1 Department of Biological Chemistry, University of Michigan Medical School, Ann Arbor, MI 48109-0660, USA
†These authors contributed equally.
Academic Editor: Graham Pawelec
Abstract
Background: The structures and activities of invertebrate sulfakinins that influence gut motility and heart rate are like the vertebrate cholecystokinin (CCK) peptides. Typical of sulfakinin precursors Drosophila melanogaster encodes non-sulfated drosulfakinin I (nsDSK I; FDDYGHMRF-NH2) and nsDSK II (GGDDQFDDYGHMRF-NH2) that bind DSK-R1 and DSK-R2. To explore the role of the nsDSK II N-terminal extension (GGDDQ) in gut we delineated its structure-activity relationship (SAR) and identified novel agonists. We then predicted the nsDSK II extension SAR is tissue specific consistent with cardiac CCK structure activity and signaling being different from gut. Methods: To evaluate our hypothesis, we tested single-substituted alanine and asparagine analogs in heart. Results: We found alanyl-substituted analogs were less active in heart than nsDSK II; in gut they include a super agonist and a protean agonist. Additionally, we discovered ns[N4]DSK II was more active than nsDSK II in pupal heart, while ns[N3]DSK II was inactive. In contrast, ns[N3]DSK II and ns[N4]DSK II were super agonists in adult heart, yet inactive in larva. Although we reported nsDSK II acts through DSK-R2 in gut, its identity in heart was unknown. Conclusions: Here we reviewed ligand-receptor interactions in conjunction with SAR data to suggest nsDSK II acts through DSK-R1 in heart consistent with sulfakinin tissue-specific signaling.
Keywords
- cholecystokinin
- FMRFamide-related peptide
Sulfakinins are known to affect invertebrate gut motility and heart rate [1], which suggests targeting their signaling pathways may be a way to influence health. A peptidergic signaling pathway may be targeted by agonists designed from structure-activity relationship (SAR) data. An agonist may mimic the activity of a naturally-occurring peptide or have an increased (super agonist) or opposite (protean agonist) effect. SAR data can also provide insight into mechanisms underlying the signaling pathway. Invertebrate sulfakinins are related by structure and activity to the vertebrate cholecystokinin (CCK) peptides [1, 2, 3, 4, 5, 6]. Cholecystokinins are processed from a polyprotein precursor to yield structurally-related peptides with identical C-terminal structures, yet different N termini [5]. Likewise, sulfakinins are encoded in a polyprotein that generates structurally-related peptides with a conserved C-terminal FDDYGH(M/L)RF-NH2 but a distinct N terminus. Typical of invertebrates Drosophila melanogaster sulfakinin encodes drosulfakinin I (DSK I; FDDYGHMRF-NH2) and DSK II (GGDDQFDDYGHMRF-NH2) which contain a sulfated or non-sulfated (ns) tyrosine [6]. The similarity to CCK extends to the presence of multiple receptors; drosulfakinin peptides bind class A G protein-coupled receptors (GPCRs) designated DSK-R1 and DSK-R2 [7]. In addition, two sulfakinin receptors are reported in the red flour beetle, Tribolium castaneum, and in the kissing bug, Rhodnius prolixus [8, 9]. Transcripts of the R. prolixus SK receptors were detected in heart in 5th instar [9], however, to our knowledge the SAR and ligand-receptor requirements for binding across development remains unknown for a cardiac sulfakinin receptor. The 5-amino acid N-terminal extension (GGDDQ) that distinguishes nsDSK II from nsDSK I is rich in hydrophilic residues of aspartic acid and glutamine with glycine at its N terminus perhaps to confer flexibility. While the structure of the extension is not as highly conserved as the C terminus, its physicochemical nature suggests it serves an important role, thus, replacing a residue may alter activity. To that end, our previous SAR analysis of the extension on gut motility across three developmental stages identified a super agonist and a protean agonist. Furthermore, ligand-receptor interactions are consistent with nsDSK II acting through DSK-R2 in gut [10]. While the nsDSK II SAR for gut motility and locomotion is reported [10], it remains unknown in heart. We predicted the extension SAR is tissue specific consistent with cardiac CCK structure activity and signaling being different from gut [11, 12]. To test our hypothesis, we used single-substituted alanyl- and aspartyl- substituted analogs discovering novel agonists. In addition, ligand-receptor interactions compared with SAR data suggested nsDSK II acts through DSK-R1 consistent with sulfakinin signaling being tissue specific in heart.
