Frontiers in Bioscience-Landmark (FBL) is published by IMR Press from Volume 26 Issue 5 (2021). Previous articles were published by another publisher on a subscription basis, and they are hosted by IMR Press on imrpress.com as a courtesy and upon agreement with Frontiers in Bioscience.
1 Hunan International Joint Laboratory of Animal Intestinal Ecology and Health, Laboratory of Animal Nutrition and Human Health, School of Life Sciences, Hunan Normal University, Changsha, Hunan, 410081, P.R. China
2 Human Engineering & Research Center of Animal and Poultry Science, Key Lab Agroecology Processing Subtropical Region, Scientific Observational and Experimental Station of Animal Nutrition and Feed Science in South-Central, Ministry of Agriculture, Institute of Subtropical Agriculture, Chinese Academy of Science, Changsha, Hunan, 410125, P.R. China
Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) is a master controller of cell growth and metabolism which integrates diverse bio-signaling inputs to coordinate various fundamental biological processes. Amino acids, especially leucine, arginine and glutamine, signal to mTORC1 activation. Classically, Rag GTPases play a crucial role in amino acids-induced mTORC1 activation in the lysosome and Golgi apparatus. More recently, multiple amino acid sensors have been identified and most of them indirectly associate with Rag GTPases. As a result, the mechanistic details on how amino acids are sensed and activate mTORC1 are rapidly evolving. This review discusses current understanding of mTORC1 activation and provides a brief and up-to-date narrative on the progress of amino acid sensors regulating mTORC1 activation.
Keywords
- Amino acid signaling
- mTORC1
- Amino acid sensors
- Review
Eukaryotic cells have evolved mechanisms for integrating signals from the environment to ensure efficiently transitioning between anabolic and catabolic states, allowing them to survive and grow. The studies of the past decades have revealed that the mechanistic/mammalian target of rapamycin complex 1 (mTORC1) has played a central role in integrating signals and regulating fundamental cell process(1-5). The mTORC1 is an evolutionary conserved atypical serine/threonine protein kinase that belongs to phosphoinositide protein kinases family (PIKKs). The mTORC1 is a key integrator of environmental cues, such as growth factors, energy status, and an array of stress(6), which control many processes from cell growth to apoptosis, and deregulated mTORC1 activation is associated with many human diseases (4, 7-9).
Among these signals, amino acids act as crucial nutrient signals to activate the mTORC1 pathway in addition to their role as protein building blocks. Amino acids are also precursor substrates to synthetize many low-molecule substances which feature enormous physiological functions. In addition, there is increasing evidence that multi-amino acids (leucine, arginine, serine, glutamine) can participate in or regulate key metabolic mTORC1 pathway to improve animal health, survival, growth, development and reproduction of organisms(10-13), even though the essential amino acid leucine has been considered as a crucial mTORC1 activator in many cell types. Therefore, in this review, we attempt to provide a brief and up-to-date narrative on key mediators of amino acid sensing pathway in the regulation of mTORC1.
The phospholipid bilayer in the cell surface is a selective barrier to signals and nutrients, and numerous receptors and transporters are embedded in cell membrane and participate in signaling and nutrient transportation(14). Amino acids do not directly diffuse across cell membrane or organelle membrane, and it require membrane-spanning transporter proteins to help transfer them into or out of cells and organelles(15, 16). Recent studies have shown that the solute carrier (SLC) superfamily has been thought to be the major group of amino acid transporters. The SLC superfamily genes include the classical transporter families which contain ion-coupled transporters, exchangers, passive transporters, etc(16, 17). The detailed information of the SLC genes is described in Table 1. Amino acid transporters have a similar structure that features several transmembrane domains organized around a central pore region which can recognize a range of structural or physico-chemical property similar amino acids(18, 19). Likewise, the monocarboxylate transporter SLC16 can transport aromatic amino acids including tyrosine, phenylalanine and tryptophan (20). In mammalian cells, some amino acid transporters are expressed in specific tissues but in most types of tissues expressing several amino acid transporters coupled with overlapping function in transporting amino acids. In epithelial cells, SLC6A19 and SLC6A14 belong to broad-scope Na+-coupled amino acid symporters that are able to transport neutral amino acids. However, in nonepithelial cells, large neutral amino acids are transported into cells by exchange or facilitative mechanisms (19, 21).
