Long noncoding RNA prostate cancer-associated transcript 1 (PCAT1) is oncogenic and causes progression of non-small cell lung cancer (NSCLC). We hypothesized that PCAT1 might be involved in the acquisition of chemoresistance of NSCLC cells to treatment with cisplatin (DDP). Here, we show that PCAT1 and ATP-binding cassette sub-family B member 1 (ABCB1) are highly expressed in NSCLC tissues and cell lines, and regulate the growth and apoptosis of these cells. Compared with those in DDP-sensitive patients, PCAT1 and ABCB1 are highly expressed in the tumors of DDP-resistant patients, and such overexpression correlates with a shorter overall survival of these patients. Knockdown of PCAT1 or upregulation of miR-129 led to apoptosis and sensitized the DDP-resistant cells to DDP. The 3’ UTR activity of PCAT1 and ABCB1, which was increased by PCAT1 overexpression, was shown to harbor an miR-129 binding site. DDP resistance is induced by elevated ABCB1 expression, which involves binding of miR-129 in DDP resistant cells. These findings suggest that the PCAT1/miR-129/ABCB1 axis may be a potential target for the treatment of DDP-resistant oat cell cancer.
Lung cancer is considered as the leading cause of cancer-related deaths worldwide, accounting for approximately 1.6 million deaths annually (1). Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, insensitive to chemotherapy, and accounting for about 85% of the total cases (2). Tobacco smoking is generally regarded as the most common etiology for NSCLC, which is associated with a poorer prognosis (3). Substantial progress has been made in the treatment of NSCLC over the last decades. The platinum-based chemotherapy drug, cisplatin (DDP), is considered as the standard therapy for patients with NSCLC, and can improve patient survival (4). However, the efficacy of DDP is hampered and, as a result, the prognosis of patients with NSCLC is very poor, mainly due to the emergence of acquired resistance of cancer cells to DDP (5). Thus, a detailed understanding of the molecular mechanisms underlying DDP resistance in NSCLC is urgently needed to improve the therapeutic outcome of patients with NSCLC.
Long noncoding RNAs (lncRNAs) are endogenous, long (>200 nucleotides), noncoding nucleotides that have a series of biological functions, such as transcriptional regulation, splicing, and imprinting (6). lncRNAs are regarded as important regulatory factors of gene expression, which is crucial for maintaining cell function. lncRNAs have been shown to function as a microRNA (miRNA) sponge to regulate the expression and functions of miRNAs, thereby modulating gene expression (7). Recent evidence has implicated lncRNAs in regulating chemoresistance in human cancers. Alterations of lncRNAs have increasingly been recognized to contribute to the occurrence of chemoresistance in cancer cells (8–10). Prostate cancer-associated transcript 1 (PCAT1) is a prostate-specific lncRNA, identified as an attractive marker for prostate cancer. PCAT1 expression has been shown to be upregulated in prostate cancer metastatic tumors and causes an elevation in DU145 cell proliferation and a reduction in DU145 cell apoptosis (11). Overexpression of PCAT1 was shown to play a tumor-promoting role in osteosarcoma, enhancing cell proliferation, migration, invasiveness, and epithelial-to-mesenchymal transition, together with a worse prognosis in patients with osteosarcoma (12). Moreover, PCAT1 knockdown was reported to enhance the sensitivity of oesophageal cancer cells to DDP (13). Importantly, a previous study showed that PCAT1 is upregulated in NSCLC tissues and cell lines and plays an oncogenic role in NSCLC progression (14). However, little is known about its functional role and regulatory mechanism in DDP resistance in NSCLC.
miRNAs are small noncoding RNAs (~22 bp nucleotides) with gene expression modulatory function, aberrantly expressed in a variety of cancer types (15). miRNAs have been shown to control various cellular processes, ranging from cell proliferation to tumor development, by regulating gene transcription through interaction with gene promoters and gene terminus sequences (16, 17). Recently, the involvement of miRNA misregulation in the acquisition of tumor cell resistance to chemotherapeutic drugs has been documented. Overexpression of miRNA-27a has been shown to be closely associated with increased sensitivity to DDP in DDP-resistant bladder cancer cells (18). However, the importance of miRNAs involved in chemotherapeutic resistance has not been adequately studied.