D. melanogaster Oregon R strain flies were maintained on cornmeal
molasses media at 24
All peptides were synthesized on a 433A Applied Biosystems peptide synthesizer
using standard Fmoc procedures and purified by reversed-phase high performance
liquid chromatography. Each synthesis purity was obtained with
Heart beat was observed in each developmental stage using a previously described
microscope- based in vivo protocol [13]. The contractions for each
animal were measured prior to application of the peptide, analog, or saline to
provide a measure of basal rate. Animals (n
The data analyzed were the maximum responses within the 10-minute recording
period reported as mean values
Three-dimensional receptor structures of DSK-R1 and DSK-R2 and ligand modeling were previously reported [10, 14, 15, 16]. The primary sequences [7] were submitted to I-TASSER [17, 18] and refined in Mod Refiner [19] after which the receptor structures were viewed in PyMOL (Schrödinger, LLC). To prepare for docking, extracellular loops (ECLs) and tails were removed and a binding pocket of less than 27000 cubic angstroms was defined using AutoDock Tools-1.5.6. Ligand models were built in PyMOL for docking in AutoDockTools [10, 14, 15, 16, 20, 21].
PyMOL and the molecular docking software AutoDock Vina were used to dock ligands to DSK-R1 and DSK-R2 to predict contact sites as previously reported [10, 14, 15, 16]. Physicochemical properties, type and number, proximity of ligand-receptor contacts, and pose overlap were used to evaluate the one group most likely to represent docking. Poses with strong contacts formed multiple favorable ligand- receptor interactions within reference distances [10, 14, 15, 16]. All poses were independently analyzed by two researchers.
We initiated our nsDSK II N-terminal extension SAR analysis by testing single,
alanyl-substituted analogs on pupal heart rate (Fig. 1; Table 1) [13]. The effect
of nsDSK II in pupa was 45
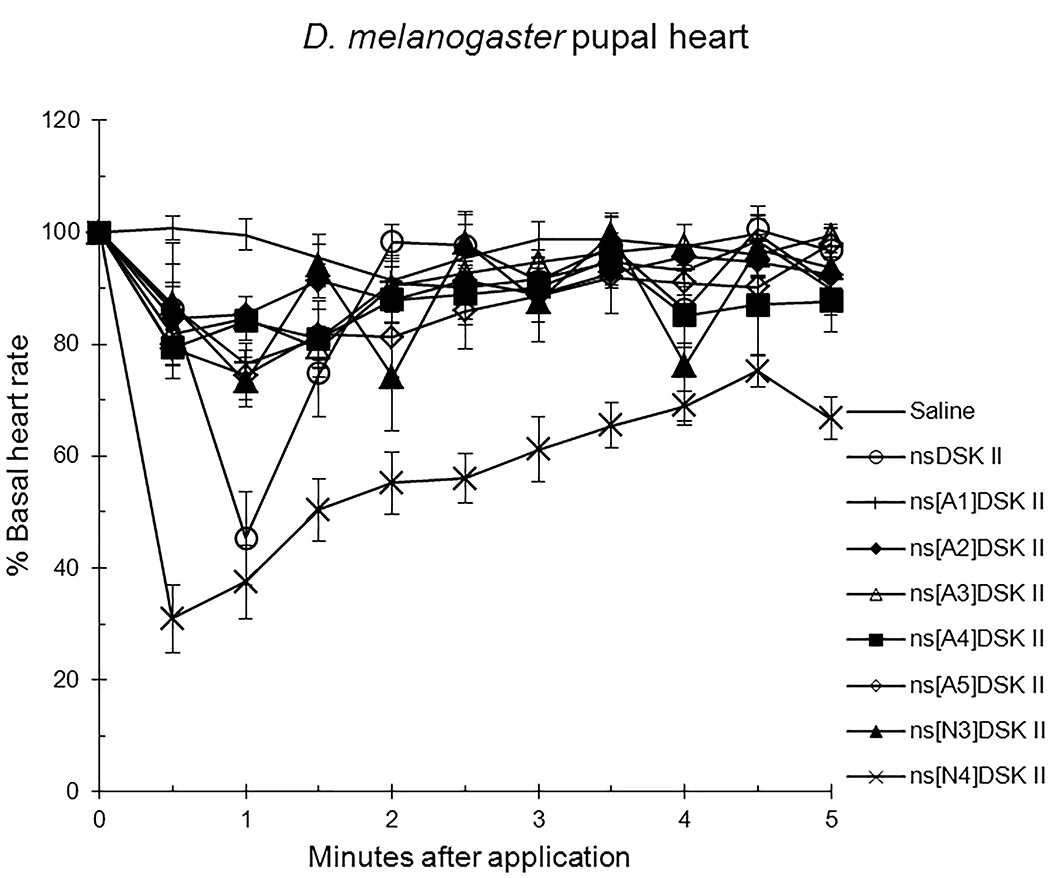 Fig. 1.
Fig. 1.
