microRNAs (miRNAs) are important in tumor suppression and oncogenesis. In this study, we aimed to explore the role of miR-4698 with its potential target, Tripartite motif-containing 59 (Trim59), a protein with oncogenic function, in hepatocellular carcinoma (HCC). The expression of miR-4698 was significantly lower in HCC tissues and HCC cell lines as compared to the levels expressed in normal tissues adjacent to tumors and in normal hepatic cell line. Overexpression of miR-4698 in HCC cells by transfection of its mimic significantly inhibited cell growth, migration, invasion and epithelial-mesenchymal transition (EMT), whereas, its antisense oligonucleotides (ASOs) exerted an opposite effect. Trim59 was identified as a target of miR-4698 in miRDB and consistent with this, the expression of Trim59 was inversely correlated with miR-4698 in HCC, and miR-4698 overexpression led to a significant decrease in luciferase activity of pRL-Trim59-3’-UTR, but not mutant pRL-Trim59-3’-UTR. Moreover, miR-4698 mimic inhibited the expression of Trim59. Overexpression of Trim59 abrogated the inhibitory effects of miR-4698. In conclusion, these data show that miR-4698-Trim59 axis plays a tumor suppressive role in HCC.
HCC is among the leading causes of cancer death worldwide (1). In recent years, much progress has been made in the diagnosis and therapy of HCC. However, the prognosis and survival rate of HCC patients still remain poor due to lack of diagnostic and therapeutic options. Thus, it is urgent to better understand the molecular mechanisms underlying HCC onset and progression so as to develop new therapeutic applications.
miRNAs are a class of small (~22 nucleotide), non-coding RNAs that participate in gene regulation by inducing degradation of target mRNA or inhibiting translational machinery (2-5). In association with Argonaute proteins, miRNAs form silencing complex to exert suppressive effect on target mRNA through binding its 3’ untranslated region (3’-UTR) (6-8). miRNAs are differentially expressed in various tissues and are closely associated with cellular processes such as proliferation, differentiation, apoptosis, motility and metabolism (3, 9-15). Recent studies reveal that miRNAs regulate initiation and develpment of cancer cells via targeting various tumor suppressors, oncogenes and molecules in signaling pathways (16-20).
Trim59 belongs to the superfamily of the tripartite motif-containing (TRIM) protein (21). Studies have shown that Trim proteins play an essential role in human tumorigenesis and progression (22-25). Trim59, a novel protein in Trim family, has been found to play a significant role in various types of human cancers including lung cancer, gastric cancer, HCC and osteosarcoma (26-29). Sun et al (29) reported that Trim59 is upregulated in HCC cell lines and contributes to growth and metastasis of HCC cells through p53 signaling pathway.
In the present study, we characterized the tumor suppressive effect of miR-4698 –Trim59 axis in HCC, which provides novel insight into the development of new approches for HCC treatment.
Human HCC tissues and the matched tumor adjacent normal tissues were obtained from 30 patients at the Affiliated Hospital of Wenzhou Medical University, Wenzhou, China. All procedures and handling of human samples were approved by the Ethic Committee at Wenzhou Medical University, Wenzhou, China.
Human Hep 3B, Hep G2, SNU-182 (ATCC, USA) and Huh-7D12 (Sigma Aldrich, USA) HCC cell lines were propagated in high-glucose Dulbecco's modified Eagle's medium (DMEM) (CellGro, USA) supplemented with 2 mM of l-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin (Invitrogen, USA) , and 10% fetal bovine serum (FBS) ( Sigma Aldrich, USA). Cells were cultured at 37 °C in an incubator with 5% CO2 and 95% humidity. For the culture of THLE-2 (ATCC) human normal liver cells, a BEGM Bullet Kit from Lonza was used as previously described (30).
Total proteins were extracted from tissues and cells by using lysis buffer (Thermo Fisher Scientific, USA), separated by SDS-PAGE and transferred onto a PVDF membrane (Bio-Rad, USA). The membrane was incubated with 5% nonfat milk and probed with primary antibodies against Trim59 (Santa Cruz, USA), E-cadherin, N-cadherin ( Abcam, USA) and β-actin (Cell Signaling Technology, USA). Proteins of interest were examined with a horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (Abcam, USA). Blots were visualized with the ECL substrate (Thermo Fisher, USA) and densitometry of the Western blot bands was measured using ImageJ software.