In this study, we present evidence that upregulation of PCAT1 contributes to DDP resistance in NSCLC through sponging miR-129, and as a result, increasing the levels of ATP-binding cassette sub-family B member 1 (ABCB1). Our findings suggest that targeting the PCAT1/miR-129/ABCB1 axis may be used as therapeutic strategy to overcome DDP resistance in patients with NSCLC.
Forty-two paired fresh-frozen NSCLC tissues and normal tissues were procured from NSCLC patients undergoing surgery at Shanxi Tumor Hospital. All participants were pathologically diagnosed with NSCLC, and patients only underwent DDP-based chemotherapy prior to surgery. Informed consent was obtained from all patients in accordance with institutional review board policies. An obvious reduction of the primary tumor was considered as effective therapy. DDP-resistant cases were distinguished when NSCLC tumors enlarged or metastasized within 12 months, otherwise, they were categorized as DDP-sensitive cases. The normalized RNA-seq data of lung adenocarcinoma were downloaded from the Cancer Genome Atlas (TCGA) data portal (https://cancergenome.nih.gov/) and analyzed by R (version 3.5.1.).
Human normal bronchial epithelial (HBE) cells, NSCLC cell lines (A549 and H1299), and 293T cells were procured from the American Type Culture Collection (Rockville, MD, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin under 5% CO2/95% air at 37°C. The DDP-resistant variants, A549/DDP and H1299/DDP, were established by stepwise selection after a 12-month treatment of A549 and H1299 cells with increasing concentrations of DDP.
miR-129, anti-miR-129, short interfering RNA targeting PCAT1 (si-PCAT1#1, si-PCAT1#2, si-PCAT1#3), and matched controls were purchased from Sangon Biotech (Shanghai, China). The full sequence of PCAT1 was amplified and cloned into a pcDNA-3.1 vector (Invitrogen, Carlsbad, CA, USA) to generate the pcDNA-PCAT1 plasmid. Cultured cells were transfected with these plasmids using Lipofectamine 2000 (Invitrogen), as per the instruction manual. Cultured cells were collected 48 h post transfection for the following experiments.
A549/DDP and H1299/DDP cells were plated in 96-well plates and cultured for 24 h. Following this, cells were incubated with different concentrations (0.1, 0.5, 1, 5, 10, 50, and 100 μM) of DDP for 48 h and then cultured with Cell Counting Kit-8 (CCK-8) solution (Beyotime, Shanghai, China) for 2 h after transfection. The absorbance of each well at 450 nm was measured with a spectrophotometer (Bio-Rad, Hercules, CA, USA), and the IC50 value (half maximal inhibitory concentration) of DDP in A549/DDP and H1299/DDP cells was calculated.
The apoptosis of A549/DDP and H1299/DDP cells was assessed using Annexin V-FITC/PI Apoptosis Detection Kit (CWBIO, Beijing, China). At 48 h post transfection, A549/DDP and H1299/DDP cells were resuspended in annexin-binding buffer and double-stained with annexin V/FITC (5 μL) and propidium iodide (PI) solution (10 μL; 20 μg/mL) at 37°C for 15 min in the dark. Following phosphate buffered saline wash, cells were analyzed by flow cytometry (FACScan; BD Biosciences, Shanghai, China).