The effects of nsDSK II and analogs on pupal heart rate are
reported as % basal heart rate (mean value
| Peptide | Basal heart rate | ||
| pupa | p | ||
| nsDSK II | GGDDQFDDYGHMRF-NH2 | 45 |
0.0004 |
| ns[A1]DSK II | AGDDQFDDYGHMRF-NH2 | 77 |
0.005 |
| ns[A2]DSK II | GADDQFDDYGHMRF-NH2 | 85 |
0.009 |
| ns[A3]DSK II | GGADQFDDYGHMRF-NH2 | 79 |
0.002 |
| ns[A4]DSK II | GGDAQFDDYGHMRF-NH2 | 84 |
0.001 |
| ns[A5]DSK II | GGDDAFDDYGHMRF-NH2 | 74 |
0.003 |
| ns[N3]DSK II | GGNDQFDDYGHMRF-NH2 | 73 |
0.001 |
| ns[N4]DSK II | GGDNQFDDYGHMRF-NH2 | 31 |
0.00005 |
| Saline | 100 |
– | |
Next, we tested analogs in adult to find ns[A5]DSK II was like nsDSK II at 78
 Fig. 2.
Fig. 2.
The effects of nsDSK II and analogs on adult heart rate are
reported as % basal heart rate (mean value
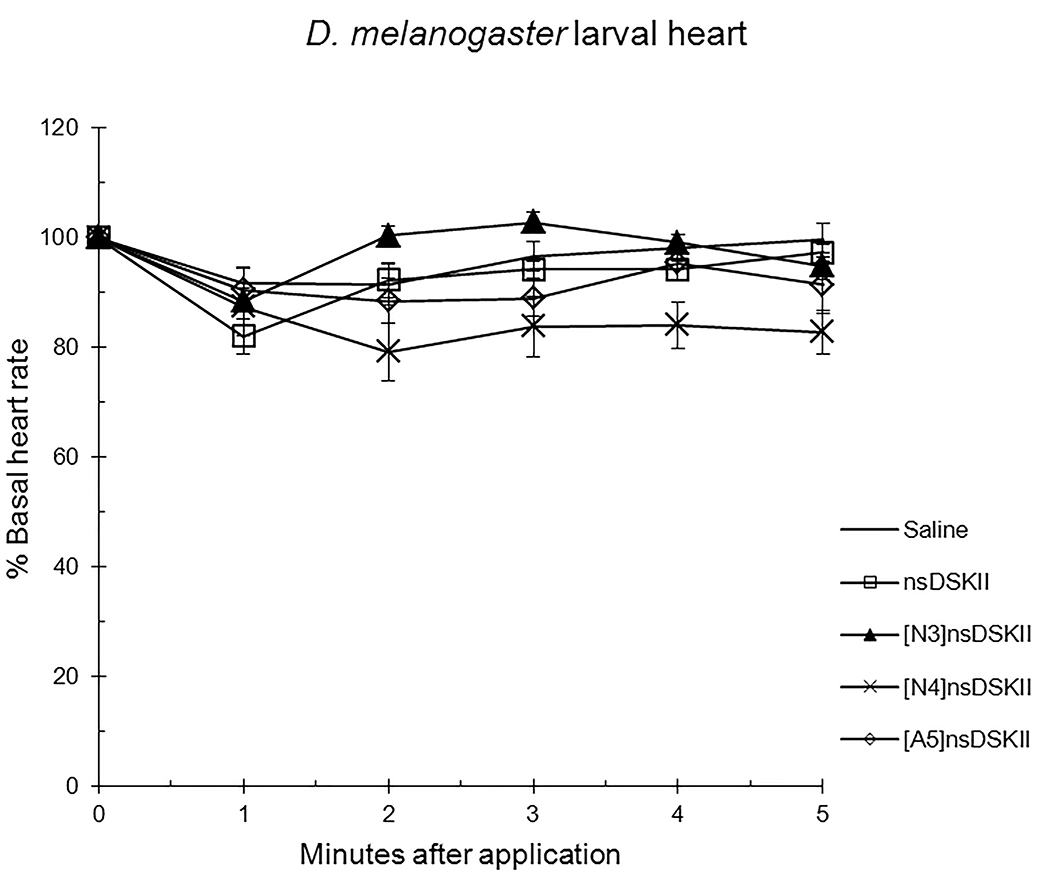 Fig. 3.
Fig. 3.
