†These authors contributed equally.
Academic Editor: Changsoo Kim
Background: Tomato is an important part of daily food, rich source of
multitude nutrients, suitable candidate for bio-pharmaceutical production due to
berry size and has numerous health benefits. Transcriptional regulation of
metalloregulatory heat shock protein-70 family plays pivotal role in plants
tolerance against abiotic stress factors including salinity, heat, cold, drought
and trace metal elements such as cadmium (Cd
Latest genetic engineering tools have increased commercial importacne of tomato
(Solanum lycopersicum L.) by increasing nutritional value via
bio-fortification, improving shelf life, developing berry size and
bio-pharmaceuticals [1, 2, 3]. Tomato is being cultivated on a large scale in
different soil types and under different biotic and abiotic stress conditions [4, 5]. Tomato crop is highly vulnerable to harsh environmental conditions [6].
Abiotic stresses including salinity, drought, trace metal elements and high
temperature cause severe yield loss upto 70% [7, 8]. Trace metal elements
stresses induce a complex signaling pathway which subsequently activates
transcription of metal-responsive genes [9, 10]. Trace metal elements stresses
also induce over-accumulation of reactive oxygen species (ROS) such as hydroxyl
(OH
Hsp70 genes are induced under sudden and rapid increase in temperature. Initiation of transcription of Hsp70 genes in plants is a protective eco-physiological adaptation and a conserved genetic response against abiotic stress factors. Initiation of Hsp70 family gene transcription in response to abiotic stressors aids acclimatisation by restoring normal confirmations of damaged metabolites and maintaining cellular homeostasis [16]. The Hsp70 gene family is very conserved and comprised of multiple genes which play key role in regulating plant developmental processes to endure abiotic stress conditions [17, 18]. Polypeptides of HSP70 family in plants differ in their molecular weights (MW) from 10 kDa to 200 kDa [19, 20]. HSP70 proteins bind with heat-denatured metabolic proteins to protect them from aggregation and refold them to their original quaterinary conformation to perform normal functions. These proteins are predominantly involved in translation, translocation and metal homeostasis as metal contents exceed normal limit [21, 22]. On the basis of C-terminal, HSP70 family protiens are divided into four subgroups, which are differnetially localized and function in almost all cell oraganells. For example, HSP70 of cytosol, plastids and mitochondria harbor EEVD, PEGDVIDADFTDSK and PEAEYEEAKK motifs, respectively [23].
Plants absrob soil-born trace metal elements such as Cadmium (Cd
To develop Cd
Whole genome of tomato (S. lycopercicum) was downloaded from Solanaceae Genomics Network (https://solgenomics.net/) database to construct a local database by employing BioEide7.0 (bioedit.software.informer.com). In order to retrieve tomato SlHsp70 gene sequences, Arabidopsis thaliana AtHsp70 gene sequences were BLAST-analyzed against newly constrctued local tomato database. The Hidden Markov Model (HMM) was employed to validate SlHsp70 gene sequences and their functional domains particularly by running against each AtHsp70 gene (Pfam: PF00012). Incomplete raw reads were excluded and assigned specific name to each gene according to Mendel database for plant gene families listed in Commission on Plant Gene Nomenclature (CPGN) (mbclserver.rutgers.edu/CPGN/), International Society of Plant Molecular Biology (ISPMB) [32]. In order to further validate retrieved SlHsp70 genes, BLAST analyses were also performed in NCBI genome database by selecting tomato whole genome (blast.ncbi.nlm.nih.gov/blast.cgi), SPud DB tomato Solanaceae Genomics Network (solgenomics.net) and phytozome (phytozome.jgi.doe.gov/).
All retrieved SlHSP70 polypeptide sequences were further validated at E-value
Arabidopsis AtHSP70 family proteins were downloaded from Arabidopsis Information Resource (TAIR) (arabidopsis.org) and potato StHSP70 family proteins from the Spud DB Potato Genomics Resources (solanaceae.plantbiology.msu.edu/). Multiple sequence allignments of HSP70 polypeptide sequences of Arabidopsis, potato and tomato were performed in ClustalX 2.0 with default parameters [35]. Phylogenetic analysis were performed to construct an outgroup rooted tree by using 20 A. thaliana AtHSP70, 19 S. tuberosum StHSP70 and 23 S. lycopercicum SlHSP70 protein sequences by performing alignment using Neighbor-Joining (NJ) method in MEGA7.0 with following parameters: 2000 bootstrap replication values, pair-wise gap deletion mode and Poisson model [36, 37].