The wild-type (WT) sequence of PCAT1 and the WT sequence of ABCB1 3’ UTR containing the putative binding region for miR-129 were amplified and fused into pGL3-control vector (Promega, Madison, WI, USA) to generate PCAT1-WT and ABCB1-WT constructs, respectively. Correspondingly, the mutant (MUT) sequences of PCAT1 and ABCB1 were introduced to create PCAT1-MUT and ABCB1-MUT constructs, respectively. These constructs were transfected into 293T cells, together with miR-129, pcDNA-PCAT1, or matched controls. The relative luciferase activity was measured 48 post transfection using the Dual-Luciferase Reporter Assay System (Promega), following the manufacturer’s specifications.
Cells were collected and lysed with ice-cold cell lysis buffer (Invitrogen), in accordance with the manufacturer’s specifications. The protein concentration was quantified with a BCA Protein Assay Kit (Pierce, Rockford, IL, USA), in accordance with the manufacturer’s specifications. Lysates were run on 14% sodium dodecyl sulfate-polyacrylamide gels and then transferred onto polyvinylidene fluoride membranes. Subsequently, membranes were rinsed with tris-buffered saline with tween, blocked for 1 h in non-fat milk, and then probed with special primary antibody against ABCB1 (Novus Biologicals, Littleton, CO, USA) overnight at 4°C. Following this, membranes were immunoblotted for 1 h at room temperature with horseradish peroxidase-conjugated secondary antibodies (Novus biologicals). Bands were developed using ECL detection reagents (Pierce) and analyzed using ImageJ software.
Total RNA extraction was carried out using TRIzol reagent (Invitrogen), as per the manual instructions. A total of 500 ng total RNA was used to synthesize cDNA using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) and One Step PrimeScript cDNA kit (Qiagen, Hilden, Germany). qRT-PCR was performed in triplicate on an ABI 7900HT Real-Time PCR system using Power SYBR Green Master Mix (Life Technologies, USA). While, TaqMan MicroRNA Assay (Applied Biosystems) was carried out to determine the expression of miR-129, following the manufacturer’s instructions. The relative expression of PCAT1, miR-129, and ABCB1 was calculated according to the 2−∆∆Ct method using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 as the housekeeping genes. The sequences of gene-specific primers used in this study are listed in Table 1.
| Gene | Forward 3’-5’ | Reverse 3’-5’ |
|---|---|---|
| PCAT1 | GCTGGCATTGGTCAACATAAC | GTGAATATGGCGGATGAGGAA |
| miR-129 | GTTGGGGAGATTTAGTTTGTT | CCTACTCCAATTCCCCCTATAATAC |
| ABCB1 | GCTGTCAAGGAAGCCAATGCCT | TGCAATGGCGATCCTCTGCTTC |
| GAPDH | GGGAAACTGTGGCGTGAT | GAGTGGGTGTCGCTGTTGA |
| U6 | CGCTTCGGCAGCACATATACTAAAATTGGAAC | GCTTCACGAATTTGCGTGTCATCCTTGC |
Statistical data are expressed as the mean ± standard error of the mean. Statistical significance was analyzed by Student’s t-test or one-way analysis of variance. The survival data were analyzed with the log-rank test. A P-value <0.05 was regarded as statistically significant.
To investigate the role of PCAT1 in NSCLC, the expression of PCAT1 was analyzed using the RNA-seq data from TCGA database, including 513 NSCLC tissues and 59 normal tissues. Publicly available data from TCGA indicated that PCAT1 expression was upregulated in NSCLC cells (Figure 1A). Then, we validated the differential expression of PCAT1 in NSCLC tissues and adjacent normal tissues from 42 patients diagnosed with NSCLC. The expression of PCAT1 was higher in NSCLC tissues than that in their adjacent normal tissues, as shown by qRT-PCR (Figure 1B). Moreover, the expression of PCAT1 was much higher in the NSCLC tissues of DDP-resistant patients than that in the NSCLC tissues of DDP-sensitive patients (Figure 1C). In line with this, PCAT1 expression was upregulated in A549 and H1299 cells as compared with that in HBE cells. Also, the expression level of PCAT1 was much higher in A549/DDP and H1299/DDP cells compared to that in their parental A549 and H1299 cells (Figure 1D and 1E). In addition, NSCLC patients with high PCAT1 expression had markedly shorter overall survival (P = 0.0049) than those with low PCAT1 expression, as shown by Kaplan-Meier survival analysis.