Total RNA was isolated using TRIzol reagent (Thermo Fisher, USA). To quantify the levels of Trim59 mRNA, equal amounts of cDNA were synthesized using a High-Capacity cDNA transcription kit (Applied Biosystems, USA) according to the manufacturer’s instructions, and mixed with Power SYBR green PCR master mix (Applied Biosystems, USA). The primers for Trim59 and β-actin used in q-PCR were shown in Table 1. β-actin served as internal control for qPCR. For q-PCR of miR-4698, QScript miRNA cDNA Synthesis Kit (Quanta Biosciences, USA) was utilized based on manufacturer’s instructions. The primers for miR-4698 were obtained from Sigma Aldrich and human small nuclear U6 RNA was amplified and served as internal control.
| Gene name | Gene sequence ( 5’-3’) | Gene sequence ( 5’-3’) |
|---|---|---|
| Trim59 | TGACTGACACACACTGGACA | CTGCTGCTCTCGTATTTCCT |
| β-actin | ACTGGAACGGTGAAGGTGAC | AGAGAAGTGGGGTGGCTTTT |
miR-4698 mimic, negative control, ASOs and scrambled control were obtained from Sigma Aldrich. Hep 3B and Hep G2 cells were seeded 24 h before transfection. The transfection was performed using 50 nM of miR-4698 mimic, inhibitor, or the negative control RNA and Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions.
Hep 3B and Hep G2 cells were seeded into a 96-well culture plate (Corning, USA) at a density of 4 × 103 cells/well. 24, 48 and 72 h later, the cells were washed and cell growth was assessed using a CCK-8 assay kit from Dojindo, Japan. 10 μl of CCK-8 solution was added and incubated for 2 h at 37°C. Optical density (OD) at 450 nm was measured by a microplate reader (Azure Biosystems, USA).
Migration and invasion assays were carried out in 12-mm transwell culture inserts with 8-μm pores (Millipore-Sigma, USA). Hep 3B and Hep G2 cells (4 × 105 cells) suspended in 250 μl of serum free medium supplemented with 0.1% BSA were seeded in the inserts. 700 μl of medium supplemented with 5% FBS was added in the lower culture well. For invasion assay, the inserts were pre-coated with matrigel (Corning, USA) diluted 1:1 in serum-free medium for 3h at 37 °C. Cell suspension was then added in the inserts and cultured for 12 h. Afterwards, the medium in the inserts was removed and the inserts were incubated with 0.5% crystal violet in 25% methanol for 30 min. The cells in the upper side of the transwell culture inserts were removed with a cotton swab. Cells that migrated or invaded were examined and counted under an Olympus CX31 microscope.
pGL3 control vector and pGL3-Trim59-3’UTR were kindly provided by Dr. Li Yang from Hong Kong University. The mutant pGL3-Trim59-3’UTR was generated using a Mutation Generation System kit (Thermo Fisher Scientific, USA). Luciferase activity was measured 48 h post transfection using a Promega dual luciferase reporter assay.
Results were shown as mean ± standard deviation (SD). The data were assessed by unpaired two-tailed Student’s t test between groups. P<0.05 was considered significant.
30 pairs of HCC tissues and the matched normal tissues were harvested. Levels of miR-4698 in the tissues were then measured by RT-PCR. miR-4698 levels were significantly lower in 79% tumor tissues as compared to the levels expressed in normal tissues (Figure 1A) consistent with this, the expression of miR-4698 was downregulated in HCC cell lines (Figure 1B) compared with normal hepatic cells .
 Figure 1
Figure 1Expression of miR-4698 in HCC tissues and HCC cell lines. A. Expression of miR-4698 in HCC tissues and the matched normal tissues analyzed by qPCR. B. Expression of miR-4698 in Hep 3B, Hep G2, Huh-7D12 and SNU-182 HCC cell lines and THLE-2 normal liver cells measured by q-PCR. Data are shown as mean±SD from three independent experiments (*p <0.05).
Downregulation of miR-4698 in HCC tissues and cells suggests that miR-4698 may play a tumor suppressive role in HCC cells. miR-4698 mimic was employed to overexpress miR-4698 in Hep 3B and Hep G2 cells. miR-4698 levels were significantly elevated in the cells transfected with the mimic (Figure 2A). miR-4698 overexpression retarded cell proliferation (Figure 2B) and inhibited migration and invasion of the cells (Figure 2C and 2D). Moreover, the expression of EMT biomarkers was significantly changed by overexpression of miR-4698, as evidenced by an increase in the expression of epithelial marker E-cadherin and a decrease in the expression of mesenchymal marker N-cadherin (Figure 2E). These findings suggest that proliferation and metastasis of HCC cells depends on a certain unknown mechanism to suppress the expression of miR-4698 in HCC cells.
