Academic Editor: Graham Pawelec
Background: Radiation-induced bystander effects are induced changes in cells that were not themselves directly irradiated but were in the vicinity of a radiation path. Such effects, which occur in the microenvironment of an irradiated tumor, remain poorly understood and depend on the cell type and irradiation quality. This study aimed to evaluate bystander effects in non-irradiated chondrocytes that received conditioned medium from irradiated chondrosarcoma cells. Methods: SW1353 chondrosarcoma cells were irradiated with X-rays and carbon ions, each at 0.1 Gy and 2 Gy, and the conditioned media of the irradiated cells were transferred to T/C-28A2 chondrocytes and Human Umbilical Venous Endothelial Cells (HUVECs). The whole proteome of bystander chondrocytes was analyzed by label-free mass spectrometry, and a comparative study was performed by dose and irradiation quality. HUVECs were evaluated for inflammatory cytokine secretion. Results: The bystander response of chondrocytes to X-ray irradiation primarily affected the protein translation pathway (DHX36, EIF3B, EIF3D, EIF3M, EIF5, RPL6, RPLP0, RPS24, SYNCRIP), IL-12 (AIP, BOLA2, MIF, GAS6, MIF, PDGFRB) and the oxidative stress pathway (MGST3, PRDX2, PXDN, SOD2, TXN, TXNL1). Following carbon-ion irradiation, the G1/S pathway (PCBP4, PSMD12, PSME, XIAP) and mitotic G2 DNA damage checkpoint pathway (MRE11, TAOK1, UIMC1) were engaged. Changes in the regulation of chromosome separation (BCL7C, BUB3, CENPF, DYNC1LI1, SMARCA4, SMC4) were associated with only low-dose X-ray and carbon-ion irradiation. Modification of the protein translation pathway represented at least 30% of bystander effects and could play a role, possibly along with stress granules, in reduction in cellular metabolism to protect proteins. Stress granules were significantly enriched according to an interaction map. Conclusions: All these accessions corresponded to a window of the proteins modulated in response to the bystander effect. Our chondrosarcoma model clarified the nature of the bystander response of chondrocytes and may suggest several interesting new mechanisms that are specific to particular irradiation doses and qualities.
Chondrosarcoma is a heterogeneous group of malignant bone tumors representing 20% of bone sarcomas and have the particularity of producing cartilaginous extracellular matrix [1, 2]. It is one of the two most common primary bone cancers in adults, the other being osteosarcoma. Chondrosarcoma has a relatively slow growth rate and is characterized by low cellularity. It has the capacity to give rise to metastases, which can have a high potential for recurrence, depending on the grade of the tumor. The first line of treatment is surgery coupled with chemotherapy. Radiation therapy is used to control the tumor locally; it is considered when the tumor cannot be completely resected, possibly due to limited access [2, 3, 4, 5, 6]. Chondrosarcoma is considered to be chemotherapy resistant; grade III chondrosarcoma has a 5-year survival prognosis of 60.6% with chemotherapy and 58.6% without chemotherapy [6]. It is also considered resistant to conventional radiotherapy, due to the properties of the tumor and the hypoxic environment. Consequently, these tumors require irradiation at high dose, especially when inoperable.
Hadrontherapy, an innovative technique of non-conventional radiotherapy, has a greater effect on cancer cells that are usually resistant to conventional radiotherapy [7]. The particles used in hadrontherapy are often protons or carbon ions (C-ions). In comparison with proton and X-ray therapy, C-ion therapy is more effective against deep and radioresistant tumors, thanks to better ballistic (dose deposition at the end of the track as a Bragg peak) and the higher linear energy transfer (LET) of C-ions [8, 9].
With this higher irradiation accuracy, it is crucial to understand the role of the microenvironment and the impact of communication between irradiated cells and nearby non-irradiated cells, i.e., the bystander effect [10]. The radiation-induced bystander effect is the biological response of non-irradiated cells in contact with or in the vicinity of directly irradiated cells [11]. This phenomenon has been observed in a diversity of model systems, both in vivo and in vitro, with primary cells, hematopoietic cells and cancer cells [12, 13, 14, 15].
Bystander effects have been assessed using endpoints usually associated with direct-radiation studies, including clonogenic survival, apoptosis, micronuclei formation and DNA damage [10, 16, 17]. However, it has also been shown that exposure of hematopoietic cells to radiation can activate inflammatory signalling pathways in bystander human umbilical venous endothelial cells (HUVECs) [18], potentially leading to the modulation of the irradiated cells’ microenvironment. This intercellular communication was highly dependent on irradiation dose and radiation quality. A non-linear dose response has often been observed, with a higher effect at lower dose [19, 20]. Between irradiated chondrosarcoma cells and bystander chondrocytes, a radiation-induced bystander effect has been observed at low dose of X-ray and, to a lesser extent, C-ion irradiation [21]. The radiation-induced bystander mechanism is still not well understood. Study results have been unclear regarding the magnitude of the effect, dose response and radiation quality, especially with C-ions [17, 22, 23].
Several lines of evidence point to intercellular bystander signalling, mediated by reactive oxygen species, cytokines, hole-junction proteins, and extracellular elements [11, 24, 25, 26, 27]. Exosomes, which are well characterized as key players in cell-to-cell communication in a variety of conditions, were altered by irradiation of head and neck cancer cells [28]. In exosomes from irradiated cells, numerous proteins were modulated, including proteins involved in radiation response, metabolism of radical oxygen species, DNA repair, chromatin packaging and protein folding [28].
In a previous analysis of the secretome of chondrosarcoma cells irradiated with low-dose X-rays, a set of forty modulated proteins was observed. These included a protein cluster related to the ribo-nucleosome compartment, cytoplasmic stress granules and proteins from exosomes involved in oxidative stress cell migration and motility [29]. In the same study, the biological response of nearby chondrocytes was assessed using a gel-based proteomic analysis. Observed bystander effects included changes in cytokine-mediated signaling pathways, interleukin-12, cell junction and motility and extracellular exosome formation [29].
In this study, we sought to explore further and to confirm some of these bystander candidates. We compared two ionizing radiation qualities (X-rays and C-ions) and specific irradiation doses, using the same cellular model (Fig. 1) and label-free direct mass spectrometry, which should display the proteome of bystander cells with high coverage [10]. In order to reproduce in vitro the communication between an irradiated tumor cell and non-irradiated normal (non-cancerous) cells during radiotherapy of bone/cartilage cancer, we selected a chondrosarcoma cell line (as irradiated tumor cells) and a chondrocyte cell line (as non-irradiated normal cells). In order to investigate bystander effects at the level of the tumor microenvironment, we analyzed the inflammatory status and response of HUVECs cultured in contact with conditioned media from irradiated chondrosarcoma cells.