 Figure 1
Figure 1lncRNA PCAT1 is upregulated in NSCLC tissues and cell lines. (A) Analysis of PCAT1 expression in TCGA database. (B) qRT-PCR analysis of PCAT1 expression showed upregulation of PCAT1 in NSCLC tissues. (C) PCAT1 expression was higher in the NSCLC tissues of DDP-resistant patients than that in the NSCLC tissues of DDP-sensitive patients. (D and E) qRT-PCR analysis of PCAT1 expression in HBE, A549, A549/DDP, H1299, and H1299/DDP cells. (F) Kaplan-Meier survival analysis of NSCLC specimens expressing high versus low levels of PCAT1. *P < 0.05, ***P < 0.001.
To determine the resistance of A549/DDP and H1299/DDP cells to DDP, A549, H1299, A549/DDP, and H1299/DDP cells were treated with increasing doses (0.1, 0.5, 1, 5, 10, 50, and 100 μM) of DDP for 48 h, and then subjected to CCK-8 assay. As shown in Figure 2A and 2B, the viability of A549/DDP and H1299/DDP cells was higher than that of A549 and H1299 cells in the presence of DDP, and the IC50 of DDP in A549/DDP and H1299/DDP cells appeared to be much higher than that in A549 and H1299 cells, suggesting that A549/DDP and H1299/DDP cells exhibited stronger resistance to DDP. To explore the potential role of PCAT1 in the resistance of NSCLC cells to DDP, we knocked down PCAT1 expression in A549/DDP and H1299/DDP cells by transfecting them with si-PCAT1#1, si-PCAT1#2, si-PCAT1#3, or si-con. The results of qRT-PCR assay showed that the expression of PCAT1 was strikingly reduced in the si-PCAT1 group, especially in the si-PCAT1#2 group, compared with that in the si-con group (Figure 2C and 2D). Therefore, si-PCAT1#2 was used to knock down PCAT1 expression in the following experiments. Furthermore, knockdown n of PCAT1 expression repressed the viability of A549/DDP and H1299/DDP cells in the presence of DDP, as evidenced by lower DDP IC50 in A549/DDP and H1299/DDP cells transfected with si-PCAT1#2 (Figure 2E and 2F). Meanwhile, flow cytometry analysis revealed that transfection of A549/DDP and H1299/DDP cells with si-PCAT1#2 led to a prominent increase in the rate of apoptosis in the presence of DDP (Figure 2G and 2H).
 Figure 2
Figure 2Knockdown of PCAT1 improves the sensitivity of NSCLC cells to DDP. (A and B) Dose-response curves indicated that A549/DDP and H1299/DDP cells showed increased cell viability following administration of DDP, compared to A549 and H1299 cells. (C and D) qRT-PCR analysis of PCAT1 expression showed reduced expression of PCAT2 in A549/DDP and H1299/DDP cells transfected with si-PCAT1#1, si-PCAT1#2, or si-PCAT1#3. (E and F) CCK-8 assay revealed that knockdown of PCAT1 strikingly inhibited the viability of A549/DDP and H1299/DDP cells in the presence of DDP. (G and H) Flow cytometry analysis of apoptosis in si-PCAT1#2- or si-con-transfected A549/DDP and H1299/DDP cells in the presence of DDP. *P < 0.05.