The phytozome plant genome database (phytozome.jgi.doe.gov/) was explored by
selecting tomato to collect information obout localization of SlHsp70
genes and a genetic map was constructed by employing MapChart tool [38]. Genes of
similar species were placed in the same group of an outgroup rooted tree, defined
as co-paralogs, which were further analyzed to identify tandem duplicatons,
segmental duplications and their coordinates. To investigate evolutionary events
such as gene duplicaiton and divergence for vigourous gene functions as well as
genetic expansion, we explored PGDD (Plant Genome Duplication Database) by
deploying circos. Co-paralogs were considered tandemly duplicated when both of
them were non-homologous and distance between their loci was
Genomic and CDS sequences of each SlHsp70 gene were analyzed to examine number and order of introns and exons with the help of Genes Structure Display Server 2.0 (GSDS 2.0) program (gsds.cbi.pku.edu.cn/index.php) by adjusting following parameters: (a) Intron: color-black, shape-line and line width-3, (b) CDS: color-dark maroon, shape-round corner rectangle and height-12 and (c) UTR: color-blue, shape-rectangle and height-10. Protein sequence of each SlHSP70 was also analyzed to identify conserved motifs with the help of Multiple EM for motif elicitation (MEME) tool (meme.nbcr.net/meme3/meme.html) with following parameters: number of motifs-10 and optimum amino acid residues per motif were 6 to 200 [41].
Phyre2 web (sbg.bio.ic.ac.uk/phyre2/) was utilised in intensive mode to undertake protein modelling of SlHSP70 polypeptides [42]. All predicted models of SlHSP70 proteins were based on c5tkyA, c5e84B, c3d2fC, c2khoA, c3c7nB and c5obuA templates with 100% identification. In-silico direct physical and indirect functional interactions among proteins were predicted by using STRING (version 11.5) database. GO enrichment analyses were performed to predict localization of SlHSP70 proteins within cellular components (red bars), expected molecular function (green bars) and participation in possbile biological processes (blue bars) (Fig. 1 & Supplementary Table 1). To perform GO enrichment analysis for each SlHSP70 protein, OmicsBox and Blast2GO v3.0.11 (www.blast2go.com) were employed [43].
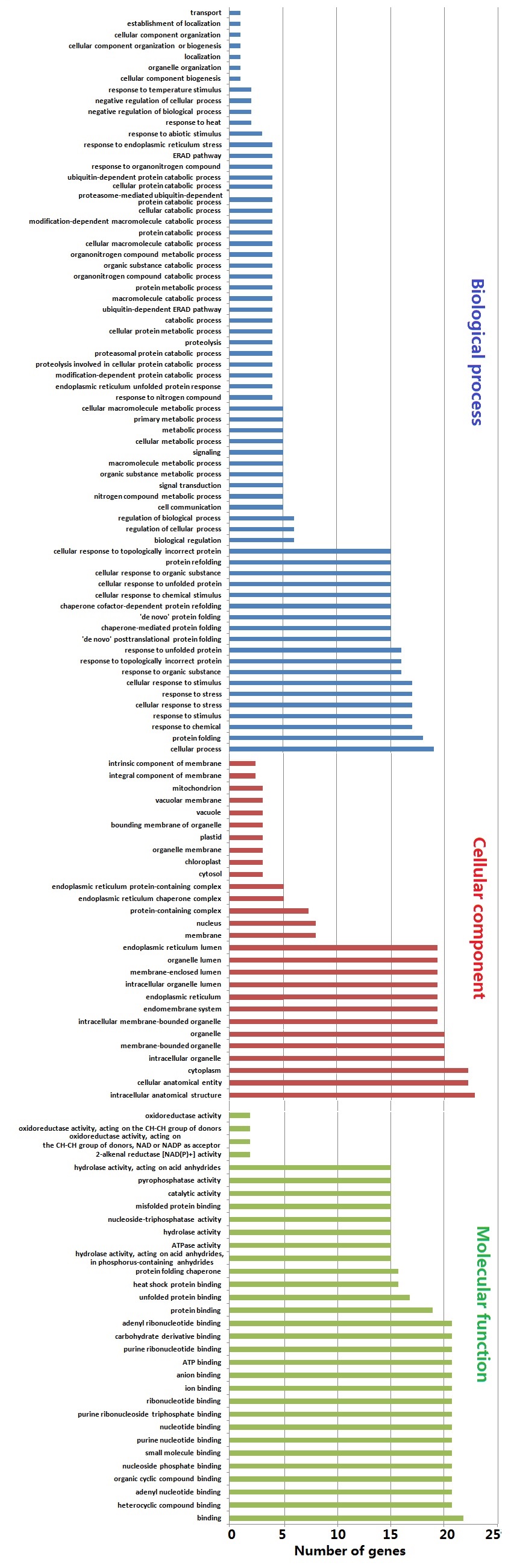 Fig. 1.