| SLC name | Transporter name | Transport types* | Substrates | Distribution |
|---|---|---|---|---|
| SLAC1A1 | EAAC1, EAAT2 | BA | L-Glu, D/L-Asp | Instestine, kidney, liver,heart, brain |
| SLC1A2 | GLT-1, EAAT2 | BA | L-Glu, D/L-Asp | Brain, Liver, Pancreas |
| SLC1A3 | GLAST, EAAT1 | BA | L-Glu, D/L-Asp | Brain, Skeletal Muscle |
| SLC1A4 | ASCT1, SATT | BC | L-Ala, L-Ser, L-Cys, L-Thr | In Most Tissue |
| SLC1A5 | ASCT2 | B | L-Ala, Ser, Cys, Thr, Gln | Lung, Skeletal Muscle, Large Intestine, |
| SLC1A6 | EAAT4 | A and B | L-Glu, D/L-Asp | Cerebellum |
| SLC1A7 | EAAT5 | A and B | L-Glu, D/L-Asp | Retina |
| SLC3A1 | rBAT | C | Cationic Amino Acids, Neutral Amino Acids | Kidney, Small Intestine, Liver, Pancreas |
| SLC3A2 | 4F2hc | C | Neutral L-Amino Acids,Cationic Amino Acids | In Most Tissue |
| SLC6A5 | UN | Glycine | Brain | |
| SLC6A9 | UN | Glycine | Liver, Spleen, Kidney, Stomach, Intestine | |
| SLC6A14 | UN | Neutral, Cationic Amino Acids | Lung, Trachea, Salivary Gland, Mammary Gland, Stomach, | |
| SLC6A15 | UN | Large, Neutral Amino Acids | Brain, Kidney | |
| SLC6A17-19 | UN | Neutral Amino Acids | Brain, Heart, Skeletal Muscle | |
| SLC7A1 | CAT-1 | C | Cationic L-Amino Acids | Ubiquitous Except For Liver, |
| SLC7A2 | CAT-2 | C | Cationic L-Amino Acids | Liver, Skeletal Muscle, Pancreas |
| SLC7A3 | CAT-3 | C | Cationic L-Amino Acids | Thymus, Ovary, Testis, Brain |
| SLC7A5 | LAT-1 | C | Neutral L-Amino Acids | Brain, Testis, Placenta, Spleen, Colon, Fetal Liver |
| SLC7A6 | y+LAT2 | C | Cationic Amino Acids, Neutral L-Amino Acids | Brain, Small Intestine, Heart, Kidney, Lung, Thymus |
| SLC7A7 | y+LAT1 | E | Cationic Amino Acids, Neutral L-Amino Acids | Small Intestine, Kidney, Spleen, Leucocytes, Placenta |
| SLC7A8 | LAT2 | C | Neutral L-Amino Acids | Small Intestine, Kidney, Lung, Heart, Spleen, Liver, Brain |
| SLC7A9 | b0,+AT | C | Cationic Amino Acids, Neutral Amino Acids | Kidney, Small Intestine, Liver, Placenta |
| SLC7A10 | Asc-1 | C | Neutral Amino Acids | Brain, Small Intestine, Muscle |
| *Note: The sequential relationship between primary, secondary and tertiary active transport systems (denoted A, B and C, respectively), the same denotation is described in Figure 1. | ||||
There is emerging evidence to show that amino acid transporters own a dual transportor/receptor function. Firstly, the amino acid transporters at the surface of cells act as gate keepers of nutrient exchange (18). Secondly, mammalian amino acid transporters may regulate nutrient signaling (eg. mTORC1 activation) and become a limiting factor for mTORC1 activation. In addition, the glutamine transporter (SLC1A5) in combination with the heterodimeric amino acid transporter CD98, solute-linked carrier (SLC)3A2-SLC7A5 (CD98hc-LAT1) can activate mTORC1 by transporting leucine into cells. SLC7A5 can directly mediate leucine-induced mTORC1 activation in fibroblasts(22). Moreover, there is emerging evidence to suggest that arginine transporters SLC7A1-4 may directly influence mTORC1 activation (23).