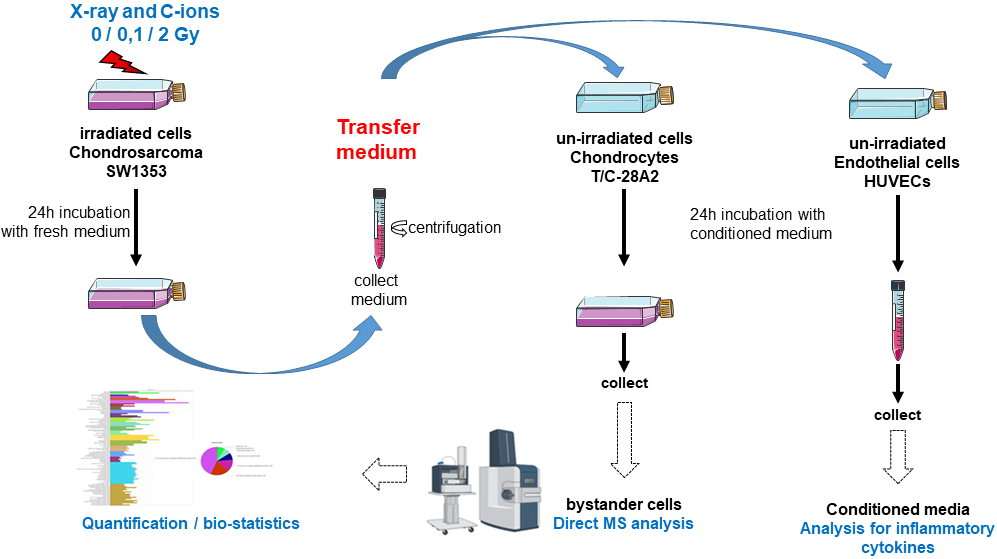 Fig. 1.
Fig. 1.Schematic representation of experiments followed for the medium transfer protocols. Chondrosarcoma cells were first irradiated with X-ray or carbon ion. Immediately after irradiation, the medium was changed with fresh new medium and these irradiated cells were incubated for 24 h. Then, the conditioned medium was collected and centrifuged. This conditioned medium was transferred to non-irradiated chondrocytes or HUVECs cells. Then, bystander cells were collected for direct MS analysis (chondrocytes) or analysis of inflammatory cytokine secretion (HUVECs).
A chondrosarcoma cell line, SW1353 (CLS Cell Lines Service GmbH, Eppelheim,
Germany), was cultured in RPMI-1640 medium (Roswell Park Institute Medium 1640,
Sigma-Aldrich) supplemented with 10% fetal calf serum, 2 mM L-glutamine and 1%
antibiotics (Penicillin-Streptomycin solution, Sigma-Aldrich). A chondrocyte cell
line, T/C-28A2 (a gift from Prof. Mary B. Goldring, Hospital for Special Surgery,
Weill Medical College of Cornell University, New York, NY, USA), was cultured in
the same medium as SW1353 cells. All experiments were performed in a humidified
atmosphere with 5% CO
X-ray irradiations were performed with a PxiXradSmart 225cX irradiator using a
tube tension of 225 kV, as previously described [21, 22, 23, 24, 25, 26, 27, 28, 29, 30], and C-ion irradiations
were performed in the GANIL facility (Caen, France) using the high-energy beam
line IRABAT, as previously described [31, 32]. For C-ions, a LET of 73
keV/
Irradiated SW1353 cells and T/C-28A2 bystander cells were plated in T25 cm
Proteins were extracted from bystander chondrocytes (dry pellet) in lysis buffer (RIPA made without protease, in order not to interfere with proteases mass spectrometry) and left on ice for 20 min [29]. Mechanical grinding was carried out using a hand-held motorized pellet pestle for 20 s on ice, followed by 20 min at rest. The sample was then centrifuged (14,000 rpm, 20 min), and the supernatant containing the proteins was transferred to a clean tube and stored at –80 °C. Protein quantification was performed using a Bradford kit (Thermo Scientific, Waltham, MA, USA).
Five micrograms of each protein extract was prepared using a modified gel-aided
sample preparation protocol [33]. Samples were digested with trypsin/Lys-C
overnight at 37 °C. For nano-LC fragmentation, protein or peptide samples were
first desalted and concentrated onto a
The chromatography step was performed on a NanoElute (Bruker Daltonics, Bremen, Germany)
ultra-high-pressure nano flow chromatography system. Approximately 200 ng of each
peptide sample was concentrated onto a C18 pepmap 100 (5 mm
Mass spectrometry experiments were carried out on a TIMS-TOF pro mass spectrometer (Bruker Daltonics, Bremen, Germany) with a modified nano electrospray ion source (CaptiveSpray, Bruker Daltonics, Bremen, Germany). A 1600 spray voltage with a capillary temperature of 180 °C was typically employed for ionizing. Mass spectra were acquired in the positive mode in the mass range from 100 to 1700 m/z. In the experiments described here, the mass spectrometer was operated in PASEF mode with exclusion of single charged peptides. A number of ten PASEF MS/MS scans were performed in 1.25 s from charge range 2–5.
Preview software (Byos TM, Protein Metrics, Cupertino, CA, USA) was employed to estimate the quality of the tryptic digestion in the samples and to predict the post-translational modifications present. The result was used for the “database search/identification” part. The sequence of the peptide was determined based on the fragmentation pattern. A search of an updated UniProt Homo sapiens database was performed in Peaks XPro software (Peaks XPro softwar Bioinformatic Solutions Inc., Waterloo, ON, Canada). The variable modifications allowed were as follows: K-acetylation, methionine oxidation, Deamidation (NQ), and Nerm-PyroGlu (Q). In addition, C-Propionoamide was set as fix modification. “Trypsin” was selected as Specific. Mass accuracy was set to 20 ppm and 0.05 Da for MS and MS/MS mode, respectively. Data were filtered according to an FDR of 5%. Protein redundancy was eliminated based on the presence of an identical set or subset of peptides.
