8 | |
| LAML | 65 | 65 | 0.779 | 0.783 | 0.796 | |
| LUAD | 254 | 255 | 0.65 | 0.616 | 0.664 | |
| LUSC | 249 | 250 | 0.574 | 0.58 | 0.594 | |
| OV | 188 | 188 | 0.636 | 0.65 | 0.703 | |
| PAAD | 88 | 88 | 0.673 | 0.798 | 0.95 | |
| READ | 78 | 79 | 0.665 | 0.715 | 0.879 | |
| SKCM | 231 | 232 | 0.699 | 0.704 | 0.7 | |
| BLCA, Bladder Urothelial Carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, Colon adenocarcinoma; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute myeloid leukemia; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; READ, Rectum adenocarcinoma; SKCM, Skin Cutaneous Melanoma. | ||||||
To generate a clinically applicable model for individual OS prediction, we first constructed a nomogram that accurately estimates the 5-, 8-, 10-, and 15-year survival probabilities in BRCA patients using the RBPLSig and other clinicopathological independent prognostic factors (Fig. 6A). The calibration curves of the nomogram for predicting patient survival at 5-,8-,10-, and 15-years are shown in Fig. 6B–E. Considering that (1) the risk score was lower for luminal A and TNBC than for HER2-enriched and luminal B subtypes, and (2) patients with a low-risk score had an improved TIME (appropriate for immunotherapy), we further constructed a nomogram for BRCA subtypes (luminal A and TNBC) appropriate