 Figure 2
Figure 2mir-4698 mimic suppressed cell growth, motility and EMT of HCC cell lines. A. Effectiveness of transfection of miR-4698 mimic on its expression in Hep 3B and Hep G2 cells. B. Cell proliferation, migration (C), invasion (D), and the expression of E-cadherin and N-cadherin in Hep 3B and Hep G2 cells transfected with miR-4698 mimic or its negative control. The results were shown as percentage changes in miR-4698 levels, cell proliferation, migration and invasion in comparison to negative control group. Densitometry results represent the ratio of E-cadherin or N-cadherin to β-actin. Data are shown as mean±SD from three independent experiments (*p <0.05 and **p<0.01).
ASOs against miR-4698 were utilized to suppress the basal expression of miR-4698 in Hep 3B and Hep G2 cells. As shown in Figure 3A, ASOs effectively silenced the expression of miR-4698 in both cell lines. Result of CCK8 analysis showed that ASOs significantly enhanced cell proliferation (Figure 3B). Migration and invasion of the cells were also promoted by miR-4698 inhibition (Figure 3C and 3D). These findings show that the low basal levels of miR-4698 still works to restrain the growth and mobility of HCC cells.
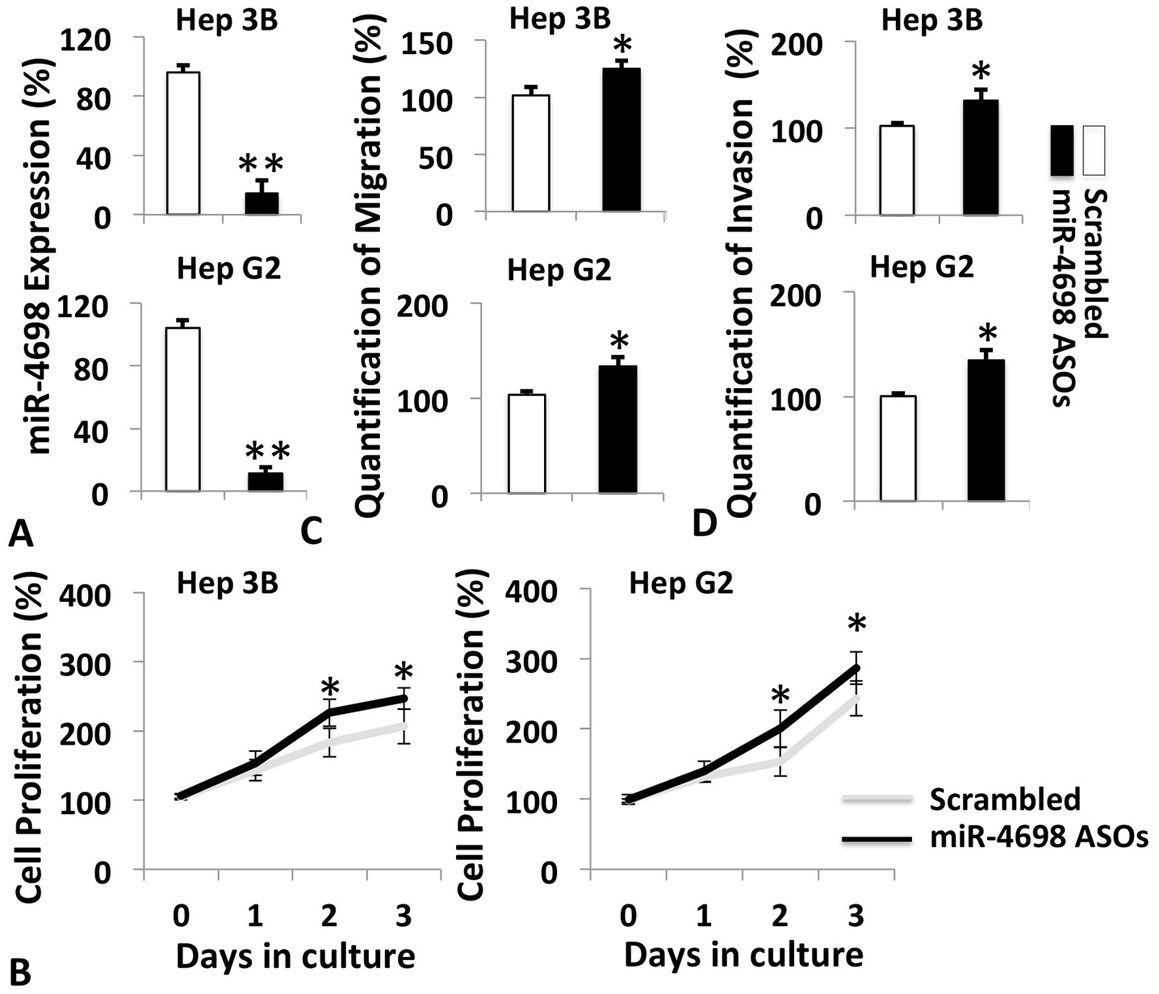 Figure 3
Figure 3ASOs against miR-4698 enhanced cell growth and motility of HCC cell lines. A. Analysis of miR-4698 levels in Hep 3B and Hep G2 cells transfected with or without miR-4698 ASOs. B. Cell proliferation, migration (C) and invasion (D) of Hep 3B and HepG2 cells with miR-4698 silenced by its ASOs. The results were shown as percentage changes in miR-4698 levels, cell growth, migration and invasion in comparison to scrambled control group. Data are presented as mean±SD from three independent experiments (*p <0.05 and **p<0.01).