The effects of nsDSK II and analogs on larval heart rate are
reported as % basal heart rate (mean value
| Structure | Basal heart rate | ||||
| adult | p | larva | p | ||
| nsDSK II | GGDDQFDDYGHMRF-NH2 | 78 |
0.01 | 82 |
0.09 |
| ns[A5]DSK II | GGDDAFDDYGHMRF-NH2 | 78 |
0.01 | 88 |
0.6 |
| ns[N3]DSK II | GGNDQFDDYGHMRF-NH2 | 50 |
0.002 | 88 |
0.7 |
| ns[N4]DSK II | GGDNQFDDYGHMRF-NH2 | 35 |
0.00001 | 79 |
0.08 |
| Saline | 96 |
– | 91 |
– | |
Alanyl-substituted analogs were less active in heart than nsDSK II. Yet in adult gut ns[A4]DSK II and ns[A5]DSK II are more effective than nsDSK II [10]. Additionally, ns[A3]DSK II increases larval gut motility compared to nsDSK II which decreases it [10]. Next, we determined ns[N4]DSK II was more active than nsDSK II while ns[N3]DSK II was inactive in pupal heart. However, ns[N3]DSK II and ns[N4]DSK II were super agonists in adult and inactive in larval heart. In contrast, in adult gut ns[N3]DSK II and ns[N4]DSK II mimic nsDSK II activity [10]. Together these data support our hypothesis the nsDSK II extension SAR is tissue specific consistent with cardiac CCK structure activity and signaling being different from gut. Additionally, the data show sulfakinin activity is under developmental regulation.
Here we review ligand-receptor interactions from nsDSK II and analogs docked to DSK-R1 and DSK-R2 [10, 15] in conjunction with our SAR data in heart. nsDSK II On DSK-R1, nsDSK II places F6 between transmembrane region (TM)2 and TM3, and F14 T-stacks with the transmission switch W443 (Fig. 1, Ref. [15]). Y9 forms weak hydrophobic contacts with TM7, and orients around the polar residues of TM2, TM3, and TM7 to generate a hydrophilic network from D7 and D8 to the 3-7 lock. N277, D281, and N450 form a polar interface between TM5 and TM6. M12 is buried in the pocket. R13 makes a salt bridge to E181 and with Q5 sequesters C184 from contacting an ECL. H11 and the C-terminal amide form polar contacts on TM5 and TM6 including a hydrogen bond to D281. Docked to DSK-R2, D3 and Q5 of nsDSK II contact the 3-7 lock and D4 forms hydrogen bonds with polar residues on TM6 and TM7 (Fig. 4, Ref. [15]). G1 is in a polar region around TM5 and TM6 and forms hydrogen bonds with the ligand backbone; D347 is not contacted. R13 forms a salt bridge with E339 near the top of TM5 and M12 points to TM6, available for ECL contact. The aromatic residues spread throughout the pocket; F6 contacts TM7, Y9 is between TM2 and TM3, and F14 makes contact between TM4 and TM5. Y9 and H11 position near TM3 and the ECL interaction with C248 is not restricted.
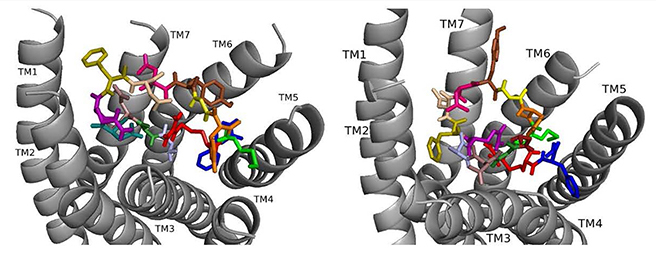 Fig. 4.
Fig. 4.ns[A1]DSK II docked to DSK-R1 (left) and to DSK-R2 (right) with A1 light blue, G2 forest, D3 dirty violet, D4 deep teal, Q5 purple, F6 olive, D7 wheat, D8 hot pink, Y9 brown, G10 yellow, H11 orange, M12 green, R13 red, F14-NH2 navy blue.
ns[A1]DSK II repositions its N terminus on DSK-R1 to weakly interact with W443 (Fig. 4). D3, D4, and R13 form a strong polar network extending to the 3-7 lock. F6 pulls away from TM2 and TM3 to the hydrophobic top of TM1, yet F14 retains a T stacking with W443. H11 forms a salt bridge to E181, while the ECL contact to C184 is restricted. M12 is at the top of TM4 and TM5 and accessible to the ECLs, and the C-terminal amide hydrogen bonds to D281. There is no contact with the transmission switch, E181, D281 and a polar network between TM3 and TM7. Aromatic contact is lost between TM2 and TM3. The ns[A1]DSK II contacts with DSK-R1 suggest it is a less effective agonist than nsDSK II consistent with the SAR data in heart.
On DSK-R2, ns[A1]DSK II rearranges to contact Y597 of the 3-7 lock, and F14 positions at the bottom of the pocket to interact with the transmission switch (Fig. 4). R13 retains a salt bridge at the top of TM5, and Y9 forms aromatic contacts between TM2 and TM3 and hydrogen bonds to polar residues around TM2, TM3, TM6 and TM7. These contacts describe ns[A1]DSK II as an agonist when it acts through DSK-R2 which is inconsistent with the SAR data in heart.