mTORC1 is comprised of mTOR, mammalian lethal with SEC13 protein 8 (mLST8, also known as GβL), the regulatory associated protein of mTOR (RAPTOR), DEP domain containing mTOR-interacting protein (DEPTOR), PRAS40 and recently identified telomere maintenance Tti1/Tel2(24-26). Among the proteins in the complex, mTOR is the core kinase, and RAPTOR and Tti1/Tel2 are scaffold proteins regulating the assembly and stability of the mTORC1. The RAPTOR interaction with mTOR can be either loosened or tightened depending on various stimuli. For example, amino acid deprivation reinforces the RAPTOR-mTOR interaction, whereas rapamycin can disrupt the interaction(26). The function of mLST8/GbL is still unknown, but it may stabilize and positively regulate mTORC1. However, this view is challenged by the observation that knockout of mLST8 does not impair the stability of mTORC1(27). RRAS40 and DEPTOR are negative regulators of mTORC1(28). PRAS40 directly interacts with RAPTOR and can block its binding to mTOR. Similarly, DEPTOR can bind to the FRAP-ATM-RRTAP (FAT) domain of mTOR(29). mTORC1 activation is regulated by two types of mechanisms: one is direct modification of mTORC1 components, and the other one is that growth factor through IRS1-PI3K or AMPK pathway signal regulation the upstream factors of mTORC1 signaling pathway (30, 31). Recent studies have showed that mTORC1 can maintain cell metabolism through regulating various key mediators or transcription factors, and this section has been well described in the review of Duan et.al(32). Thus, in this review, we will focus on the proteins that participate in amino acid sensing to mTORc1.
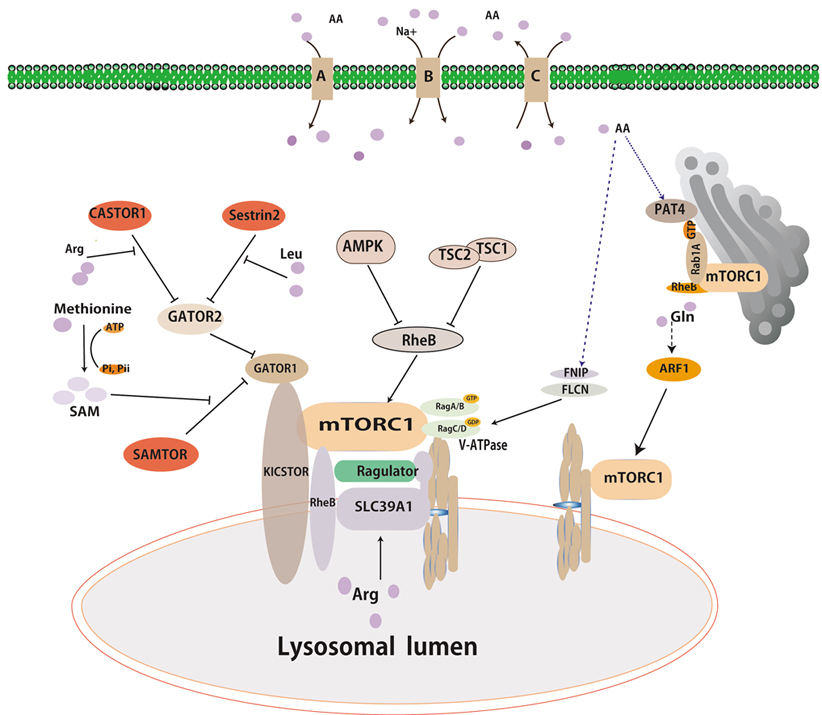
Recent studies have suggested that the lysosome has been recognized as a major effector and serves as a platform for mTORC1 activation by amino acids or growth factors. Intracellular amino acids can induce the movement of mTORC1 to the lysosome membranes and lead to mTORC1 activation(33-35). Rags and Rheb, two different types of small GTPases cooperate to accurately regulate mTORC1 activity. The Rags is involved in the translocalization of mTORC1 to the lysosome surface in response to amino acids. Full activation of mTORC1 requires Rheb, which is activated under conditions of high energy status and growth factor signals(36, 37). Together, the GTPases act as coincidence detectors, which integrate signals relating to nutrients, energy status, and growth factors.