miRanda analysis suggested that PCAT1 harbors an miR-129 binding site (Figure 3A). To identify whether miR-129 is a direct target of PCAT1, we performed luciferase reporter assay in 293T cells. Transfection with miR-129, but not miR-con reduced the relative luciferase activity of PCAT1-WT construct in 293T cells. Meanwhile, transfection with either miR-129 or miR-con did not alter the relative luciferase activity of PCAT1-MUT construct in 293T cells (Figure 3B). Furthermore, a remarkable elevation in miR-129 expression was observed in A549/DDP and H1299/DDP cells transfected with si-PCAT1#2 as compared with that in cells transfected with si-con (Figure 3C). In addition, upregulation of miR-129 strikingly decreased the IC50 of DDP in A549/DDP cells. Moreover, knockdown of PCAT1 in A549/DDP cells obviously reduced DDP IC50, which was markedly mitigated by silencing of miR-129, and similar results were obtained in H1299/DDP cells (Figure 3D and 3E). In parallel, flow cytometry analysis revealed that transfection of A549/DDP and H1299/DDP cells with miR-129 led to a prominent increase in the rate of apoptosis in the presence of DDP. Transfection of A549/DDP and H1299/DDP cells with si-PCAT1#2 prior to DDP treatment caused a dramatic increase in the rate of apoptosis; however, this effect could be mitigated by transfection with anti-miR-129 (Figure 3F and 3G).
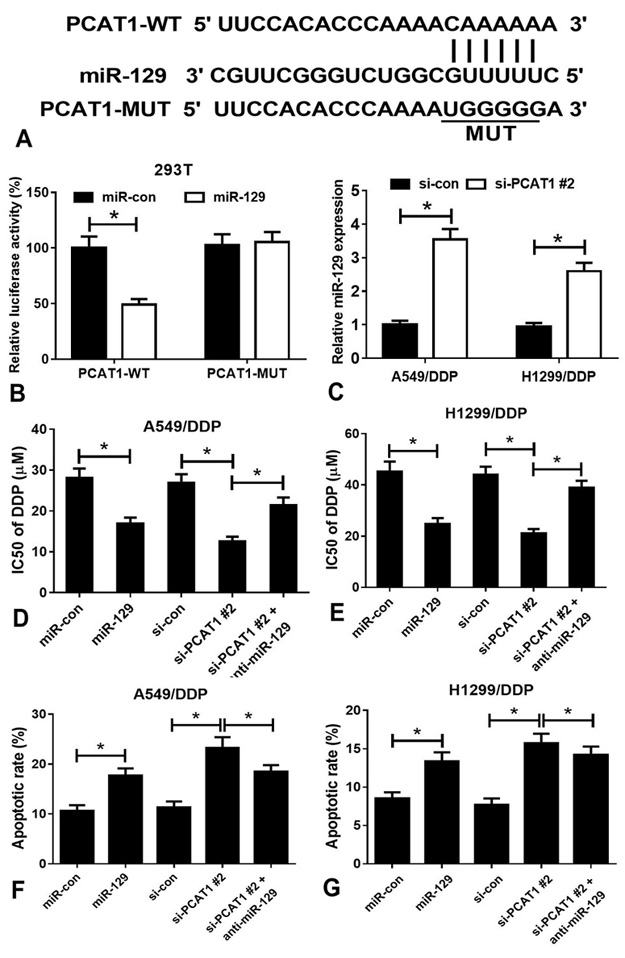 Figure 3
Figure 3PCAT1 acts as a ‘sponge’ for miR-129 in NSCLC cells. (A) A schematic representation of the functional interaction between miR-129 and PCAT1 as predicted by miRanda. (B) Luciferase reporter assay of 293T cells co-transfected with reporter constructs (PCAT1-WT or PCAT1-MUT) and miR-129 or miR-con. (C) qRT-PCR analysis showed that transfection with si-PCAT1#2 increased the expression of miR-129 in A549/DDP and H1299/DDP cells. (D) IC50 values of DDP in miR-con-, miR-129-, si-con-, si-PCAT1#2-, and anti-miR-129-transfected A549/DDP cells. (E) IC50 values of DDP in miR-con-, miR-129-, si-con-, si-PCAT1#2-, and anti-miR-129-transfected H1299/DDP cells. (F) Flow cytometry analysis of apoptosis in miR-con-, miR-129-, si-con-, si-PCAT1#2-, and anti-miR-129-transfected A549/DDP cells treated with DDP. (G) Flow cytometry analysis of apoptosis in miR-con-, miR-129-, si-con-, si-PCAT1#2-, and anti-miR-129-transfected H1299/DDP cells treated with DDP. *P < 0.05.