Fig. 1.GO enrichment analysis of SlHsp70 genes in S. lycopercicum. Red columns represent cellular components in which SlHsp70 genes displayed expression, blue columns represent biological processes in which SlHsp70 genes played their specific roles and green columns represent expected molecular function of SlHsp70 genes.
We analyzed RNA-seq data under normal conditions and constructed a heatmap to explore expression profiles of all SlHsp70 family genes in different tissues of S. lycopercium including flower bud, unopened flowers, fully opened flowers, fruits (1 cm, 2 cm and 3 cm), mature green fruits, breaker fruits, breaker + 10 fruits, leaves and roots (Fig. 2 and Supplementary Table 2). RNA-seq data and PFKM values were downloaded from tomato functional genomics database (ted.bti.cornell.edu/pgsc_download.shtml) and analyzed using cufflinks v2.2.1. FPKM values were divided by their mean, transformed into log2 ratio and clustered into expression data in the form of heat map (heatmapper.ca/) with the help of MeV4.5 with default parameters [44, 45].
 Fig. 2.
Fig. 2.Expression level of of all 23 SlHsp70 in different tissues including roots, leaves, buds, flowers and fruits of different stages of S. lycopercicum based on RNA-seq (http://ted.bti.cornell.edu/).
The seeds of tomato (line M82) were cultured in greenhouse of Yibin University
in mid Autumn 2020. In order to perform surface sterilization, seeds were soaked
in 10% hypochlorous acid for 5 mins and then washed thrice with ddH
We used TRIzol™ Reagent (Thermo Fisher Scientific, USA) for total
RNA extraction and SuperMix Kit (TransGen, Beijing) was used for cDNA synthesis.
Gene specific primers were manually designed (Supplementary Table 3) and
Hsp70 genes sequences were retrieved by downloading and subsequently analyzing S. lycopercicum genome database, incomplete raw reads were excluded and finally 23 complete candidate Hsp70 gene sequences were selected. All genes were assigned scientific name given as SlHsp70-1 to SlHsp70-23 (Table 1). Gene synteny analysis revealed that all 12 tomoto chromosomes harbored Hsp70 genes except chromosome 5. Total number of amino acids in each SlHSP70 polypeptide were 186 to 890, and molecular weight (MW) was 21273.47 Da to 98787.40 Da (Table 1). Comparatively, high contents of negative and acidic amino acids such as L-glutamic acid and L-alpha-aspartyl residues (Asp + Glu) were observed in almost all SlHSP70 proteins except SlHSP70-12 (Table 1). Isoelectric point (pI) of all 23 genes was also acidic (Table 1).