To quantify the relative levels of protein abundance between different groups,
samples were analyzed using the label-free quantification feature in the PEAKS
XPro software. Feature detection was performed separately on each sample by the
expectation-maximization-based algorithm. The features of the same peptide from
all replicates of each sample were aligned through the retention time alignment
algorithms. Mass error tolerance was set at 30 ppm, ion mobility tolerance (1/k0)
at 0.07 and retention time tolerance at 7 min. Normalization factors of the
samples were obtained by the total ion current (TIC) of each sample. The level of
protein abundance was calculated as the summed area of the top three unique
peptides. A significance
Early-passage HUVECs from a single donor (#798) were a gift from Dr. N.
Cherradi (Biology and Biotechnologies for Health Laboratory, CEA-Grenoble,
France). For routine culture, HUVECs were passaged twice per week, seeded at 3500
cells/cm
For bystander studies, conditioned media were obtained as described above, at 6 and 24 h after irradiation (or not) with X-ray or C-ions. To match HUVEC optimal culture conditions as closely as possible, the conditioned media were diluted by half using 2 X M199 medium. HUVEC cultures, after passages 5 to 8, were expanded for 3 days in 12-well plates and then incubated at 37 °C for 24 h in a humidified incubator with 5% CO2 without or with LPS (100 ng/mL, cat # L3024, Sigma-Aldrich, Saint-Quentin-Fallavier, France). Then supernatants were harvested to quantify cytokine secretion. Bystander HUVEC supernatants were centrifuged (10 min at 1400 rpm) and filtered to eliminate cells fragments, and aliquots were then frozen at –20 °C until use.
We simultaneously measured the secretion of IL-6, IL-8, IP-10, GM-CSF, MCP-1,
VEGF and TNF
The radiation-induced bystander effect was evaluated at the proteome level in bystander chondrocytes that had received the conditioned medium of chondrosarcoma cells irradiated with 0 Gy (control), 0.1 Gy or 2 Gy of X-ray or C-ions. The experiments were performed in biological triplicates. The medium-transfer protocol was selected for consistency with our previous characterization of bystander cells and their secretome and proteome [21, 29] (Supplementary Fig. 1). This protocol allowed us to compare the cell responses of non-irradiated cells (bystander response) across irradiation doses (0 vs. 0.1 vs. 2 Gy) and irradiation quality (X-ray vs. C-ions). Immediately after irradiation treatment, chondrosarcoma cells were cultured in fresh medium, which was itself never irradiated (Fig. 1). Chondrocytes were incubated for 24 h with the conditioned medium. Proteins from total cellular lysates of the chondrocytes were extracted and digested in solution, and the resulting peptides were analyzed using LC-MS/MS.
In total, 64,517 peptides, assembled into 6104 proteins, were identified from the 18 samples analyzed. Differential expression of proteins was considered significant when the p-value exceeded 0.05 (see Methods).
Control samples from X-ray and C-ion experiments were grouped together as biological replicates to reduce biological variability between experiments and to allow direct comparison between doses and irradiation qualities. These control samples corresponded to chondrocytes that had received conditioned medium from sham-irradiated (X-ray and C-ion) chondrosarcoma cells. Controls were compared with chondrocytes cells that had received conditioned medium from chondrosarcoma cells irradiated with X-rays (Comparison 1) or with C-ions (Comparison 2). Finally, a third comparison was performed between chondrocytes in medium conditioned by chondrosarcoma cells irradiated with X-rays (0.1 Gy) and chondrocytes in medium conditioned by chondrosarcoma cells irra diated with C-ions (0.1 Gy) (Table 1).
| Comparison 1 | Comparison 2 | Comparison 3 | |
| X-ray effects | C-ions effect | Low dose effect | |
| X-ray 0 Gy (1; 2; 3) C-ions 0 Gy (1; 2; 3) | X | X | X |
| X-ray 0, 1 Gy (1; 2; 3) | X | X | |
| X-ray 2 Gy (1; 2; 3) | X | ||
| C-ions 0, 1 Gy (1; 2; 3) | X | X | |
| C-ions 2 Gy (1; 2; 3) | X | ||
| Comparison 1 = control (0 Gy) vs. X-ray (0.1 Gy) vs. X-ray 2 Gy; Comparison 2 = control (0 Gy) vs. carbon ion (0.1 Gy) vs. carbon ion 2 Gy; Comparison 3 = control (0 Gy) vs. X-ray (0.1 Gy) vs. carbon ion 0.1 Gy. (1; 2; 3) = 3 biological replicates. | |||
The first comparison identified proteome changes in bystander cells that had
received medium from X-ray irradiated cells. A total of 179 accessions (protein
groups) were found to be differentially expressed (p
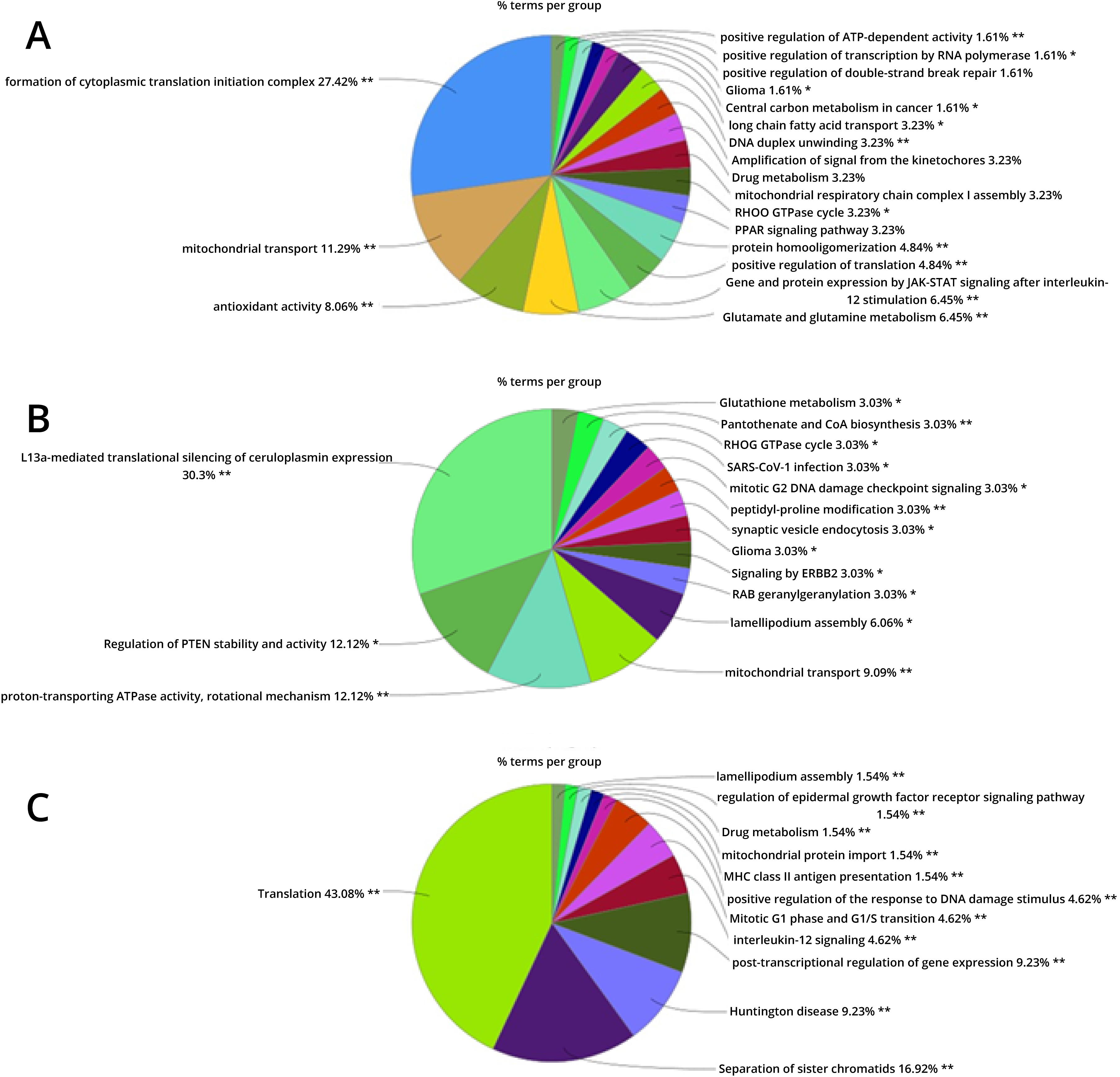 Fig. 2.