The Rag proteins are obligate heterodimers of functionally redundant small GTPases composed of either RagA or RagB and RagC or RagD. The heterodimers RagA/B in the GTP bound state (RagA/BGTP) and RagC/D in GDP bound state (RagC/DGDP) are conceived as active forms in terms of mTORC1 activation(38-40). Multiple studies have provided insights into the complex machinery implicated in the regulation of Rag nucleotide status that convey amino acid signals to mTORC1(41). The activity of Rag GTPases is regulated by GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs)(42). GEFs activate Rag GTPases by exchanging their nucleotide status, while GAPs can inactivate Rag GTPases by stimulating GTP hydrolysis (43).
Extensive works have suggested that a large protein complex activates Rag GTPases’s activity on the lysosome surfaces in response to amino acids(44-46). Within this complex, Vacuolar H+-adenosine triphosphatase ATPase (v-ATPase) interactes with Ragulator(47), which is anchored in the lysosomal surfaces and functions as a guanine nucleotide exchange factor (GEF) for RagA/B to facilitate the exchange of the heterodimer RagA/BGDP to the active form (RagA/BGTP)(10, 48). V-ATPase plays a role in positively regulating the mTORC1 pathway through an unknow mechanism, while it is believed to regulate Ragulator’s GEF activity towards RagA/B.
A multiprotein complex known as GAP activity toward Rags 1 (GATOR1) was identified by immunoprecipitation and mass spectrum (IP-MS) analysis using RagB as a bait(49). GATOR1 is a trimeric protein complex composed with DEPDC5, Nprl2, and Nprl3, and functions as a GTPase-activating protein toward RagA/B. In this study, a second complex named CASTOR2 was also identified by IP-MS analysis. GATOR2 interacts with GATOR1 and negatively regulates GATOR1 activity (50). Recently, the KICSTOR complex is composed with KPTN, C12orf66,ITFG2 and SZT2-containing regulator of mTORC1, and function as a scaffold protein for GATOR1 binding to the lysosome(51). As mentioned above, FNIP1/2 complex with Folliculin acts as a GTPase-activating protein for RagC/D(52) that can regulate Rag GTPases’ activity. Thus, FlCN-FINP1/2 is a positive regulator of mTORC1 in a Rag GTPase-dependent manner(53).
Overall, multiple regulatory proteins, including GAPs, GEFs, and scaffold proteins, have been characterized to directly or indirectly modulate the activity of Rag GTPases, which play a central role in the activation of mTORC1(54).
The discovery of Rag GTPases is a milestone in understanding of mTORC1 activation by nutrients. However, recent studies have shown that anther two small GTPases are also essential for mTORC1 activation in cells. Glutamine, a nitrogen sources and energy substance, can promotes the translocation of mTORC1 to the lysosome and its activation in a Rag GTPases-independent mechanism. A small GTPase named ribosylation factor1 (ARF1) has been characterized to have a similar function as Rag which involve glutamine-induced mTORC1 activation(10, 55, 56). Knockdown of ARF1 in mammalian or Drosophila S2 cells can block glutamine signaling to mTORC1 in the absence of Rags(10). Overexpression of constitutively GTP-bound Arf or treatment with brefeldin A, an inhibitor for ARF1, can inhibit mTORC1 translocalization and activation even in the presence of glutamine(57, 58). Overall, ARF1 seems to be specific to glutamine-induced mTORC1 activation(58).
Recent studies have revealed that amino acids can activate mTORC1 via Rab GTPase in the surface of Golgi apparatus(59, 60). Thomas et al have identified Rab1 as a conserved regulator of mTORC1 and elucidated a Golgi-based mechanism by which Rab1 engages mTORC1 interaction with RheB(61, 62). In addition, overexpression of Rab1A promotes mTORC1 signaling. Furthermore, Fan et al have revealed that SLC36A4, known as PAT4, is predominantly localized in the Golgi apparatus and engages mTORC1 interaction with Rab1A(63).