When ns[A2]DSK II docks to DSK-R1 it interacts weakly with W443 in the bottom of the pocket (Fig. 5). F6 remains between TM2 and TM3, but the N terminus is near the transmission switch resulting in F14 being higher between TM4 and TM5. M12 interacts with the ECLs. H11 retains the salt bridge with E181 and with Q5 blocks access to C184. R13 is between the polar residues of TM5 and TM6 and forms a hydrogen bond to D281. D7 and D8 point to the ECLs and are involved in a strong network of hydrogen bonds propagating to the 3-7 lock. ns[A2]DSK II lacks contact to the transmission switch to the extent of the T-stacking in nsDSK II which is consistent with being a less effective agonist in heart.
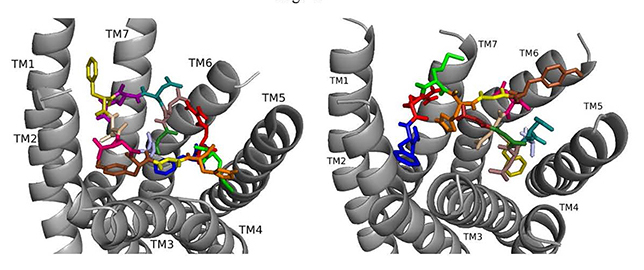 Fig. 5.
Fig. 5.ns[A2]DSK II docked to DSK-R1 (left) and to DSK-R2 (right) with G1 firebrick, A2 light blue, D3 dirty violet, D4 deep teal, Q5 purple, F6 olive, D7 wheat, D8 hot pink, Y9 brown, G10 yellow, H11 orange, M12 green, R13 red, F14-NH2 navy blue.
On DSK-R2, ns[A2]DSK II maintains critical contacts including an aromatic ring positioned between TM2 and TM3 and another located at the top of TM4 and TM5, as a salt bridge to TM5 (Fig. 5). A polar network formed by D3, D4, Q5, D7, R13, and the C-terminal amide directly contacts the 3-7 lock but does not extend to the ECLs. Its interactions with DSK-R2 suggest ns[A2]DSK II is a strong agonist, yet, in heart, the analog is less effective.
Docked to DSK-R1 ns[A3]DSK II loses strong aromatic interaction with the transmission switch resulting in a weakened polar network between TM3 and TM7 (Fig. 6). F6 remains between TM2 and TM3, but the N terminus is at the base of the pocket and F14 pi-stacks with Y276 higher on TM5. R13 forms a salt bridge with E181, limits access to C184, and M12 points out of the pocket. The N terminus and H11 retain polar contact to TM5 and TM6 with hydrogen bonding to D281 propagating into the pocket, yet A3 blocks it from reaching the 3-7 lock. No charged residues contact the ECLs. These interactions suggest that ns[A3]DSK II is less active than nsDSK II which is like the SAR data in heart.
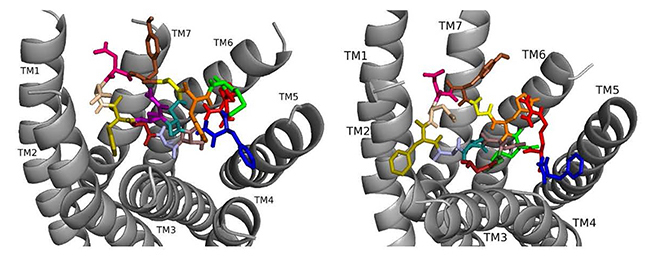 Fig. 6.
Fig. 6.ns[A3]DSK II docked to DSK-R1 (left) and to DSK-R2 (right) with G1 firebrick, G2 forest, A3 light blue, D4 deep teal, Q5 purple, F6 olive, D7 wheat, D8 hot pink, Y9 brown, G10 yellow, H11 orange, M12 green, R13 red, F14-NH2 navy blue.
On DSK-R2, R13 is in the polar region around TM5 and TM6 to interact with D347 but not E339 (Fig. 6). The C-terminal amide and D8 form strong contacts to the 3-7 lock, and Q5 interacts with polar residues. G1, G2, and A3 dock near TM2 in a folded manner to contact D7 rather than the receptor. F14 sits low in the pocket pointing to TM4 and Y9 docks between TM2 and TM3. The analog retains hydrophobic interactions to TM2 and TM3, polar interaction with the 3-7 lock, and contacts to D347 and W563 which nsDSK II does not. These contacts suggest ns[A3]DSK II is an effective agonist acting through DSK-R2 which is inconsistent with the SAR data in heart.
ns[A4]DSK II docked to DSK-R1 does not sustain the hydrogen bond to Q473 (Fig. 7). F6 remains between TM2 and TM3, and Y9 T-stacks with W443, which displaces F14 upward to contact Y476 on TM5. M12 points out of the pocket, R13 forms a salt bridge to E181, and C184 is accessible to ECL contact. Contact to TM5 and TM6 polar residues is minimal and only through the ligand backbone. A strong polar network stretches from the top of TM3 across the pocket and down TM7 where D7 makes direct contact to Y477 of the 3-7 lock. These interactions define reduced agonistic activity consistent with the SAR data in heart.