Fig. 2.ClueGO functional analysis (% terms per group) of the modulated accessions from. (A) The comparison 1 (0 Gy vs. X-ray 0.1 Gy vs. X-ray 2 Gy). (B) The comparison 2 (0 Gy vs. carbon ion 0.1 Gy vs. carbon ion 2 Gy). (C) The comparison 3 (0 Gy vs. X-ray 0.1 Gy vs. carbon ion 0.1 Gy). See the method section for the ClueGO settings.
The second comparison identified changes in the proteome of bystander samples
that had received medium conditioned by C-ion-irradiated cells. A total of 126
accessions were found to be modulated (p
The third comparison identified differences in the bystander effect after
low-dose X-ray versus low-dose C-ions; the proteome of bystander samples in
medium conditioned by cells irradiated by low-dose X-ray were compared to those
in medium from cells irradiated by low-dose C-ions. A total of 182 accessions
were found to be modulated (p
In parallel to the proteomic analysis, the same conditioned media were applied
to HUVECs to look for an inflammatory response, potentially indicated by cytokine
levels (Fig. 1). First, HUVECS were tested for their capacity to secrete several
cytokines, with or without LPS activation (Supplementary Fig.
8). Treatment with LPS induced a significant increase in the concentration of
all the cytokines tested (IL-8, IP-10, IL-6, MCP-1, GM-CSF, TNF
Two independent experiments were performed for the two different radiation types, X-rays and C-ions. In each experiment, conditioned media from chondrosarcoma cells irradiated at different doses was applied to HUVECs, and the level of each cytokine was observed. The sample with sham-irradiated conditioned media was used to normalize measured cytokine levels.
Depending on the dose, the irradiation quality and the cytokine tested, a
non-explained variability was observed between the two experiments
(Supplementary Fig. 9). In the C-ion experiment, the levels of
IL-8, IL-6, GM-CSF, MCP-1 and TNF
This study follows on a previous study of the bystander response in chondrocytes receiving conditioned medium from chondrosarcoma cells that had been irradiated with low-dose X-rays [29]. In the previous study, a gel-based analysis of the proteome of bystander cells indicated several potential effectors, pathways and mechanisms. These included oxidative stress response, cellular motility, exosome pathways and stress granules [29]. For the present study, we maintained the same protocol for cellular conditions [21, 29]. However, label-free mass spectrometry-based analysis allowed a deeper and broader investigation of the cellular response of bystander chondrocytes (Supplementary Fig. 10).
We observed that several GO pathways were enriched, including the cytoplasmic translation complex, mitochondrial transport, antioxidant activity and JAK-STAT signaling after interleukin-12 stimulation. Some of these pathways were also observed in our previous studies. For example, the interleukin-12 signaling pathway was proposed to be involved in the propagation of the bystander effect, through the modulation of cyclophylin A [29, 34]. Pathways connecting oxidative stress and translation complex stimulation proteins were impacted as well [29]. The modulated proteins were different, but the impacted pathways were similar.
Our study indicated that several pathways typically altered following direct irradiation of cells (i.e., homologous recombination and double-strand break repair) also experience bystander effects [10, 17, 20].
We constructed an in silico functional protein association network, using the program String, to reveal the connections between all modulated accessions (Fig. 3). Five processes with high probability of interaction emerged from this network, including the ribonucleoprotein granule (blue) and complex (green), cellular response to stress (yellow), the oxidation-reduction process (magenta) and the interleukin-12-mediated signaling pathway (red) (Table 2). We confirmed that stress granules were enriched [29] by a connection with several accessions that had previously been implicated in the cytoplasmic translation complex.