Sestrins have been initially identified as a family of stress-inducible proteins, which are capable of attenuating various stresses, stimulating autophagy, and regulating cell metabolism(64, 65) (Figure 1). Sestrins family contains three homologs (Sestrin1-3) sharing nearly 50% amino acid sequence similarity. Apart from the function of redox regulation, Sestrin 1-3 have been characterized as negative regulators of mTORC1 and positive regulators of AMPK through TSC2(66-69). The Sestrins have been proposed to interact with and function as guanine nucleotide dissociation inhibitor for the Rag GTPases(70). More recently, Sestrin1 and Sestrin2 showed a high affinity (Kd) of 10~15 and ~20 μM to physically bind to leucine(71). Moreover, Sestrin2 which purified from prokaryotic cell, has ability to directly bind to leucine but not arginine in vitro. And the assay of mutation Sestrin2 and crystalized Sestrin2 strongly indicated residues of Sestrin2 directly bind leucine and importance for binding leucine capacity of the protein. Sestrin2 interacts with GATOR2 to inhibit mTORC1 under leucine deprivation, not arginine deprivation(72, 73). When leucine added in media, it binds to Sestrin2 to dissociate the complex of Sestrin2-GATOR2. Leucine must bind to Sestrin2 in order for leucine to activate mTORC1 in mammalian cells. Overall, Sestrin2 acts as a leucine sensor in the cytoplasm(74).
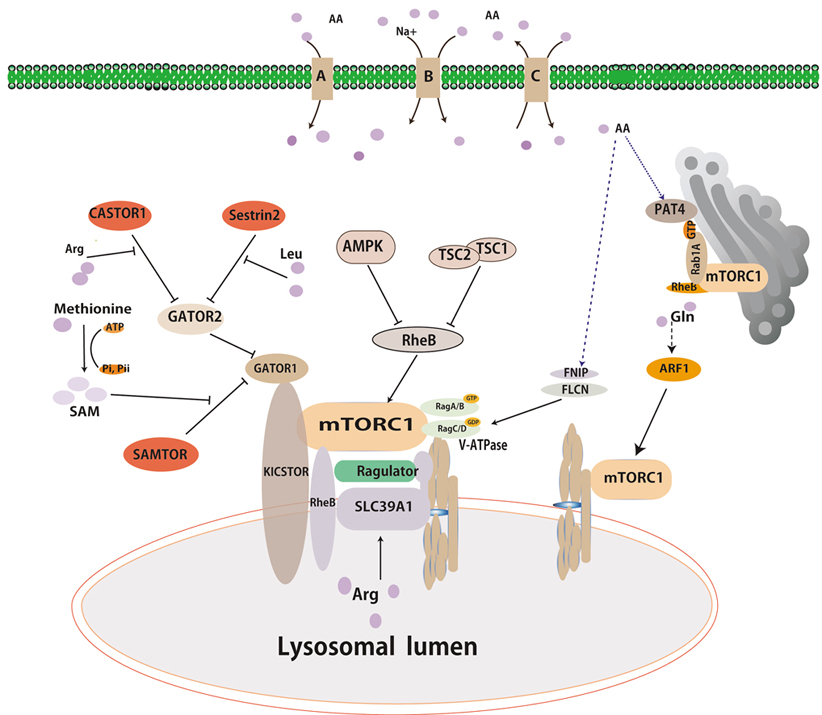 Figure 1.
Figure 1.Overviews of multiple signals activating mTORC1. Amino acids, growth factors and energy signals can lead to mTORc1 activation. Growth factors and energy signals activate mTORC1 primarily through the PI3K pathway and AMPK pathway, respectively. Amino acids signal to mTORC1 in Rag-dependent and Rag-independent pathway. Sestrin2, CASTOR1 and SMATOR (shown in red box) are reported to be cytosolic amino acid sensors for leucine, arginine, and s-adenosyl-L-methionine, respectively. Arrows and bars represent activation and inhibition, respectively, of downstream proteins.