Trim59 was identified as a target of miR-4698 in miRDB database. High levels of Trim59 at mRNA and protein levels were detected in the tumor tissues (Figure 4A and 4B) and HCC cell lines (data not shown) as compared to the levels in matched adjacent normal tissues or normal hepatic cells. Luciferase reporter assays were then utilized to further confirm this result. Transfection of miR-4698 mimic in Hep 3B and Hep G2 cells resulted in a remarkable suppression of luciferase activity of pGL-Trim59-3’-UTR, but not mutant pGL3-Trim59-3’UTR (Figure 4C). Furthermore, miR-4698 overexpression in Hep 3B and Hep G2 cells significantly downregulated the expression of Trim59 mRNA and protein. (Figure 4D).
 Figure 4
Figure 4Trim59 was a target of miR-4698. A. qPCR and Western blot analysis (B) of the expression of Trim59 in HCC tissues and HCC cell lines in comparison to the matched adjacent normal tissues and THLE-2 cells. β-actin served as internal control. The results were shown as percentage changes in miR-4698 and mRNA levels of Trim59 in comparison to normal control group (normal tissues or THLE-2 cell line) (100%). C. Luciferase activity in Hep 3B and Hep G2 cells co-transfected with miR-4698 mimic and wild-type pGL3-Trim59-3’UTR or mutant pGL3-Trim59-3’UTR. pGL control plasmid was transfected for normalization. miR-4698 mimic reduced luciferase activity of wild-type pRL-Trim59-3’-UTR. Luciferase activity of mutant pRL-Trim59-3’-UTR remained unchanged following transfection of miR-4698 mimic. The results were shown as percentage changes in luciferase activity in comparison to control cells co-transfected with wild-type pGL3-Trim59-3’UTR and miR-4698 negative control. D. The expression of Trim59 mRNA and protein in Hep 3B and Hep G2 cells transfected with miR-4698 mimic. The q-PCR result was shown as percentage changes in mRNA levels of Trim59 in comparison to control group (100%). Densitometry results represent the ratio of Trim59 to β-actin. Data are presented as mean±SD from three independent experiments (*p <0.05 and **p<0.01).
To confirm whether targeting Trim59 mediated the inhibitory effect of miR-4698 in HCC cells, transfection of pcDNA3.1-FL-Trim59 (full-length Trim59, stored in our lab) was performed to overexpress Trim59 in Hep 3B and Hep G2 cells. As shown in Figure 5A, forced Trim-59 overexpressio was confirmed by Westerb blot. Overexpression of Trim59 abrogated the suppressive effect of miR-4698 mimic on cell proliferation (Figure 5B), migration (Figure 5C) and invasion (Figure 5D) in both cell lines. Moreover, forced overexpression of Trim59 by transfection in both cell lines did not alter growth and mobility of the cells without co-transfection with miR4698 mimic, whereas Trim59 siRNA significantly suppressed the proliferation and motility of the cells (data not shown). Thus, these results suggest that miR-4698 plays tumor suppressive role HCC cells by targeting Trim59.