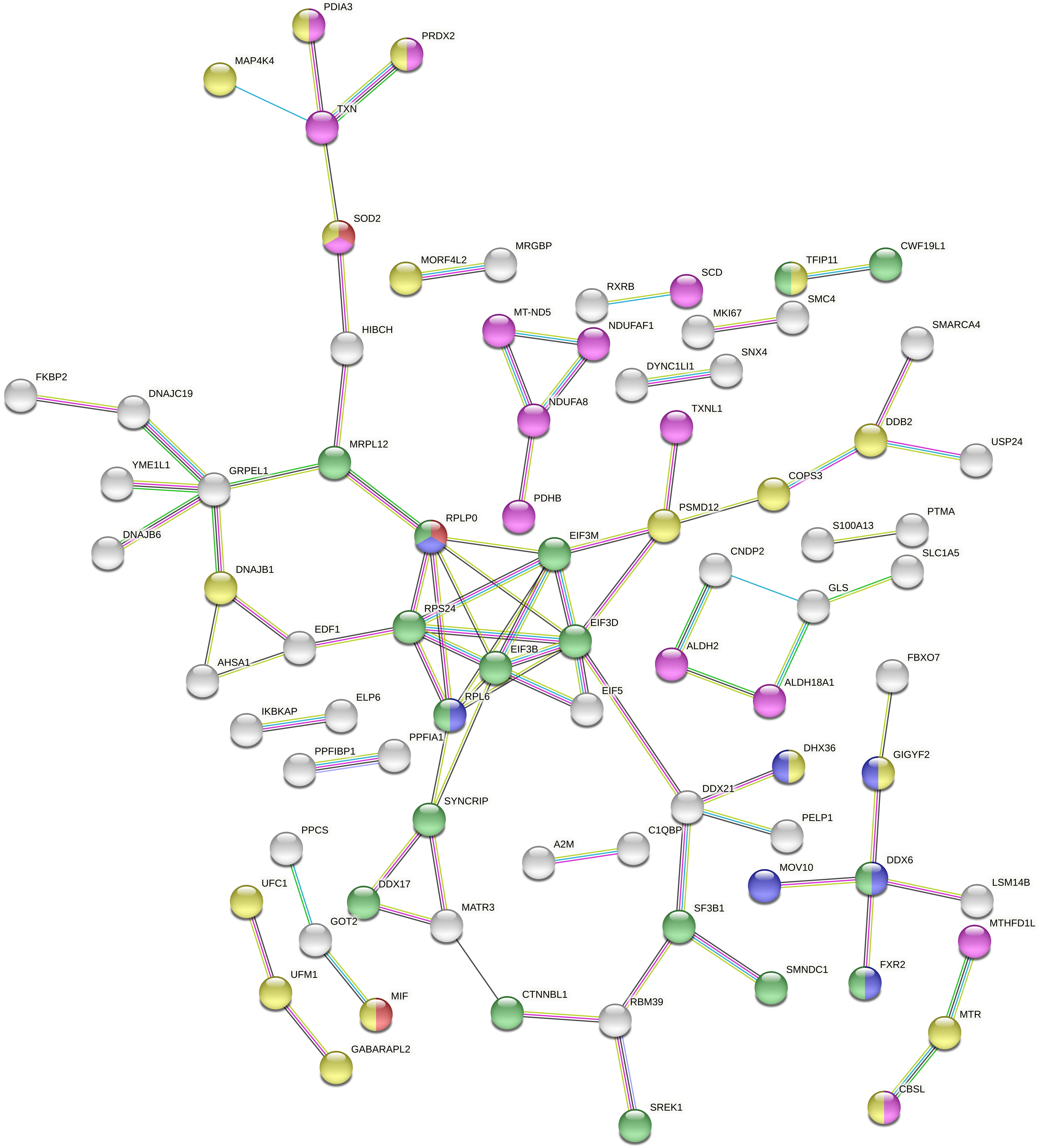 Fig. 3.
Fig. 3.Full String network of the modulated accessions from the comparison 1 (0 Gy vs. X-ray 0.1 Gy vs. X-ray 2 Gy). The minimum interaction score was set to high confidence (0.700) score, and disconnected nodes were hidden in the network. The corresponding colour and GO were displayed in Table 2 (https://string-db.org/).
| #Color | Term ID | Term description | Observed gene count | Strength | False discovery rate | Matching proteins in your network (labels) |
| Comparison 1 | ||||||
| Blue | GO:0035770 | Ribonucleoprotein granule | 10 | 0.66 | 0.0108 | YTHDC2, FXR2, POLR2G, HABP4, GIGYF2, MOV10, RPL6, DHX36, RPLP0, DDX6 |
| Limegreen | GO:1990904 | Ribonucleoprotein complex | 17 | 0.44 | 0.0162 | EIF3D, FXR2, CWF19L1, MRPL12, SREK1, SF3B1, EIF3B, CTNNBL1, SMNDC1, SYNCRIP, DDX17, TFIP11, RPL6, RPS24, EIF3M, RPLP0, DDX6 |
| Yellow | GO:0033554 | Cellular response to stress | 33 | 0.32 | 0.0144 | GABARAPL2, MIF, UNG, DNAJB1, DDB2, BRIP1, THBS1, PDGFRB, C19orf10, GET4, COPS3, PDIA3, PRDX2, POLR2G, WDR48, GAS6, MAP4K4, MAP2K3, PSMD12, BABAM1, MTR, UFC1, UFM1, CHD2, AGAP3, CBSL, TFIP11, DYSF, GIGYF2, MORF4L2, USP19, DHX36, SOD2 |
| Red | GO:0035722 | interleukin-12-mediated signaling pathway | 5 | 1.07 | 0.0285 | MIF, AIP, BOLA2B, SOD2, RPLP0 |
| Magenta | GO:0055114 | Oxidation-reduction process | 21 | 0.39 | 0.0406 | TXNL1, SPR, PXDN, NDUFAF1, ALDH2, PDIA3, PRDX2, PDHB, IMPDH2, MT-ND5, MGST3, SCD, ALDH18A1, NDUFA8, TXN, CBSL, CRYZ, SLC25A13, CROT, SOD2, MTHFD1L |
| Comparison 2 | ||||||
| Blue | GO:0070062 | Extracellular exosome | 32 | 0.38 | 0.00086 | ATP6V1D, TMEM109, MOGS, CANX, PXDN, MPP5, TAOK1, ALDH2, RAB2A, RAB7A, ATP6V1B2, ATP1B3, AP2M1, ARPC2, VPS35, CALML3, SNX18, RPLP2, HNRNPM, HSP90AA1, PSMD12, ECE1, ANXA1, PTPRA, GMDS, PSME1, FAH, FAM49B, GSTK1, APPL2, RPLP0, COASY |
| Red | KW-0687 | Ribonucleoprotein | 8 | 0.65 | 0.0412 | MRPL3, HNRNPH3, RPL38, RPLP2, HNRNPM, HNRNPUL1, PCBP4, RPLP0 |
| Comparison 3 | ||||||
| Yellow | GOCC:0070062 | Extracellular exosome | 13 | 0.58 | 0.0046 | MIF, CANX, THBS1, RAB2A, RAB7A, VPS35, FASN, A2M, ANXA1, ANXA4, DHX36, SLC1A5, SOD2 |
| Magenta | GO:0007005 | Mitochondrion organization | 19 | 0.66 | 4.81e-05 | GABARAPL2, PPIF, ATP5B, NDUFS3, C19orf52, AIP, WIPI2, VPS35, YME1L1, NDUFAF3, MT-ND5, NDUFA8, SLC25A6, DNAJC19, TIMM9, PNPT1, SSBP1, SOD2, ATPAF1 |
| Blue | GO:0022613 | Ribonucleoprotein complex biogenesis | 18 | 0.66 | 7.59e-05 | RRP9, ERAL1, LUC7L, UTP15, RPL38, RUVBL1, EIF3C, SF3B1, RPS2, DDX21, RPL10A, CRNKL1, SRPK2, DDX17, RPL6, EIF3F, EIF3M, RPLP0 |
| Red | GO:0033554 | Cellular response to stress | 33 | 0.32 | 0.0117 | GABARAPL2, MIF, POLR2I, CDC37, PPIF, CANX, ARHGEF6, RRM2B, THBS1, PDGFRB, PSMD3, TNC, EGFR, WIPI2, POLR2G, WDR48, HSPA6, RUVBL1, POLE, HNRNPM, BRAT1, PSMD12, ANXA1, PSME1, LARS, LAMB2, MORF4L2, PNPT1, USP19, DHX36, PCBP4, SOD2, TIMELESS |
| Limegreen | HSA-447115 | Interleukin-12 family signaling | 6 | 1.06 | 0.0037 | MIF, CANX, AIP, BOLA2B, SOD2, RPLP0 |
The radiation-induced bystander effect began rapidly, within several minutes after radiation [35]. The mechanism of the rapid cellular response likely involves stress granule formation and protein translation modulation.