for immunotherapy evaluation (Supplementary Fig. 4). To further explore the value of predictive nomograms, we developed an online analysis tools to predict the survival rate and recommended medication for BRCA patients (https://www.origingenetic.com/BreastCancerModel). With the aid of the prediction online tools, clinicians can assist in evaluating patient survival rates to optimize treatment plans.
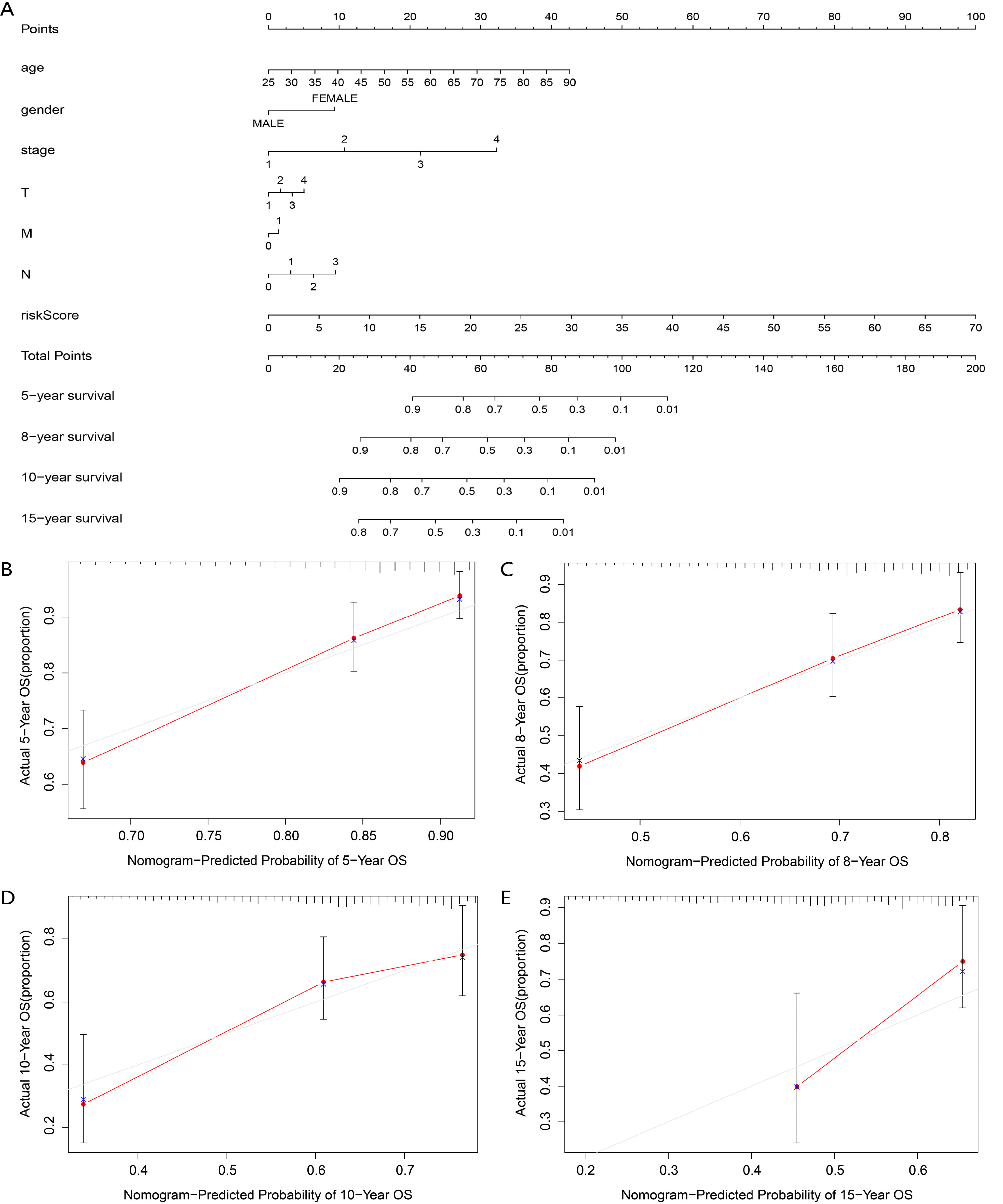 Fig. 6.
Fig. 6.Construction of a nomogram for survival prediction. (A) The clinical prognostic nomogram developed to predict 5-, 8-, 10-, and 15-year survival. (B–E) Calibration curves showing nomogram predictions for (B) 5-, (C) 8-, (D) 10- and (E) 15-year survival.
For further study on the potential function of these lncRNAs and to verify the aforementioned results, we focused on the upregulated WAC-AS1, which is associated with metastasis and poor prognosis for OS in selective patient subgroups from TCGA and Gene Expression Omnibus (GEO) datasets (Fig. 7A–C).
 Fig. 7.
Fig. 7.WAC-AS1 was upregulated in TNBC tissues and
WAC-AS1 silencing induced TNBC cells apoptosis. (A) Microarray analysis
of WAC-AS1 in TCGA-TNBC tissues corresponding normal tissues. (B,C)
Kaplan-Meier survival curves of patients with different level of WAC-AS1