 Figure 5
Figure 5Inhibitory effect of miR-4698 on HCC cells was mediated by suppressing Trim59. A. Confirmation of overexpression of Trim59 in Hep 3B and Hep G2 cells transfected with pcDNA3.1-FL-Trim59. Overexpression of Trim59 abrogated the suppressive effect of miR-4698 mimic on cell proliferation (B), migration (C) and invasion (D) of Hep3B and HepG2 cells. The results were shown as percentage changes in cell proliferation, migration and invasion in HCC cells in comparison to negative control group co-transfected with pcDNA3.1 control plasmid and negative control of miR-4698 mimic. Densitometry results represent the ratio of Trim59 to β-actin. Data are presented as mean±SD from three independent experiments (*p <0.05 and **p<0.01).
miRNAs play a dual role in tumor onset and development by exhibiting either tumor suppressive or oncogenic properties. Tumor suppressive miRNAs can inhibit tumorigenesis and growth through negative regulating proliferation, metastasis, angiogenesis and metabolism, and enhancing apoptosis (31-33). However, the role of miR-4698 in HCC has not been reported. This is the first study identifying tumor suppressive role of miR-4698 –Trim59 axis in HCC.
In the present study, we first identified that the expression of miR-4698 was significantly lower in HCC tissues and HCC cell lines as compared to the levels expressed in normal tissues adjacent to tumors and in norma hepatic cell line. This result led us to speculate about whether the expression of miR-4698 could be influential on HCC cells. As expected, miR-4698 overexpression by its mimic effectively inhibited cell growth and motility of HCC cells. EMT confers tumor metastasis by promoting migration and invasion of tumor cells (34). We found that overexpression of miR-4698 by transfection of its mimic upregulated the expression of epithelial biomarker E-cadherin and downregulated mesenchymal biomarker N-cadherin, suggesting that miR-4698 affects mobility of HCC cells by suppressing EMT. Although basal expression of miR-4698 remained at low level in HCC cells, it could still retard growth and motility of HCC cells. Further studies may be needed to elucidate how HCC cells downregulate miR-4698 during the processes of tumor growth and metastasis.
It is well known that miRNAs can induce mRNA degradation or suppress translation of target mRNA. In the present study, we identified Trim59 as a target gene of miR-4698. Trim59 is a biomarker of various cancer, and exhibits oncogenic properties. Previous study showed that Trim59 is highly expressed in BEL7402, Hep3B, HepG2, Huh7, SMMC7721 and SK-Hep-1 HCC cell lines (29). In our study, we confirmed this finding in Hep 3B, Hep G2, Huh-7D12 and SNU-182 HCC cells (data not shown). Moreover, HCC tissues showed high levels of Trim59, which was correlated inversely with the levels of miR-4698. So far, the molecular mechanism by which the expression of Trim59 is regulated in HCC still remains undefined. Trim59 was predicted as a target of miR-4698 in miRDB database, which was validated by luciferase assay. Regulation of the expression of Trim59 by miR-4698 in HCC cells may occur at mRNA level in a degradation-dependent manner or by translation silencing. It is likely that the mechanism that Trim59 is upregulated via downregulating miR-4698 is employed by HCC cells in the tumor microenvironment.
Next, we speculated whether the inhibitory effect of miR-4698 on HCC cells was directly mediated by targeting Trim59. As expected, restoration of Trim59 in HCC cells effectively abrogated the inhibitory effect of miR-4698 mimic. It is interesting that in HCC cells with forced overexpression of Trim59 but without co-transfection with miR-4698 mimic, there was no significant increase in the capability of cells to grow, migrate and invade, suggesting that the endogenous expression of Trim59 in HCC cells might reach a relative high or saturation level that could be sufficient for regulating the growth and mobility of the cells. Since Trim59 is a signaling protein of Ras signaling pathway that could mediate tumor angiogenesis (35), downregulation of miR-4698 in HCC cells could also indirectly facilitate the crosstalk between tumor cells and other cells such as endothelial cells in tumor microenvironment. It will also be interesting to investigate the role of miR-4698-Trim59 axis in other pathological processes in HCC cells such as DNA damage, drug resistance, and aberrant metabolism. This study demonstrates a tumor suppressive role for miR-4698-Trim59 axis in HCC (Figure 6). On the basis of our findings, we suggest that miR-4698 acts as a tumor suppressor in HCC cells by targeting Trim59. Thus, manipulation of miR-4698 may represent a potential new strategy for HCC treatment.
 Figure 6
Figure 6Graphic demonstration of tumor suppressive role of miR-4698 – Trim59 axis in HCC .
We appreciate RCBC, LLC., USA for English language editing services. The authors declare no conflict of interest.
miRNAs
microRNAs
hepatocellular carcinoma
Tripartite motif-containing 59
antisense oligonucleotides
3’ untranslated region
epithelial mesenchymal transition
fetal bovine serum
horseradish peroxidase
Dulbecco's modified Eagle's medium
optical density