We observed bystander effects on oxidative stress responses (or oxidation-reduction processes) with their 21 involved proteins (Table 2). Bystander activation of these responses has been observed in other model systems, recently in HepG2 and normal liver cells [36], human hematopoietic progenitor and stem cells [37] and mice exosomes [38].
The activation of JAK-STAT signaling after interleukin-12 stimulation was less expected. According to our proteomic study, the proteins MIF, AIP, BOLA2B, SOD2 and RPLP0 were modulated in bystander chondrocytes. These proteins are all involved in the interleukin-12-mediated signaling pathway. The interleukin-12 cytokine likely plays a central role in regulating immune responses, synergizing with several other cytokines for increased immune-regulatory activities in inflammation responses [39].
To further explore how signals from the bystander effect might influence the immune microenvironment, we also analyzed whether chondrosarcoma-conditioned culture media could modulate inflammatory signaling in HUVECs, as has been described for monocytes and macrophage cells. Our supernatant transfer experiments provide preliminary evidence that irradiated chondrosarcoma cells release soluble factors able to modulate the secretion of several cytokines from resting or LPS-stimulated HUVEC cells.
Some bystander responses of chondrocytes seem to be similarly stimulated across different types of irradiation. Initiation of cytoplasmic translation was the most pronounced GO enriched in the bystander response to C-ion irradiation and was likewise observed following X-ray irradiation. Change in mitochondrial transport was also observed after both kinds of irradiation.
In contrast, the regulation of intracellular pH and G1/S DNA damage checkpoints appeared to be bystander responses specific to C-ion irradiation. Modulation was observed in proteins PCBP4, PSMD12, PSME1 and XIAP, which are all involved in G1/S DNA damage response. This bystander response to C-ion irradiation has not been reported previously and could represent a specific signature of this biological effect. Some of these proteins were also associated with the extracellular exosome (Fig. 4, Table 2).
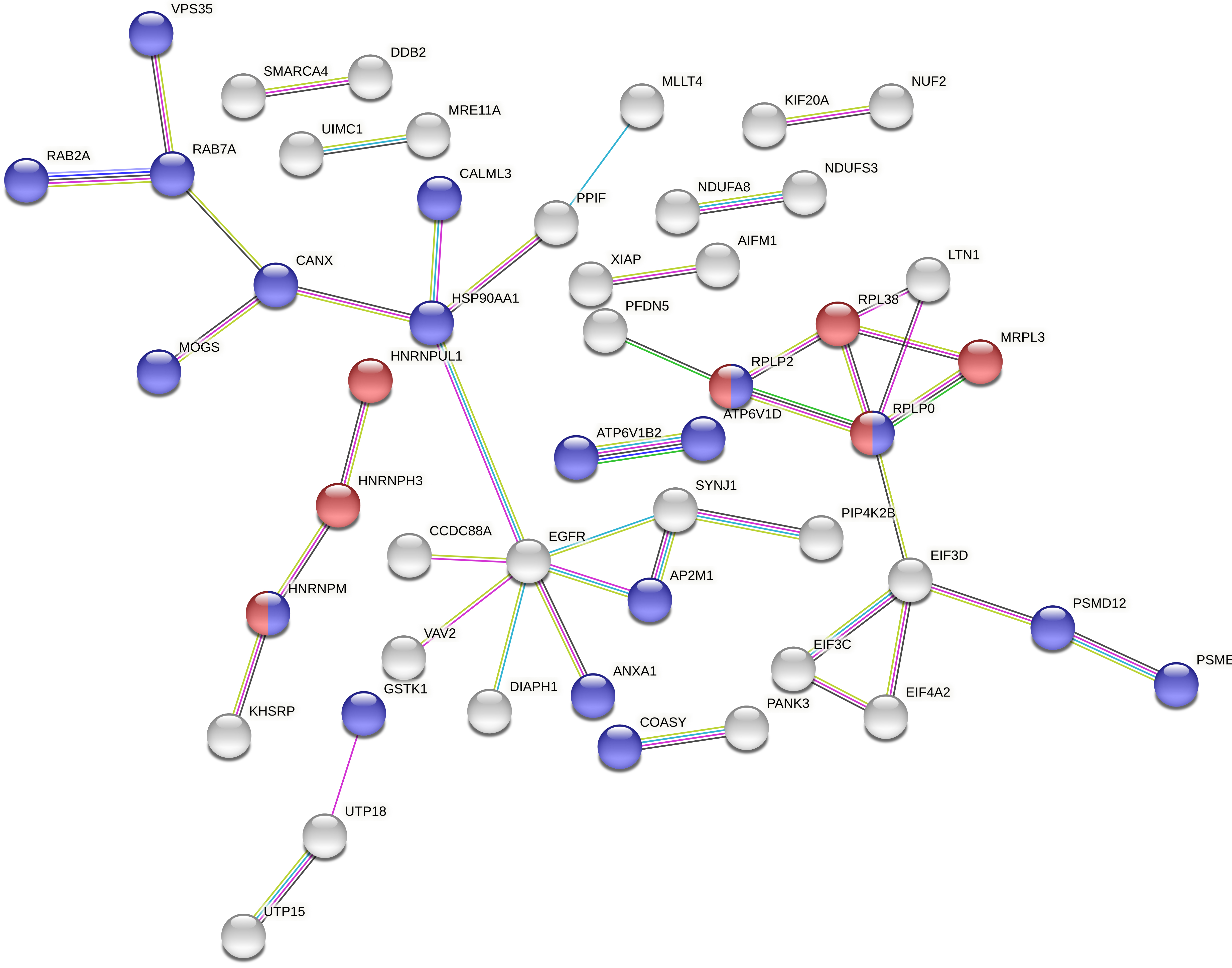 Fig. 4.