in TCGA (B) and GEO (C) datasets. (D) TNBC cell lines (MDA-MB-231and HS578T) was
transfected with si-WAC-AS1. The expression of WAC-AS1 in TNBC
cell lines was analyzed by qRT-PCR. (E–G) The Effects of si-WAC-AS1 on
the proliferation and apoptosis of TNBC cell lines was detected by CCK-8 assay
(E,F) and TUNEL assay (G). * p
Next, we synthesized a specific smart RNA silencer (including small interfering
RNA and small nucleolar RNAs) against WAC-AS1. The RT-qPCR indicates
that smart RNA silencer-WAC-AS1 (si-WAC-AS1) could abolish the
expression of WAC-AS1 in TNBC cell lines (MDA-MB-231 and HST578T cells;
Fig. 7D). The CCK-8 assay indicates that cell viability was diminished by
silencer-WAC-AS1 transfection compared to that with si-NT (Fig. 7E,F,
p
Currently, lncRNAs are usually used as prognostic or diagnostic biomarkers and
lncRNA-based drug targets [24], the latter of which, while reported in several
studies, still requires more research [25]. Accordingly, the search for new
lncRNA-based drugs is considered a promising field. Targeting the binding
partners (pro-oncogenic RBPs) may indirectly affect the function of these related
pro-oncogenic lncRNAs. Drug sensitivity analysis was performed on five breast
cancer cell lines, including MCF7, MDA-MB-231, HS 578T, BT-549, and T-47D
(
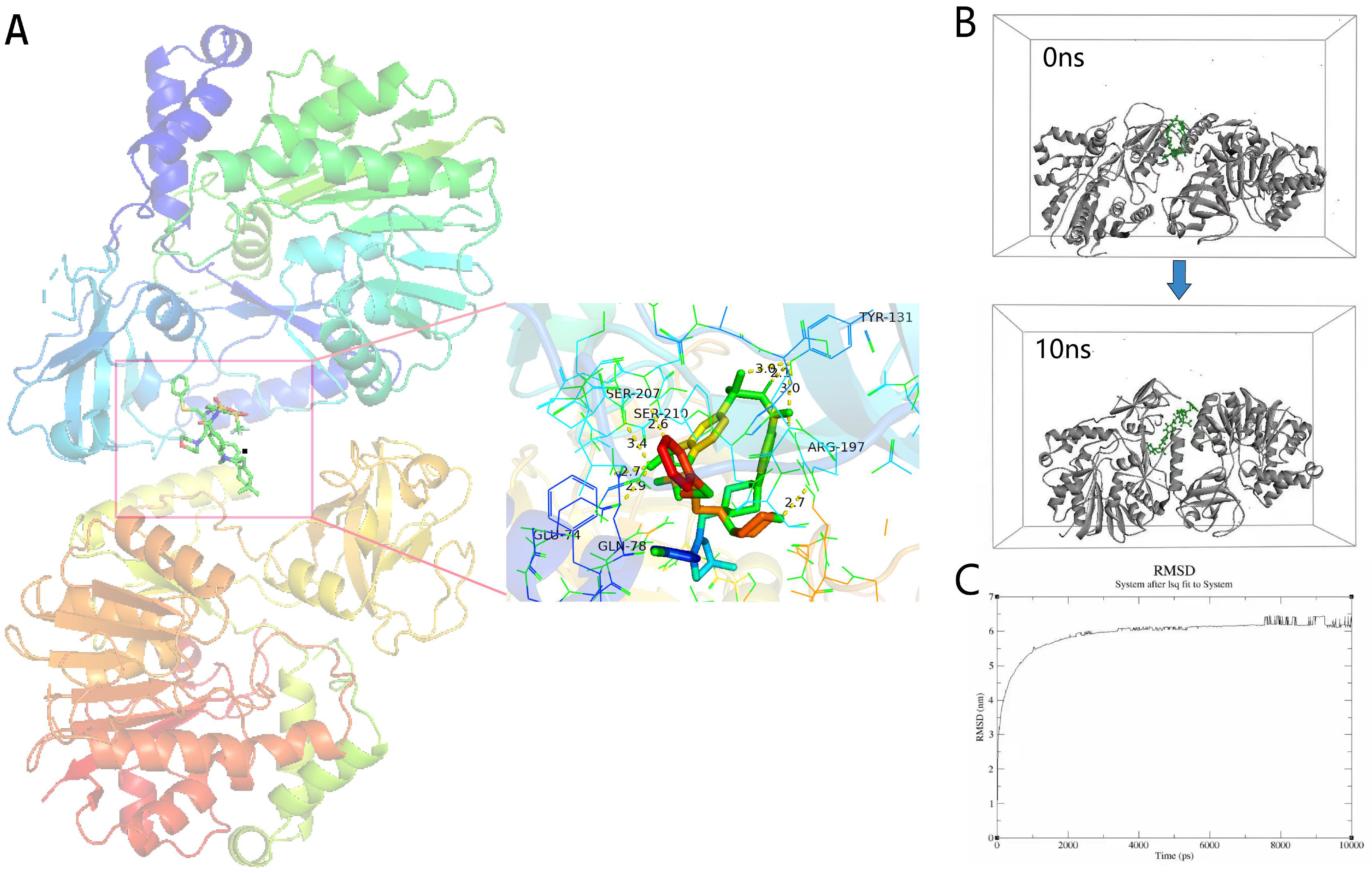 Fig. 8.