TargetScan software version 7.1 (www.targetscan.org) predicts that ABCB1 is a potential molecular target of miR-129 (Figure 4A). As determined by luciferase reporter assay, overexpression of miR-129 reduced the relative luciferase activity of ABCB1-WT construct in 293T cells, which was reversed by overexpression of PCAT1. Moreover, the relative luciferase activity of ABCB1-MUT construct was unaffected in 293T cells transfected with miR-129 alone or in combination with pcDNA-PCAT1 (Figure 4B). Additionally, the mRNA and protein expression levels of ABCB1 were remarkably decreased when A549/DDP and H1299/DDP cells were transfected with miR-129 or si-PCAT1#2. Notably, the effect of PCAT1 knockdown on ABCB1 expression was obviously blocked by silencing of miR-129 (Figure 4C–4E). Transient knockdown of ABCB1 in A549/DDP cells strikingly decreased the expression of ABCB1 and IC50 of DDP as compared with those in control cells, and similar results were observed in H1299/DDP cells (Figure 4F and 4G). Flow cytometry analysis revealed that ABCB1 silencing by si-ABCB1 gradually promoted the apoptosis of A549/DDP and H1299/DDP cells as compared with respective controls (Figure 4H).
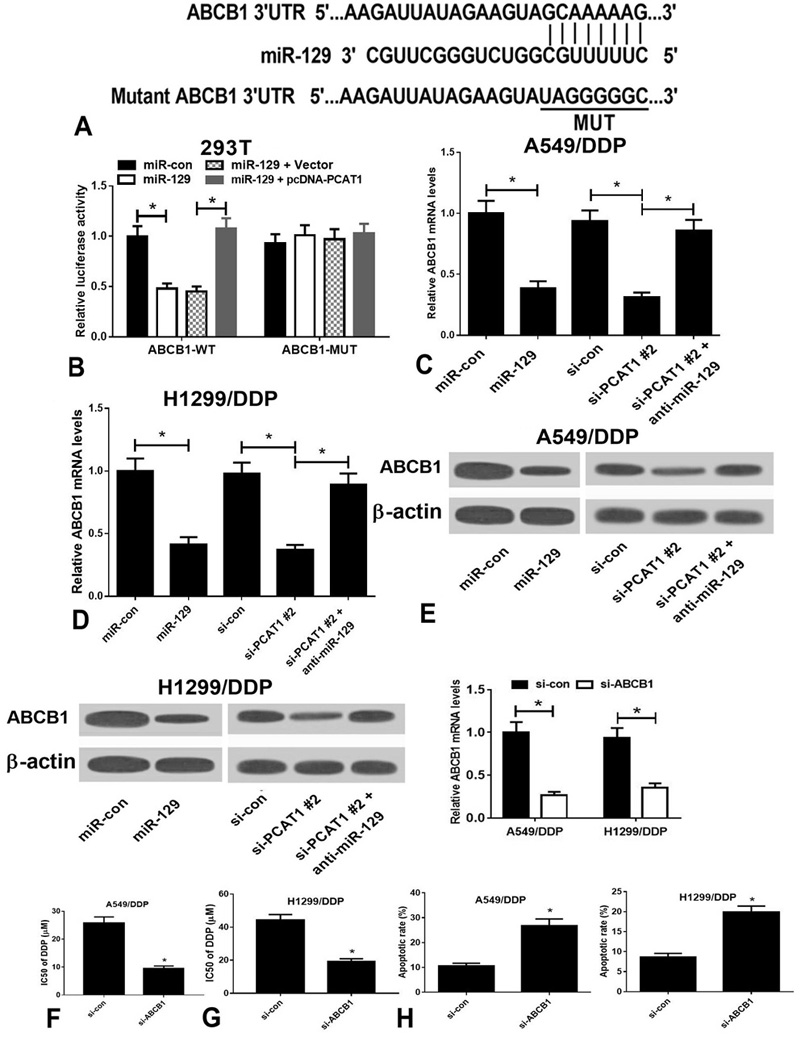 Figure 4
Figure 4ABCB1 expression upregulated by PCAT1/miR-129 axis contributes to DDP resistance in A549/DDP and H1299/DDP cells. (A) The predicted binding site of miR-129 within the ABCB1 3’ UTR and the mutated sites are shown. (B) Luciferase reporter assay of 293T cells co-transfected with reporter constructs (ABCB1-WT or ABCB1-MUT) and miR-con, miR-129, vector, or pcDNA-PCAT1. (C) qRT-PCR assay was performed to analyze the expression of miR-129 in A549/DDP and H1299/DDP cells transfected with miR-con, miR-129, si-con, si-PCAT1#2, or anti-miR-129. (D and E) Western blot analysis of miR-129 expression in A549/DDP and H1299/DDP cells transfected with miR-con, miR-129, si-con, si-PCAT1#2, or anti-miR-129. (F) qRT-PCR analysis showed that transfection with si-PCAT1#2 downregulated the expression of ABCB1 in A549/DDP and H1299/DDP cells. (G) IC50 values of DDP in si-con- and si-ABCB1-transfected cells. (H) Flow cytometry analysis of apoptosis in si-con- and si-ABCB1-transfected cells. *P < 0.05.