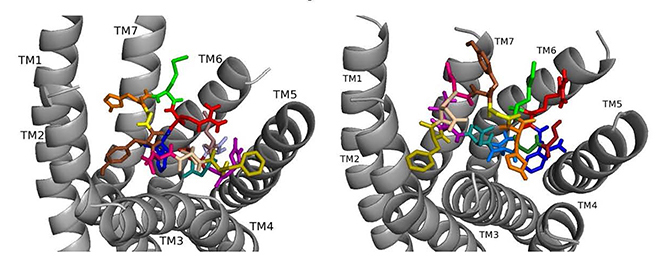 Fig. 7.
Fig. 7.ns[A4]DSK II docked to DSK-R1 (left) and to DSK-R2 (right) with G1 firebrick, G2 forest, D3 dirty violet, D4 light blue, Q5 purple, F6 olive, D7 wheat, D8 hot pink, Y9 brown, G10 yellow, H11 orange, M12 green, R13 red, F14-NH2 navy blue.
On DSK-R2, ns[A4]DSK II positions R13 to form strong interactions with Q255 and Y597 of the 3-7 lock (Fig. 7). D7 makes strong contact in the polar region from TM7 to TM2 and M12 docks near TM5 with its sidechain toward the intracellular side of the receptor. It does not contact D347, but H11 forms a salt bridge with E339. F14 is near hydrophobic residues between TM4 and TM5 and the C-terminal amide only makes intramolecular contact. F6 contacts TM2 and TM3, and Y4 pi-stacks with Y586 of TM7. D3 is high on TM3 to contact C248, which limits its ability to form a disulfide bond with ECL2. These contacts describe an agonist with strong ligand-receptor interactions not reflective of the SAR data in heart.
Docked to DSK-R1 ns[A5]DSK II does not retain the strong hydrogen bonds made by Q5 (Fig. 8). F6 T-stacks with the transmission switch and F14 docks between TM2 and TM3. It does not make polar contact with TM5 and lacks a salt bridge to E181. The N terminus blocks access to C184 leaving only D4 and M12 accessible to the ECLs. Extensive hydrogen bond propagation is present, yet no ligand residue docks near the 3-7 lock. These ligand-receptor interactions are suggestive of an agonist with reduced activity similar to the SAR data in heart.
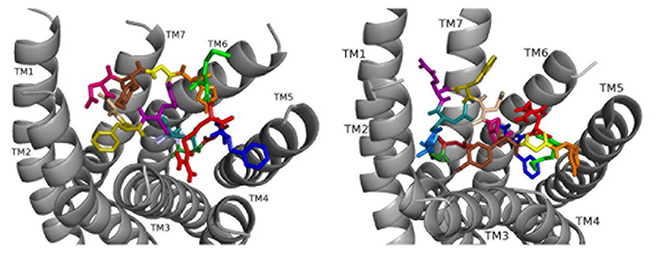 Fig. 8.
Fig. 8.ns[A5]DSK II docked to DSK-R1 (left) and DSK-R2 (right) with G1 firebrick, G2 forest, D3 dirty violet, D4 deep teal, A5 light blue, F6 olive, D7 wheat, D8 hot pink, Y9 brown, G10 yellow, H11 orange, M12 green, R13 red, F14-NH2 navy blue.
On DSK-R2, ns[A5]DSK II retains most of the contacts of the parent peptide nsDSK II, but D7 and D8 point up toward the ECLs (Fig. 8). The N-terminal extension rearranges to contact hydrophobic residues of TM3 and reduce a polar network between TM3 and TM7 like nsDSK II. While the additional strong, charged interactions may explain ns[A5]DSK II being a super agonist on gut, they are inconsistent with the SAR data in heart.
Docking ns[N3]DSK II to DSK-R1 places F6 between TM2 and TM3 where F14 T-stacks to the transmission switch and H11 forms a salt bridge to TM3 (Fig. 9). It restricts C184 contact to ECL and M12 is high in the pocket to interact with the ECLs. N3 forms strong polar contact to TM5 and TM6 along with G1, R3, and the C-terminal amide form a hydrogen bond to D281. D7 and D8 are at the top of the pocket and form a strong polar network extending down TM7 to the 3-7 lock. It takes on a different conformation but retains strong contact to the transmission switch and hydrogen bond to D281 consistent with it being an effective agonist in heart.