| Gene name | Sequence ID | Location | (-) | (+) | MW | aa | Total no. of atoms | Instability | Aliphatic index | Intron | pI |
| SlHsp70-1 | Solyc01g106210 | SL2.50ch01:94157485..94161397 | 89 | 84 | 72969.84 | 681 | 10334 | 38.21 | 87.27 | 5 | 5.75 |
| SlHsp70-2 | Solyc06g076020 | SL2.50ch06:47192489..47195586 | 102 | 82 | 71008.48 | 648 | 9984 | 32.79 | 82.02 | 1 | 5.04 |
| SlHsp70-3 | Solyc03g082920 | SL2.50ch03:52794869..52798836 | 114 | 92 | 73457.21 | 667 | 10382 | 29.90 | 85.95 | 6 | 5.07 |
| SlHsp70-4 | Solyc10g086410 | SL2.50ch10:65236863..65240232 | 100 | 81 | 70779.21 | 644 | 9949 | 35.11 | 82.39 | 1 | 5.07 |
| SlHsp70-5 | Solyc01g106260 | SL2.50ch01:94215968..94220340 | 86 | 81 | 71876.57 | 670 | 10151 | 38.91 | 85.04 | 5 | 5.95 |
| SlHsp70-6 | Solyc07g043560 | SL2.50ch07:57457649..57465996 | 134 | 121 | 98787.40 | 890 | 13932 | 39.17 | 80.89 | 12 | 5.91 |
| SlHsp70-7 | Solyc02g080470 | SL2.50ch02:44673835..44685301 | 110 | 98 | 84108.25 | 753 | 11771 | 42.99 | 78.62 | 8 | 6.02 |
| SlHsp70-8 | Solyc06g052050 | SL2.50ch06:35713191..35716219 | 102 | 82 | 67513.37 | 619 | 9543 | 29.09 | 87.74 | 8 | 5.04 |
| SlHsp70-9 | Solyc03g117630 | SL2.50ch03:66724560..66726524 | 99 | 83 | 71849.40 | 654 | 10088 | 31.01 | 87.72 | 0 | 5.21 |
| SlHsp70-10 | Solyc01g099660 | SL2.50ch01:89839013..89842124 | 112 | 98 | 74641.87 | 669 | 10568 | 34.03 | 87.16 | 6 | 5.36 |
| SlHsp70-11 | Solyc07g005820 | SL2.50ch07:655717..659235 | 103 | 86 | 71953.43 | 654 | 10115 | 33.75 | 81.10 | 1 | 5.15 |
| SlHsp70-12 | Solyc03g117620 | SL2.50ch03:66722304..66723457 | 18 | 30 | 21273.47 | 186 | 2997 | 46.72 | 73.33 | 1 | 9.37 |
| SlHsp70-13 | Solyc09g075950 | SL2.50ch09:67581791..67583521 | 69 | 53 | 62723.17 | 576 | 8852 | 42.31 | 98.49 | 0 | 5.56 |
| SlHsp70-14 | Solyc11g020040 | SL2.50ch11:10015582..10019521 | 101 | 90 | 74493.21 | 692 | 10544 | 27.93 | 84.36 | 7 | 5.36 |
| SlHsp70-15 | Solyc11g066100 | SL2.50ch11:51773141..51775439 | 100 | 82 | 71458.91 | 654 | 10036 | 33.10 | 80.69 | 1 | 5.10 |
| SlHsp70-16 | Solyc04g011440 | SL2.50ch04:3894918..3898067 | 100 | 82 | 71389.83 | 651 | 10030 | 32.91 | 80.78 | 1 | 5.13 |
| SlHsp70-17 | Solyc12g043110 | SL2.50ch12:39110693..39115806 | 130 | 106 | 93882.82 | 852 | 13211 | 42.35 | 80.06 | 8 | 5.23 |
| SlHsp70-18 | Solyc12g043120 | SL2.50ch12:39096307..39100382 | 130 | 105 | 92996.38 | 846 | 13051 | 42.45 | 77.74 | 8 | 5.22 |
| SlHsp70-19 | Solyc08g082820 | SL2.50ch08:65489311..65493585 | 113 | 93 | 73200.96 | 666 | 10357 | 30.84 | 87.85 | 7 | 5.10 |
| SlHsp70-20 | Solyc08g079170 | SL2.50ch08:62804339..62810456 | 98 | 91 | 65165.56 | 579 | 9124 | 36.18 | 67.03 | 6 | 5.99 |
| SlHsp70-21 | Solyc01g103450 | SL2.50ch01:92060728..92065237 | 98 | 85 | 74896.54 | 703 | 10598 | 25.83 | 86.13 | 7 | 5.20 |
| SlHsp70-22 | Solyc11g066060 | SL2.50ch11:51740558..51743431 | 101 | 90 | 77141.77 | 698 | 10869 | 33.98 | 84.14 | 2 | 5.51 |
| SlHsp70-23 | Solyc09g010630 | SL2.50ch09:3965253..3968837 | 100 | 82 | 71224.69 | 649 | 10008 | 35.04 | 82.53 | 1 | 5.13 |
| (-), negatively charged amino acid residues (Asp + Glu); (+), positively charged amino acid residues (Arg + Lys); MW, Molecular Weight; aa, Total amino acid residues; pI, isoelectric points. | |||||||||||
Phylogenetic analysis revealed distribution of all SlHsp70 genes in four groups I-IV (Fig. 3). Highest number of Hsp70 genes were placed in Group I of phylogenetic tree which comprised of 6, 9 and 8 Hsp70 genes of Arabidopsis, potato and tomato, respectively. Group II was smallest which comprised of five tomato SlHsp70 genes, two potato Hsp70 genes and three Arabidopsis AtHsp70 genes, respecitvely. Group III comprised of four tomato SlHsp70 genes, three potato Hsp70 genes and five Arabidopsis AtHsp70 genes. Group IV was second largest group which compried of six tomato SlHsp70 genes, five potato Hsp70 genes and five Arabidopsis AtHsp70 genes (Fig. 3).