Recently, the significance of lncRNAs in the development of DDP resistance has been widely discussed in many types of cancers. As a novel lncRNA, PCAT1 has been shown to act as a crucial factor in the acquisition of tumor cell resistance to chemotherapeutic drugs. PCAT1 can be predictive of infiltration depth, lymph node metastasis, and clinicopathologic stage (19). Upregulation of PCAT1 was previously reported in oesophageal cancer tissues. Furthermore, targeted deletion of PCAT1 enhanced cell proliferation, increased the sensitivity of oesophageal cancer cells to DDP in vitro, and repressed oesophageal tumor growth in vivo (13). Functional studies in Caco-2 and HT-29 cells showed that silencing of PCAT1 suppressed cell motility, migration, and invasiveness, induced cell apoptosis, and increased cell sensitivity to 5-fluorouracil by upregulating the expression of c-Myc (20). These studies suggest that upregulation of PCAT1 in cancer cells leads to phenotypes that contribute to chemotherapeutic resistance. However, the regulatory mechanism of PCAT1 in the acquisition of NSCLC cell resistance to DDP remains unknown. In this study, upregulation of PCAT1 expression was observed in NSCLC tissues, DDP-resistant patients, A549, and H1299 cells, as well as in A549/DDP and H1299/DDP cells. Knockdown of PCAT1 enhanced the sensitivity of A549/DDP and H1299/DDP cells to DDP. Hence, our work establishes a role for PCAT1 in regulating responsiveness to DDP in NSCLC cells.
Previous studies have reported the significance of miRNAs. They serve as tumor suppressors or oncogenes in tumorigenesis, and also function as master regulators of chemotherapeutic drug resistance. miR-129 has been shown to be aberrantly expressed in a variety of cancer types. More importantly, its dysregulation is closely linked with the acquisition of cancer cell resistance to DDP. Previously, miR-129-5p was shown to be underexpressed in trastuzumab-resistant breast cancer cells (JIMT-1) compared with that in SKBR-3 cells. Overexpression of miR-129-5p enhanced the sensitivity of JIMT-1 cells to trastuzumab through targeting ribosomal protein S6 (21). Moreover, miR-129 was found to be downregulated in colorectal cancer. Overexpression of miR-129 induced cell apoptosis, suppressed cell proliferation, and increased the cytotoxic effect of 5-fluorouracil in vitro and in vivo (22). In addition, silencing of miR-129-3p increased the sensitivity of MDA-MB-231/Doc and MCF-7 cells to docetaxel by inhibiting the expression of centrosomal protein of 110 kDa (23). These studies suggest that miR-129 serves as a key player in chemotherapeutic resistance. However, the functional importance of miR-129 in the acquisition of NSCLC cell resistance to DDP remains poorly understood. In this work, we found that PCAT1 functions as a sink for miR-129 to contribute to DDP resistance in A549/DDP and H1299/DDP cells, suggesting the involvement of miR-129 in the acquisition of NSCLC cell resistance to DDP.