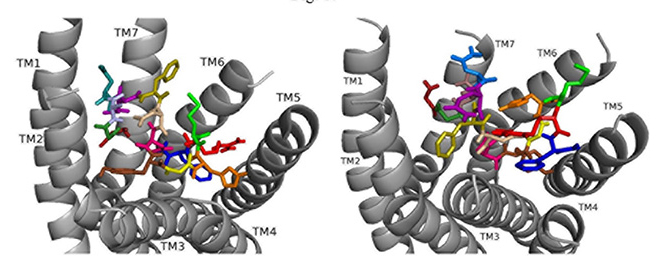 Fig. 9.
Fig. 9.ns[N3]DSK II docked to DSK-R1 (left) and to DSK-R2 (right) with G1 firebrick, G2 forest, N3 light blue, D4 deep teal, Q5 purple, F6 olive, D7 wheat, D8 hot pink, Y9 brown, G10 yellow, H11 orange, M12 green, R13 red, F14-NH2 navy blue.
ns[N3]DSK II docks to DSK-R2 with F14 low in the pocket between TM3 and TM5 (Fig. 9). It points to TM5 allowing the C-terminal amide to interact with polar residues on TM6 and TM7 but prevents contact with the transmission switch. R13 forms a salt bridge with E339 on TM5 but does not contact D347. F6 pi-stacks with Y586 of TM7 and G1, G2, N3, and D4 interact intramolecularly. Y9 docks to TM3 distinct from between TM2 andm TM3 and no contact is made with the 3-7 lock or polar residues nearby. It shares ionic contact with TM5 as in nsDSK II but lacks substantial polar contact to the lock. These contacts describe a less effective agonist inconsistent with the SAR data in heart.
ns[N4]DSK II docks DSK-R1 making intramolecular hydrogen bonds and changes in its conformation (Fig. 10). D3, D7, and D8 are deep in the pocket and form a strong polar network that propagates hydrogen bonds to the 3-7 lock. N4 is at the top of the pocket, F6 between TM2 and TM3, and Y9 T-stacks with the transmission switch; yet it lacks polar interaction with TM5. F14 is between TM3 and TM4, but the C-terminal amide fails to form polar contact with D281. R13 forms a salt bridge with TM3 blocking access to C184 and presents polar but not negatively- charged residues to the ECLs. H11 and R13 are available to interact with E265 of ECL2. These interactions including F6 around TM2 and TM3 are in accord with acting as a super agonist consistent with the SAR data in heart.
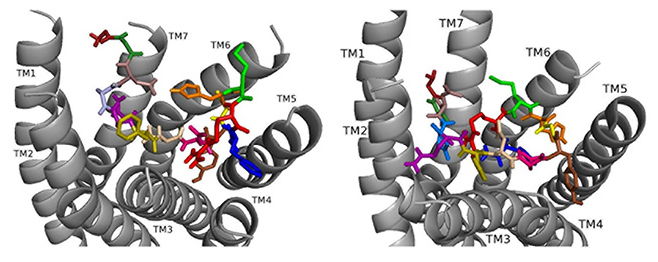 Fig. 10.
Fig. 10.ns[N4]DSK II docked to DSK-R1 (left) and to DSK-R2 (right) with G1 firebrick, G2 forest, D3 dirty violet, D4 deep teal, Q5 purple, F6 olive, D7 wheat, D8 hot pink, Y9 brown, G10 yellow, H11 orange, M12 green, R13 red, F14-NH2 navy blue.
Docked to DSK-R2, ns[N4]DSK II places H11 between TM5 and TM6 to form a salt bridge with D347 out of range of E339 (Fig. 10). R13 is above the 3-7 lock to form hydrogen bonds with Q255, Q593, and Y597. G1, G2, and N4 near the 3-7 lock form strong polar contacts and hydrogen bond propagation. F14 interacts with the transmission switch; its aromatic ring points to TM6 allowing the C-terminal amide to contact Q255 and Y597 of the 3-7 lock. F6 contacts TM3 close to C248 hindering it from forming a disulfide bond with ECL2. Thus, contacts are distinct from nsDSK II including a lack of contact to TM7, E339, and TM2 and TM3, and loss of a strong polar contact to the 3-7 lock, yet its gains strong contact to the transmission switch and a salt bridge to D347. The ligand-receptor interactions determined with molecular docking software [10, 14, 15, 16] compared to our SAR data in heart suggest nsDSK II and alanyl- and asparagyl-substituted analogs examined act through DSK-R1 in heart. This finding is different from the conclusion reached in gut where we predicted the peptides signal through DSK-R2 adding further support for our hypothesis.