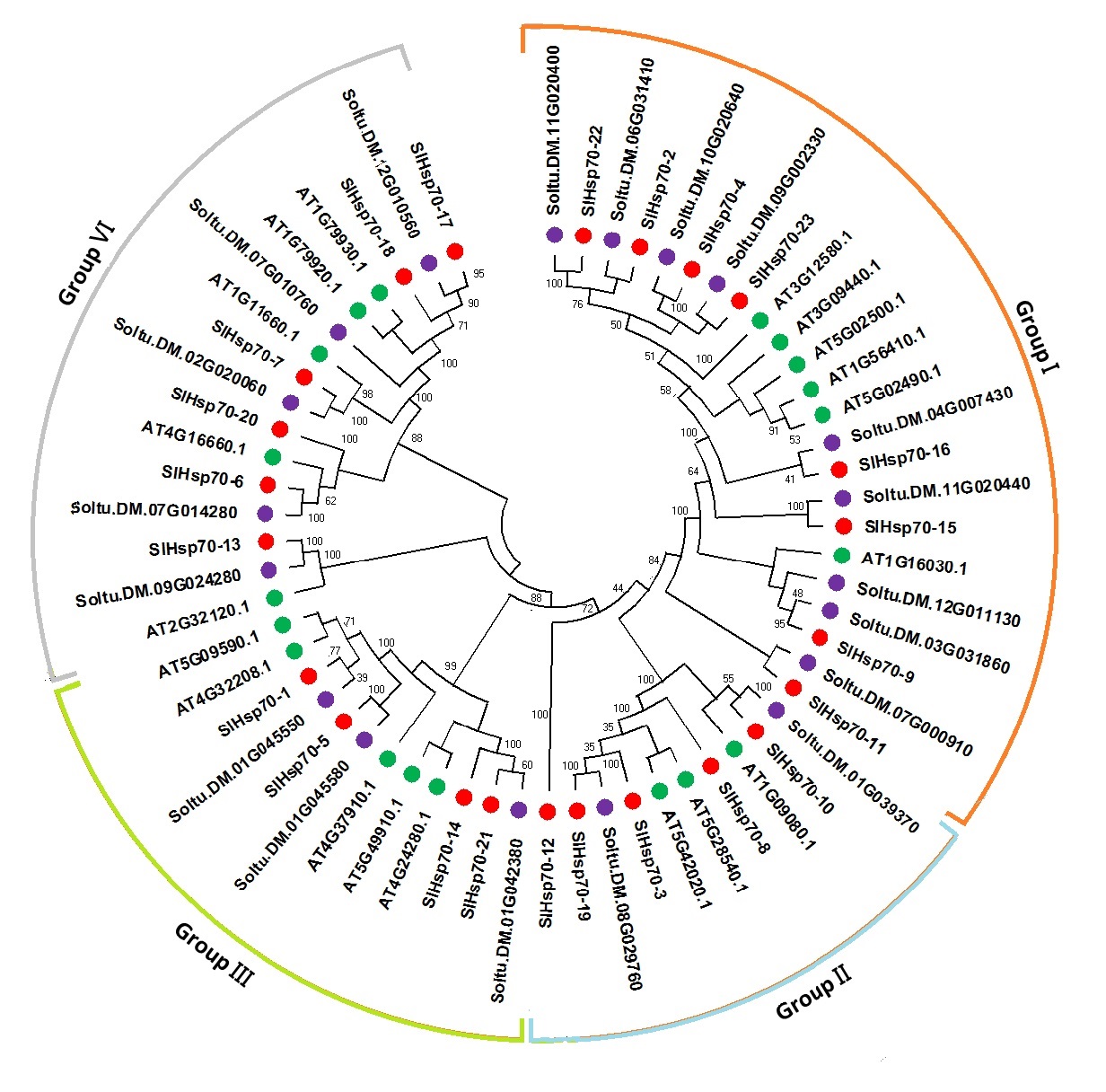 Fig. 3.