Chemotherapeutic resistance is still a major obstacle in the treatment of patients with NSCLC (24). Elucidating and understanding mechanisms of chemotherapeutic resistance are of great significance in improving the therapeutic effect of DDP in NSCLC. ABCB1, also known as p-glycoprotein 1, is an ATP-dependent membrane efflux pump and has been shown to be a major player in chemotherapeutic resistance (25). Of note, overexpression of ABCB1 was shown to be responsible for drug resistance (26). As an example, overexpression of ABCB1 contributed to the development of nab-paclitaxel resistance, and ABCB1 inhibition by cabozantinib and crizotinib potentiated the sensitivity of ABCB1-overexpressing urothelial bladder cancer cells to nab-paclitaxel and paclitaxel (27). In prostate cancer, docetaxel-resistant prostate cancer cells exhibited cross-resistance to cabazitaxel, which was mediated by ABCB1. Inhibition of ABCB1 in docetaxel-resistant TaxR cells repressed cell proliferation, promoted cell apoptosis, and sensitized TaxR cells to cabazitaxel (28). While ABCB1 has been characterized in various cancers, its expression and function in the acquisition of NSCLC cell resistance to DDP remain to be investigated. Here, we found a novel mechanism of direct regulation of ABCB1 expression by the PCAT1/miR-129 axis, contributing to increased sensitivity towards DDP in A549/DDP and H1299/DDP cells. The competitive endogenous RNA (ceRNA) regulatory network underlying DDP resistance is shown in Figure 5. Apart from our study, some previous studies also reported that PCAT1 could act as an miRNA sponge to elevate miRNA target gene expression in cancers. For example, PCAT1 promoted the invasion and migration of HCC hepatocellular carcinoma cells through de-repressing high mobility group box 1 expression by sponging miR-129 (29). PCAT1 overexpression promoted proliferation, migration, and invasion, and inhibited apoptosis of prostate cancer cells through acting as an miR-145 sponge to release fascin-1 (30). Moreover, PCAT1 promoted extrahepatic cholangiocarcinoma cell growth, migration, and invasion, and suppressed apoptosis through miR-122 repression and WNT1 upregulation (31). All these data demonstrated that PCAT1 exerts an oncogenic role in cancers through a ceRNA mechanism.
 Figure 5
Figure 5The graphical abstract is shown. (–) negatively regulate, (+) positively regulate.
Taken together, our work identified a novel lncRNA-miRNA-mRNA regulatory mechanism of DDP resistance in NSCLC cells, providing a thorough understanding of the mechanisms of chemotherapeutic resistance in patients with NSCLC. Therefore, targeting the PCAT1/miR-129/ABCB1 axis can be used as a potential therapeutic approach to overcome DDP resistance in patients with NSCLC.
Drs Ruifen Tian and Congjun Zhang contributed equally to this work. This research was carried out without using a grant from funding agencies in the public, commercial, or not-for-profit sectors.
lncRNA
Long noncoding RNA
prostate cancer-associated transcript 1
non-small cell lung cancer
microRNAs
ATP-binding cassette sub-family B member 1
cisplatin
Lung adenocarcinoma
high mobility group box 1
competitive endogenous RNA