Fig. 8.Molecular docking analysis. (A) Molecular docking results of navitoclax-NSUN6 (NOP2/Sun RNA methyltransferase 6 NSUN6; PDBID: 5WWQ). Hydrogen bonds are shown as yellow dotted lines. (B) Snapshots of navitoclax-NSUN6 system after 10 ns. (C) RMSD of navitoclax-NSUN6 systems. RBP, RNA-binding protein.
Several studies have revealed that the aberrant expression of lncRNAs is a key regulator of many cellular processes. The specificity of these processes, which contribute to carcinogenesis and tumor progression, may greatly depend on their protein interactors, including classical RBPs and unconventional RBPs [26]. RNA functions greatly depend on RNA-protein interactors, including those with RBPs. In particular, several emerging RBPs are well recognized in regulating lncRNA stability, transport, and localization [27]. Some RBP-related lncRNAs in the RBPLSig have been reported to be beneficial prognostic indicators in BRCA. However, many questions about how RBP-related lncRNAs affect the occurrence and development of BRCA remain to be elucidated.
Among the 20 RBP-related lncRNAs (AL136368.1, AL136531.1, AL138724.1, LINC01871, MAPT-AS1, AL357054.4, AC061992.1, AL122010.1, USP30-AS1, AL359752.1, LINC00987, AC245297.3, LINC00667, SEMA3B-AS1, AC009119.1, AP003119.3, WAC-AS1, AP003469.4, AC004585.1, and Z68871.1), 14 were associated with a good prognosis, whereas six were associated with a poor prognosis. MAPT-AS1, LINC00667, and SEMA3B-AS1 have been confirmed to play roles in the pathogenesis and prognosis of various cancer types. For example, high expression of MAPT-AS1 is associated with better survival rates in patients with BRCA [28]. LINC00667 has been reported to be responsible for non-small cell lung cancer [29] and colorectal cancer progression [30]. Consistently, overexpression of oncogenic SEMA3B-AS1 is associated with poor survival outcomes in hepatocellular carcinoma and gastric cancer [31, 32]. Furthermore, recent studies revealed that seven RBP-related lncRNAs (LINC01871 [33], AC061992.1 [34], AL122010.1 [35], USP30-AS1 [36], Z68871.1 [33, 35], AC245297.3 [35], and AP003119.3 [35]) included in the RBPLSig could be prognostic factors in some cancers. Nevertheless, the biological functions and the underlying molecular mechanisms of these lncRNAs in cancer have not yet been clarified. The prognostic roles of the remaining 10 lncRNAs (AL136368.1, AL136531.1, AL138724.1, AL357054.4, AL359752.1, LINC00987, AC009119.1, WAC-AS1, AP003469.4, and AC004585.1) in cancers have not been previously reported. We further used in vitro experiments to initially validate the results, focusing on the expression, prognostic value, and associated pathways of the lncRNA WAC-AS1 in selective TNBC patient subgroups from TCGA-TNBC and GEO datasets. We then conducted an in vitro experiment using TNBC cell lines. The results demonstrated that lncRNA WAC-AS1 silencing induced TNBC cell apoptosis.
In studies of lncRNA functions, it is important to determine how many such lncRNAs exist in the cell type and, more importantly, whether the lncRNA is correctly transported to and located at its site of action. Therefore, the deregulation of RBPs in cancer has a profound impact on the associated lncRNAs because it affects the localization and in turn affects their function. Interestingly, drug sensitivity analysis and molecular docking analysis of a dysregulated RBP, NSUN6, in the TNBC cell line found that it is associated with sensitivity to the drug navitoclax. Previous studies have demonstrated that navitoclax can target epidermal growth factor receptor (EGFR) in TNBC [37]. Our results showed that NSUN6 may be a target of navitoclax. NSUN6 is associated with WAC-AS1, which are both oncogenes. Based on this, we speculated that inhibiting these RBPs may affect the subcellular localization of lncRNA or even affect its function. However, additional experiments are required to validate these hypotheses. It is worth noting that the experimental verification of lncRNA-protein interactions is still time-consuming and expensive, which is the main technical bottleneck in the field of lncRNA-protein interactions.