Fig. 3.Phylogenetic tree comprised of 23 S. lycopercicum (red circle), 19 Arabidopsis thaliana (green circle) and 19 S. tuberosum (purple circle) HSP70 protein sequences. ClustalX 2.0 was employed for protein alignment. Neighbor-Joining (NJ) method was used at 2000 bootstrap value to construct a phylogenetic tree in MEGA 7.0.
An uneven distribution of all SlHsp70 genes on different chromosomes was detected except chromosome number 5 which did not harbor any SlHsp70 gene. The highest number of SlHsp70 family genes localized on any chromosome were following four: SlHsp70-1, SlHsp70-5, SlHsp70-10 and SlHsp70-21, which were present on chromosome 1 (Fig. 4). Both segment and tandem duplication revealed 120 collinearity gene pairs with 50–100% duplication events (Supplementary Table 4). We noted 4 sister pairs among all 23 SlHsp70 genes. Multiple segment duplication events of SlHsp70-3, SlHsp70-10 and SlHsp70-13 genes were also observed. On the contrary, four parlog pairs were observed having distance less then 5 kb which were tandemly duplicated gene clusters and localized on chromosome 1, 3, 8, 11, and 12 (Fig. 4 and Supplementary Table 5).
 Fig. 4.
Fig. 4.Gene synteny analysis of SlHsp70 genes in S. lycopercicum. Blue lines represent orthologs and paralogs to express segmental duplication while red boxes represent tandem duplication.
On the base of structure, all 23 SlHsp70 gene family members were sub-divided into A, B, C, D and E sub-families (Fig. 5a). Largest subfamily was A containing 8 genes, subfamily E was second largest with 6 genes and subfamily D was smallest with only 1 gene (Fig. 5a). Except for SlHsp70-9 and -13, which had no introns, the number of exons and introns varied across all sub-families. Highest degree of similarity in exon and intron numbers was observed with in sub-families (Fig. 5c). Each motif of SlHPS70 proteins comprised of 50 amino acids except motif 4, 6, and 8 which contained only 41, 33 and 29 amino acids, respectively. The motifs 1, 3 and 8 were found in all subfamilies except D, and were followed by motifs 4, 6 and 7, which were likewise found in all subfamilies but two members of E. Noticeably, number, order and types of motifs were similar within a subfamily but different among subfamilies (Fig. 5b and Table 2).
 Fig. 5.
Fig. 5.Phylogenetic analysis, gene structure analysis and conserved motifs of SlHsp70 genes in S. lycopercicum. (a) The neighbor-joining (NJ) method was used at 2000 bootstrap value to construct a phylogenetic tree of SlHSP70 amino acid sequences in MEGA 7.0. (b) Ten conserved motifs of all SlHSP70 proteins were presented in unique colour symbol shown in box. (c) Dark lines represent exons, black lines represent introns annd blue lines represent UTRs. Specific size scale is given below.