Fig. 4.Full String network of the modulated accessions from the comparison 2 (0 Gy vs. carbon ion 0.1 Gy vs. carbon ion 2 Gy). The minimum interaction score was set to high confidence (0.700) score, and disconnected nodes were hidden in the network. The corresponding colour and GO were displayed in Table 2 (https://string-db.org/).
Other affected accessions were related to translation processes. Mitotic G2 DNA damage checkpoint signaling was also impacted, through proteins MRE11, TAOK1 and UIMC1. A modulation of cell cycle progression that would reduce protein translation could explain a decrease in cell division and motility that has been observed in other studies [21].
Finally, in our analysis of low-dose bystander response, we observed changes in a pathway that was related to the regulation of chromosome separation. This step in mitosis, between metaphase and anaphase, seemed to be impacted by low-dose irradiation with both X-rays and C-ions. Proteins such as BCL7C, BUB3, CENPF, DYNC1LI1, SMARCA4 or SMC4 were modulated. Some of these accessions are involved in separation and exchange of sister chromatids (Supplementary Table 4).
The induction of sister chromatid exchange has been observed in CHO cells irradiated in the G1 phase of the cell cycle with alpha-particles [40]. Although 30% of the cells showed an increased frequency of sister chromatid exchanges after exposure to 0.31 mGy, less than 1% of cell nuclei were actually traversed by an alpha-particle; this phenomenon was not observed in cells defective in homologous recombination [41]. In addition to such modification of chromosome separation, other of the pathways previously impacted by X-ray irradiation (translation, stress granules, extracellular vesicles, and interleukin-12) were still observed within the corresponding String network (Fig. 5 and Table 2).
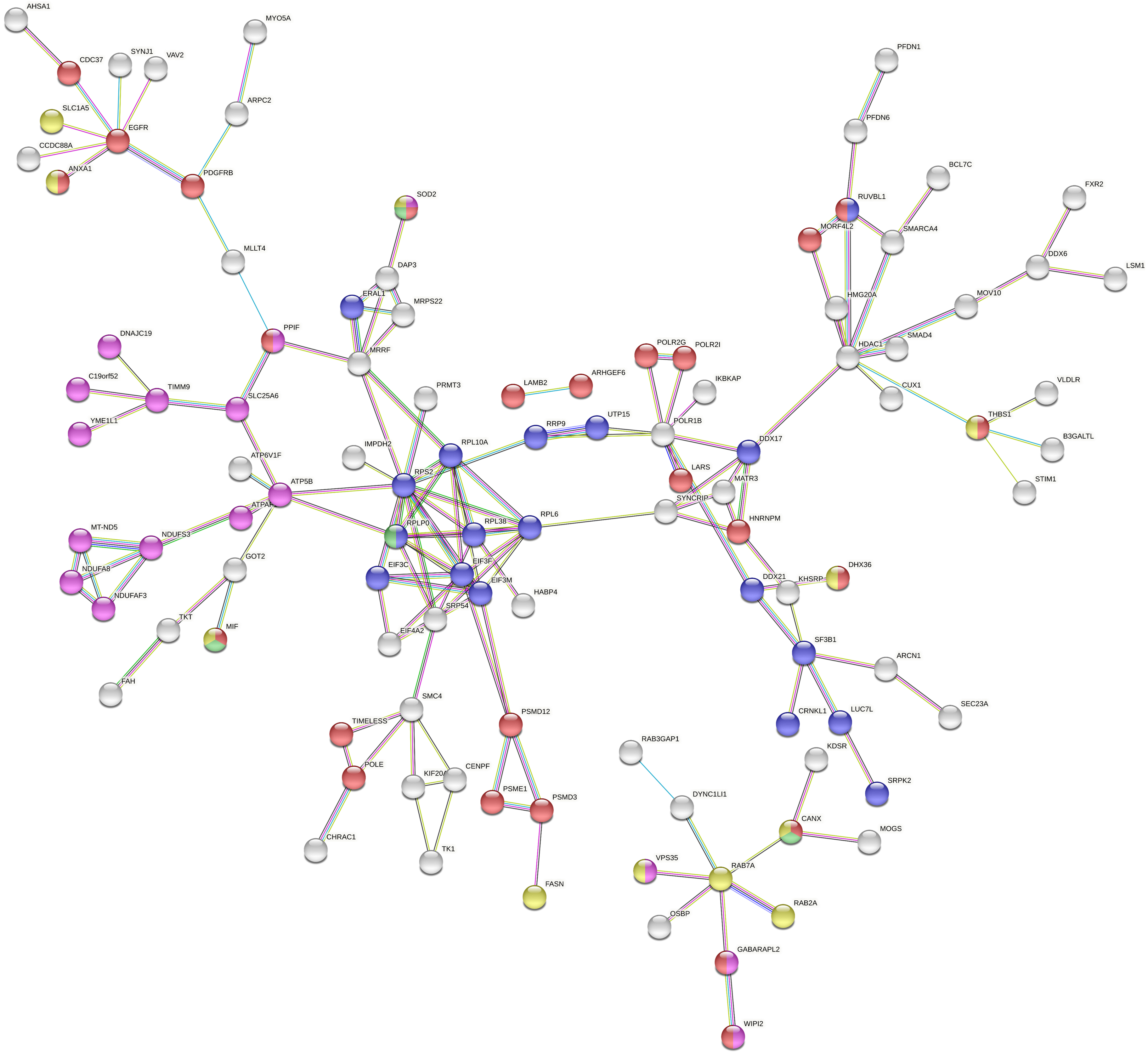 Fig. 5.