The most significant contribution of this research was demonstrating the relationship between the RBPLSig and TIME. Some studies indicate that manipulating the level of citrate can affect the behaviors of both cancer and immune cells, resulting in the induction of cancer cell apoptosis, boosting immune responses, and enhancing cancer immunotherapy [38, 39, 40, 41, 42, 43]. The complex interplay between tumor cells and TIME not only plays a pivotal role during tumor development but also has significant effects on immunotherapeutic efficacy and patient OS [44]. Furthermore, CD8 T cell infiltration has been reported to be associated with better OS in BRCA patients [45, 46]. Our results showed that the score of immune cell infiltration in the low-risk group was higher, whereas the expression of CD8 T cells and memory-activated CD4 T cells in the low-risk group was higher, indicating that the infiltration of specific immune cells could expedite tumor progression and predict BRCA survival rates. Meanwhile, immune checkpoint blockade therapies have been proposed as a promising approach to treat a variety of malignancies. The increased expression level of immune checkpoint ligands and tumor-associated antigens on tumor cells is correlated with good ICI treatment outcomes [47]. Notably, when detecting ICIs, we found that the risk score was associated with the expression of ICIs. CTLA-4, PD-1, and PD-L1 were highly expressed in the low-risk group, indicating that patients with low-risk scores might benefit more from anti-CTLA-4, PD-1, and PD-L1 immunotherapy. As a novel antitumor target comparable to PD-L1, Siglec-15 has also been implicated in immune tolerance regulation and might play an essential role in autoimmune and auto-inflammatory diseases and tumorigenesis [48]. For patients who do not respond to PD-1/PD-L1 antibodies, targeting Siglec-15 could be an effective alternative therapy. Patients with high-risk scores might also benefit more from anti-Siglec-15 immunotherapy, which provides new insights into BRCA immunotherapy.
As a highly heterogeneous tumor, BRCA has four distinct molecular subtypes that are more important for survival prognosis and guide individual therapeutic decisions [49, 50]. Generally, TNBCs in young patients are much larger than those in the elderly, and therefore, the former might prove to be an appropriate cohort for cancer immunotherapy. Our studies illustrated that the low-risk score appeared to be maintained in patients with luminal A and TNBC. To provide an individualized and accurate prediction, a nomogram for BRCA subtype patients (luminal A and TNBC) that was appropriate for immunotherapy evaluation was constructed. Results of the calibration curve demonstrate that it performed well.
In summary, the RBPLSig could provide promising evidence for OS prediction. Furthermore, the RBPLSig is associated with immune infiltration levels and could be a new biomarker to predict the therapeutic response of the BRCA patient. However, further studies are necessary to evaluate the accuracy of our nomogram and prediction online tools for patients with different breast cancer subtypes. More experiments are also necessary to confirm the potential role of RBP-related lncRNAs.
The datasets analyzed during the current study are available in the TCGA repository (https://tcga-data.nci.nih.gov/tcga/) and GEO repository (https://www.ncbi.nlm.nih.gov/geo/).
QX, JZ and HZ conceived and designed the study. JZ and HZ analyzed and interpreted the data. YG, KH, QD, and WS verified the data. JZ associated all the statistical analysis. QX, HZ, and JZ prepared figures and wrote the manuscript. All authors have read and approved the final version.
Not applicable.
Not applicable.
The present study was supported by the Natural Science Foundation of China (#81701185), the High School Key Research Project of Henan Province (#22A320039), and the ZHONGYUAN QIANREN JIHUA (#ZYQR20191205).
HZ is an employee of Zhengzhou Revogene Ltd. QX was an employee of Zhengzhou Revogene Ltd at the time this research was conducted. All authors declare that they have no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