| Motif | Logo | Most probable matches | Width |
| 1 |  |
VKBAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAJAYGLDKK | 50 |
| 2 |  |
LLDVTPLSLGJETAGGVMTKLIPRNTTIPTKKEQVFSTYSDNQPGVLIQV | 50 |
| 3 |  |
EKNVLVFDLGGGTFDVSJLTIEEGIFEVKATAGDTHLGGEDFDNRLVNHF | 50 |
| 4 |  |
TRARFEELNMDLFRKCMEPVEKCLRDAKLDKSDIHEVVLVGGSTRIPKVQ | 41 |
| 5 |  |
EGERARTKDNNLLGKFELSGIPPAPRGVPQIEVCFDIDANGILNVSAEDK | 50 |
| 6 |  |
FNGKEPCKSINPDEAVAYGAAVQAAILSG | 33 |
| 7 |  |
ERLIGDAAKNQAAMNPENTVFDAKRLIGRRFSDP | 50 |
| 8 |  |
FKRKHKKDISGBPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFY | 29 |
| 9 |  |
FKRKHKKDISGBPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDFY | 21 |
| 10 |  |
YKGEEKQFSPEEISAMVLTKMKEIAEAFL | 29 |
Amino acid sequences of SlHSP70 were analyzed in-silico to predict 3D protein structures as 3D conformation guarantees specific function of any protein (Fig. 6 & Supplementary Table 6). Except for SlHSP70-1 and -5, the rest of the SlHSP70 proteins were modelled using the c3d2fC template at a confidence level of 100 percent., while modeling of SlHSP70-12, -13 and -20 proteins was performed using c2khoA, c5gjjA, c5tkyA and C5nnrD templates. STRING database analysis revealed 22 nodes, 97 edges and following 13 local network clusters: 4141, 4411, 4655, 4661, 4020, 4021, 4023, 4025, 4084, 3997, 4339, 4682 and 4138. Among all, the biggest cluster was 3997 which comprised of 12 SlHsp70 proteins (Supplementary Table 7). Protein analysis also revealed existence of two common protien domains (PF00012 in all 23 SlHSP70 proteins and PF06723 in 19 SlHSP70 proteins) and five KEGG pathways (such as sly04141 in 14 SlHSP70 proteins, sly03060 in 3 SlHSP70 proteins, sly04144 in 10 SlHSP70 proteins, sly03040 in 10 SlHSP70 proteins and sly03018 in 2 proteins) (Supplementary Table 8). Sub-cellular distribution percentages of SlHSP70 proteins were 2/4% in vaculoar membranes, 3.5/8% in chloroplast and 9/42% in endoplasmic reticulum (ER). SlHsp70-8 was localized in 21/28 sub-cellular compartments. Overall percentages of SlHSP70 proteins in different biological processes were as follow: heat and cadmium resistance response was 3/4%, cellular stress response was 17/23% and chemical stress response was 15/23%.
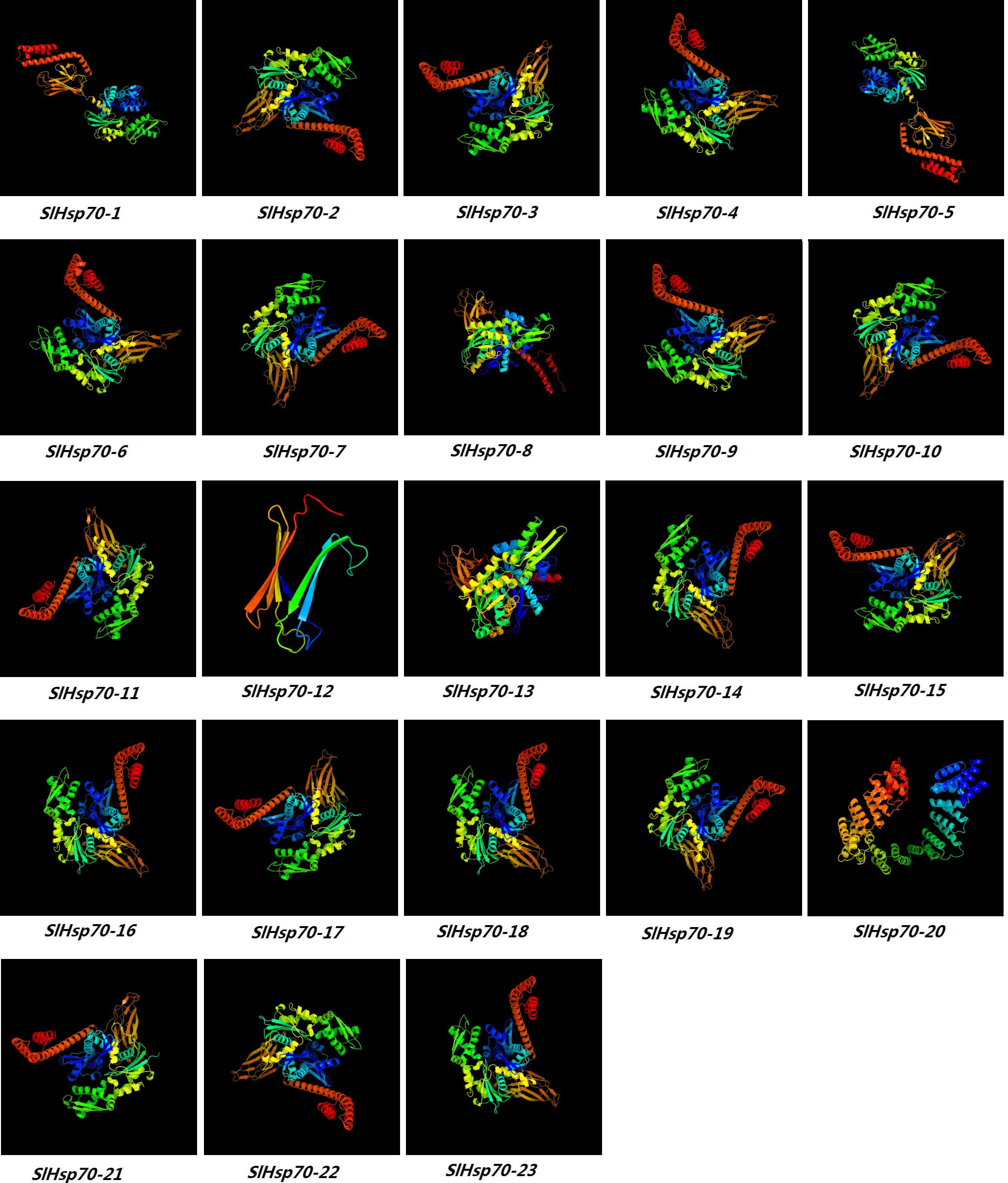 Fig. 6.