Our SAR analysis and the docking of alanyl- and aspartyl-substituted nsDSK II analogs provide insight into sulfakinin activity and signaling across development in heart. The effect of alanyl- and aspartyl-substituted analogs in pupa, adult, and larval heart compared to gut [10] indicates sulfakinin signaling in heart is tissue-specific. A mechanism involved in degradation of the peptide injected or sulfakinin activity may also vary in development. Reviewing ligand-receptor interactions of nsDSK II and analogs [10, 15] along with SAR data suggest nsDSK II acts through DSK-R1 in heart further support for the prediction sulfakinin signaling is tissue specific. Comparing SAR and signaling in heart and gut in pupa, adult, and larva demonstrates the complexity of the sulfakinins reminiscent of the vertebrate CCK peptides [3, 4, 5, 6, 10, 11, 12, 15]. ns[A1]DSK II, ns[A2]DSK II, ns[A3]DSK II, ns[A4]DSK II, and ns[A5]DSK II showed reduced activity in pupa with weaker or different receptor interactions compared to nsDSK II [10, 15]. Additionally, the effect of ns[A5]DSK II was like nsDSK II in adult and larva. We propose based on SAR data and ligand- receptor interactions the salt bridge to E181 and/or a salt bridge to D281 may be critical for DSK-R1 activation by nsDSK II in pupa but not adult or larval heart. In addition, the frequency of peptide ionic contact to E339 suggests it may be involved in DSK-R1 activation. The ns[N3]DSK II was partially effective in pupa, a super agonist in adult, and like nsDSK II inactive in larval heart. When the analog docks to DSK-R1 it retains most of the contacts made by nsDSK II but shifts the polar Q5 sidechain to decrease the potential for polar interactions with the ECLs and increase the polar network between TM3 and TM7. That ns[N3]DSK II retains contacts of the parent peptide but increases the strength of polar contact around the 3-7 lock on DSK-R1 is consistent with it being a super agonist in adult heart, whereas the loss of some contacts on DSK-R2 is not. These interactions also predict nsDSK II signals through DSK-R1 in adult heart and a strong polar network between TM3 and TM7 can influence receptor activation. ns[N4]DSK II docks to DSK-R1 without polar contact between TM5 and TM6 but with a stronger polar network between TM3 and TM7 than nsDSK II, like ns[N3]DSK II. Docked to DSK-R2 ns[N4]DSK II gains contact to the transmission switch and a salt bridge to TM5 but lacks hydrophobic contact between TM2 and TM3 interactions that are distinct from ns[N3]DSK II. ns[N4]DSK II was an agonist in pupa heart, a super agonist in adult, and inactive like nsDSK II in larva. The greater similarity of contacts between ns[N3]DSK II and ns[N4]DSK II on DSK-R1 than DSK-R2 was consistent with their activity in adult heart. The stronger polar network between TM3 and TM7 formed by ns[N3]DSK II and ns[N4]DSK II with DSK-R1 is consistent with their super agonist effect on adult heart. However, ns[A5]DSK II, ns[N3]DSK II, ns[N4]DSK II and nsDSK II were all saline-like in larva. Docking data suggest the unusual conformation of the 3-7 lock in DSK- R1 may propagate hydrogen bonds to the tyrosine toggle switch, thus, ligands with a stronger network of hydrogen bonds between TM3 and TM7 showed super agonist activity in adult heart. Yet, nsDSK II, ns[A5]DSK II, ns[N3]DSK II, and ns[N4]DSK II do not make direct polar contact to the 3-7 lock, a molecular switch known to be critical for class A GPCR activation [22]. We predict a source of the inactivity in larval heart is the failure to break the 3-7 lock but polar propagation in the region is sufficient to activate the DSK-R1 signaling pathway in adult.
Our study identified novel agonists designed to the nsDSK II N-terminal extension that influenced the effect of sulfakinin on heart rate in a tissue-specific manner. We also presented data for the first time that are consistent with sulfakinin signaling through a tissue-specific pathway in heart. The effects of nsDSK II structural analogs were distinct in larva, pupa, and adult which suggests the sulfakinin signaling pathway or a mechanism associated with the peptide is developmentally regulated. Finally, this study further demonstrates the complexity of sulfakinin biology.
CCK, cholecystokinin; DSK, drosulfakinin; ECL, extracellular loop; GPCR, G protein-coupled receptor; SAR, structure-activity relationship; TM, transmembrane.
RN conceptualized, designed, directed, and funded the research study. CB and CK performed the research, RN, CB and CK critically analyzed the data. RN and CB wrote the manuscript. All authors contributed to editorial changes in the manuscript, and read and approved the manuscript.
Not applicable.
The authors acknowledge and thank their colleagues Megan Baker, Amanda Duttlinger, Kathy Berry, and Melissa Mispelon who provided technical assistance in collecting data.
This research was supported by internal funds to RN, but received no external funding.
The authors declare no conflict of interest.