Fig. 5.Full String network of the modulated accessions from the comparison 3 (0 Gy vs. X-ray 0.1 Gy vs. carbon ion 0.1 Gy). The minimum interaction score was set to high confidence (0.700) score, and disconnected nodes were hidden in the network. The corresponding colour and GO were displayed in Table 2 (https://string-db.org/).
In interpreting the results of C-ion irradiation, one must consider the relative biological effects (RBEs), especially when comparing C-ions and X-rays [7, 42]. Our analyses included no correction factors, only physical doses of C-ions, as has been recently recommended [43]. Our biological samples were not directly irradiated with C-ions; bystander chondrocytes received only conditioned media from irradiated chondrosarcoma cells. In previous experiments of clonogenic survival, when a comparison of direct effect was performed with X-rays [32], a significant RBE was observed on chondrosaroma cells irradiated with C-ions (RBE ~3), depending on the cell lines and the LET of C-ions. When bystander cells were analyzed with the same approach, a reduction in clonogenic survival was observed at low doses of X-rays and C-ions. However, no RBE of C-ions was observed, as the effect was stronger with X-rays in comparison with C-ions [21]. For this reason, it seems unlikely that the proteomic differences observed in our study between X-rays and C-ions would be related to the RBE of C-ions observed when cells were directly irradiated. The observed differences were more likely related to a qualitative effect of C-ions, which induced different types of DNA damage, such as multiple localized breaks, that are more difficult repair for the cells [44]. Factors secreted by damaged cells could induce a differential response in bystander cells, when compared with X-ray irradiation, as observed in this study. If RBE-corrected doses were applied for C-ion irradiation, as is done during carbon therapy, this could modify the bystander response [45]. In our study, the bystander effect was observed at low dose (about 0.1 Gy). If an RBE-correction were applied, the dose would be divided by three, or about 30 mGy. Such an analysis would be interesting in the context of clinical C-ion therapy.
Irradiation stimulates a DNA damage response in the directly affected cells. It is now clear that cell-cell communication in the microenvironment of irradiated cells also plays a significant role in the cell- and tissue-level resроnse tо rаdiаtiоn exроsure. However, it is challenging to define the impact of this bystander effect in a clinical context because the extent of the effect depends on the tissue and the irradiation dose. Previous studies have reported possible impact of intercellular communication in the biological dose distribution of a treatment plan, with a signal range of 15 mm and an increased dose, up to 30% [46].
We observed bystander effects in non-cancerous cells at low doses of both X-ray and C-ions. This finding suggests important potential impact on nearby non-targeted regions. We report bystander effects on protein translation and oxidative stress homeostasis, which could alter normal cell functioning and potentially delay tissue reconstruction. Several factors involved in G1/S and G2 DNA damage checkpoints were affected, which could also reduce cells’ capacity to divide or to repair DNA damage.
C-ion irradiation is considered more accurate than X-ray irradiation, based on the physical properties of the ions. However, we observed bystander effects on DNA damage checkpoints following C-ion irradiation. Thus, the bystander effect could induced a kind of “biological inaccuracy” of C-ions, for which the magnitude is still to be defined. Our results indicate a necessity to consider the impact on normal tissue close to the tumor during C-ion therapy.
On the other hand, a modification of the microenvironment within the tumor bed could benefit to the treatment, by recruiting immune cells and/or promoting anti-tumor immunity [47]. C-ion irradiation has been associated with a possible increase in inflammation. In a mouse glioma model, C-ion irradiation induced an antiangiogenic and immune-permissive niche in association with a reduced viability of hypoxic and stem cell-like tumor cells [48]. Furthermore, in a human osteosarcoma cell model, an upregulation of PD-L1 expression after high-LET carbon-ion irradiation was greater than that induced by X-rays at the same biological dose [49]. However, such an inflammation response was not observed as the first mechanism involved in the C-ion-related bystander effect. Moreover, in our inflammation analysis on endothelial cells in medium from C-ion-irradiated chondrosarcoma cells, we observed a decline in the secretion of selected inflammatory cytokines.
Taken together, all these results showed a very complex and intricate bystander response in chondrocytes, which depends on irradiation quality. Label-free mass spectrometry analysis furthered our understanding of the molecular mechanisms involved in this bystander response. Many potential bystander effectors were proposed, some specific to either X-ray or C-ions and some common to both forms of irradiation. The translation of proteins was associated with both irradiation qualities; IL12 and antioxidant activity pathways seemed to be specific to X-rays; G1/S mitotic and G2 DNA damage checkpoints were specific to C-ion irradiation. In agreement with our previous study, stress granules seemed to be involved in the bystander response of chondrocytes.
FC designed the study and the experiments; AG and VP performed the cell culturing and protein extractions; BB performed the mass spectrometry experiments and the bio-statistics; IT, ECG and SC performed the inflammation experiments; FC, BB and SC analyzed and interpreted the results; FC wrote the manuscript. All authors read and approved the final manuscript.
Not applicable.
We would like to thank the CIRIL platform (Cimap Caen, France) for C-ions dosimetry and the GANIL facility (Caen, France) for providing us the C-ion beam. The C-ion experiments were performed according to projects P1146-H and P1243-H (F. Chevalier) of the iPAC committee proposal for High Energy beamline on interdisciplinary research at GANIL.
This work was supported in large part by a grant from EDF (Electricité de France), by funding from the Life Sciences group of the four-way national agreement CEA–EDF–IRSN–FRAMATOME (2019–2022) and by funding of the Master 2 internships of A. G. and V. P. Further support was provided by “Agence Nationale de la Recherche”, Equipex Rec-Hadron (ANR-10-EQPX-1401). This work was also supported by a grant from the Region of Normandy, the RIN Tremplin CHOXTRACC (2021–2022) “Recherche de combinaisons thérapeutiques hadronthérapie - thérapies ciblant la radiorésistance biologique liée à l’hypoxie et au stress oxydant tumoral”, co-funded by the Normandy County Council and the French State, in the framework of the interregional development contract, “Vallée de la Seine” 2015–2020 (HAdronBIOlogie en NORmandie: Programme de recherche en Radiobiologie pour l’hadronthérapie au centre ARCHADE). This work was also supported by GRAL, a program from the Chemistry Biology Health (CBH) Graduate School of University Grenoble Alpes (grant number ANR-17-EURE-0003), and in part by the LABEX PRIMES (grant number ANR-11-LABX-0063).
Given his role as Guest Editor, François Chevalier had no involvement in the peer-review of this article and has no access to information regarding its peer-review. Full responsibility for the editorial process for this article was delegated to Graham Pawelec. The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