Fig. 6.Prediction of 3D structures of SlHSP70 proteins in S. lycopercicum. Phyre 2 server was used in an intensive mode to generat protein models, visualized by rainbow colours in direction from N to C terminus.
Tissue specific expression of the gene SlHsp70-1, -5, -9, -10, -13, -14, -15, -16, -17, -18, -20 and -21 diplayed upregulated expression level in breaker + 10 fruits while SlHsp70-8 exhibited highest expression level in breaker fruits. SlHsp70-11 displayed upregulated expression level in fully opened flowers and mature green fruits while SlHsp70-22 showed mild expression level in 2 cm fruit and upregulated expression level in fully opened flowers. Only single gene with upregulated expression level in leaves was SlHsp70-12 but it also displayed very mild expression in unopened flower bud. SlHsp70-6 exhibited mild expression level in roots, 2 cm fruit, breaker + 10 fruits and mature green fruits. SlHsp70-19 displayed mild expression in roots, 1 cm fruit, 2 cm fruit and unopened flower bud. Finally, SlHsp70-23 showed mild expression in mature green fruit and 3 cm fruit (Fig. 2).
Each SlHsp70 gene exhibited differential expression level on treatment
of different trace metal elements in both leaves and roots. The roots of plants
treated with Cd
 Fig. 7.
Fig. 7.qRT-PCR analysis of all 23 SlHsp70 genes in root and
leaf tissues under different metal stresses.
Trace metal element stress, such as Cd
Commercial cultivation of tomato is being predominantly performed in greenhouse,
where it is irrigated with recycled water containing very high Cd
 Fig. 8.
Fig. 8.Protein-protein interaction network building among SlHSP70 proteins in S. lycopercicum. Empty nodes: unknown 3D structure, fillled nodes: known or predicted 3D structure, colored node: query proteins and first shell of interactors and white nodes: second shell of interactors.
Cd
In order to evade deleterious effects of Cd
Cd
Conceptualization—MA (Manzar Abbas), YL, RGE and AHEl-S; Data acquision—AHEl-S, MA (Manzar Abbas), RGE and JL; Formal analysis—MA (Manzar Abbas), AHEl-S, SAS, MMI, VY, SZ, MA (Mubashir Abbas), NS, SSH, SA and ZN; Methodology—AHEl-S, RGE, MA (Mubashir Abbas), SZ, MMI, VY, JL, and AJR; Writing-original draft—MA (Manzar Abbas), and AHEl-S; Editing and proof-reading—MA (Mubashir Abbas), AHEl-S, AJR, KY and YL; Corresponding—AHEl-S, YL, and JL.
Not applicable.
The authors are grateful and acknowledge Sichuan Province Government to provide such a well-equipped platform to do research work, management of Yibin University for their support and providing us a pleasant environment of research, Chinese Government and Chinse Public in particular for their love of Science and Research.
This study was supported by Scientific Research Project of Yibin University (Grant no. 2020RC09 & Grant No. XJ2020007601), the Sichuan Provincial Department of Science and Technology Project (Grant No. 18ZDYF0293), National Nature Science Foundation (No. 32060679) and projects of Guizhou University (No. GuidapeiYU[2019]52.
The authors declare no conflict of interest.