GATOR2 have been characterized as integrating multiple amino acid inputs to mTORC1, and thus specific amino acid sensors may interact with GATOR2, analogous to that of SESN2 (Figure 1). Chantranupong and his colleagues searched a protein interaction database named BioPlex to identify potential GATOR2-interacting partners, and they found that CASTOR1 encoded by GATS protein-like 3 (GATSL3) gene is one of putative GATOR2 interactors(75). Subsequently, CASTOR1 has been characterized as a cytosolic arginine sensor. The homodimeric CASTOR1 protein can interact with GATOR2 under the arginine deprivation condition to inhibit mTORC1 activation. In vitro binding assays showed that CASTOR1 can directly and specifically bind to arginine at a highly affinity of ~30 μm(76). Arginine-bound CASTOR1 can lead to dissociation of CASTOR1 from GATOR2, leading to mTORC1 activation. Thus, CASTOR1 acts as an arginine sensor in the cytoplasm(75, 77).
As the mentioned above, GATOR2 plays a role in integrating multiple amino acid inputs to mTORC1. The function of GATOR2 remains known, however it might be upstream of CATOR1 which are GAPs for RagA/B(49), and the KICSTOR complex bound with GATOR1 is necessary for amino acid depravation to inhibit mTORC1 activation (51)(Figure 1). Xin Gu et, al found that SAMTOR, previously named C7orf60, interacts with GATOR1 and KICSTOR through searching the BioPlex database and co-immunoprecipitation assays(78). Methyl donor S-adenosylmethionine (SAM) can directly bind to GATOR1 and then disrupt the SAMTOR-GATOR1 at the constant of approximately 7 μM in media (78). SAMTOR senses SAM to signal methionine sufficiency to mTORC1, while methionine may translate into SAM to activate mTORC1. Thus, SAMTOR acts as a SAM sensor in the cytoplasm (78).
SLC38A9, an uncharacterized protein with sequence homology to amino acid transporters, functions as a positive regulator of mTORC1 in an amino acid-sensitive manner (79, 80) (Figure 1). SLC38A9 has been characterized as a lysosomal transmembrane protein that interacts with GTPases and Ragulator which can regulate mTORC1 activation (12). SLC38A9 binds to arginine with a high Michaelis constant (81). Knockout of SLC38A9 inhibits mTORC1 activation by arginine, and overexpression of SLC38A9 makes mTORC1 activation more sensitive by arginine. Thus, SLC38A9 is an excellent candidate for being an arginine sensor in the lysosome for activating mTORC1(82).
Amino acids act as protein building blocks, precursor substrates for signaling molecule, and crucial nutrient signals to mTORC1 activation. In the past few years, our understanding of amino acid sensing to mTORC1 has increased tremendously(8, 28, 29, 33, 51). Of note, mTORC1 acts as a critical regulatory node that controls cell metabolism, including cell proliferation, cycle and death. Dysfunction of mTORC1 is highly associated with many diseases such as insulin resistance(83-85), diabetes(86) and various types of cancer(87-89). the discovery of Rag proteins is a breakthrough in understanding of mTORC1 activation by nutrients, and it helps researchers identify other key components in the mTORC1 signaling pathway, especially amino acid sensors. Recently, Sestrin2, CASTOR1, SMATOR and SLC39A1 have been identified as amino acid sensors. However, sensors for other amino acid such as serine and glutamine remain to be identified. Overall, the molecular mechanisms of amino acid sensing to mTORC1 is quite complex, and more regulatory components of mTORC1 require further investigation.
This article does not contain any studies with human participants or animals performed by any of the authors. Hence, no informed consent is required for any part of this review. Authors declare no conflict of interest. This study was supported by the China Postdoctoral Science Foundation (2017M612562), the Key Research Project of Frontier Sciences of Chinese Academy of Sciences (QYZDY-SSW-SMC008), the Hunan Science and Technology Project (2017XK2020), the Xiaoxiang Scholar Distinguished Professor Fund of Hunan Normal University, the National Thousand Young Talents Program and the Hunan Hundred Talents Program.