- Academic Editor
†These authors contributed equally.
Both classic epigenetic modifications and microRNAs can impact a range of bodily processes, from metabolism to brain function, and may contribute to the development of diseases such as cancer, cardiovascular disorders, and psychiatric disorders. Numerous studies suggest a connection between epigenetic changes and mood disorders. In this study, we performed a comprehensive search using PubMed and Google for the terms “epigenetics”, “ageing”, “miRNA”, “schizophrenia”, and “mood disorders” in the titles and abstracts of articles. Epigenetic changes during early life may play a crucial role in triggering severe mental disorders and shaping their clinical trajectory. Although these alterations can take place at any age, their impact may not be immediately evident or observable until later in life. Epigenetic modifications play a crucial role in the ageing process and challenge the prevailing belief that mutations are the primary driver of ageing. However, it is plausible that these epigenetic changes are a consequence of the disorder rather than its root cause. Moreover, both the disorder and the epigenetic alterations may be influenced by shared environmental or genetic factors. In the near future, we might be able to replace chronological age with biological age, based on the epigenetic clock, with the promise of providing greater therapeutic benefits. A wide range of epigenetic drugs are currently under development at various stages. Although their full effectiveness is yet to be realized, they show great potential in the treatment of cancer, psychiatric disorders, and other complex diseases.
The field of medical genetics has made remarkable advancements, mainly in terms of diagnoses, and more recently in uncovering innovative treatment possibilities, particularly in relation to cancer [1]. Currently, medical genetics encompasses aspects of both Mendelian genetics and epigenetics, acknowledging the importance of both in the onset of genetic disorders. Simultaneously, epigenetics provides valuable understanding into how environmental factors influence gene expression and contribute to the development of diseases. Indeed, epigenetic modifications play a pivotal role in modulating gene expression in response to environmental signals, all while preserving the DNA sequence intact [2].
Throughout an individual’s developmental journey, epigenetic alterations continue to arise as cells divide and specialize. Epigenetic alterations can occur from embryonic stages all the way through adulthood. In adulthood, these changes result from the ongoing interplay between genetic factors and the surrounding environment. Dietary habits, exposure to toxins, stress, and other lifestyle elements can all influence the epigenetic regulation of genes [3].
Epigenetic changes in gene expression are not caused by changes in the DNA sequence itself, but rather by modifications to the chromatin. Classic epigenetic modifications include DNA methylation (DNAm), histone acetylation, deacetylation, and phosphorylation, which can either increase or decrease gene expression by controlling the accessibility of the DNA to the transcription machinery [4]. Recently, non-coding RNAs (ncRNAs), such as microRNAs (miRNAs), have been shown to modulate and shape the complexities of our genomic blueprint beyond the simple DNA sequence. Their primary role involves the post-transcriptional control of gene expression by binding to complementary sequences in messenger RNAs (mRNAs) and interfering with the translation machinery [5]. Both classic epigenetic modifications and miRNAs can impact a range of bodily processes, from metabolism to brain function, and may contribute to the development of diseases such as cancer, cardiovascular disorders, and neurological conditions [6].
In this study, we performed a comprehensive search using PubMed and Google for the terms “epigenetics”, “ageing”, “miRNA”, “schizophrenia”, and “mood disorders” in the titles and abstracts of recently published articles. After applying inclusion criteria, a total of 75 articles, both original contributions and reviews, were deemed suitable for further analysis. We adhered to a wide criteria for article selection, focusing exclusively on English-language, original research articles to ensure clarity and methodological stringency. Titles and abstracts were first screened for direct relevance to specific topics. Shortlisted articles then underwent a quality check for appropriate experimental design and statistical rigor and studies that were transparent and reproducible were prioritized. High-impact and frequently cited articles received additional consideration, although this was not a requirement for inclusion. Finally, ethical compliance, such as informed consent or ethical animal research approval, was a non-negotiable criterion. This approach aimed to ensure the credibility, coherence, and ethical integrity of our review.
Data extraction and analysis were then conducted on the relevant articles. Articles written in languages other than English, off-topic studies, duplicates, and those that were inaccessible were excluded. The search was performed on July 9th, 2023, and a total of 52 studies met the quality criteria, listed in detail below.
To maintain a stringent focus on uncovering new insights, this review was intentionally limited mostly to original research articles. By doing so, the aim was to draw upon primary, empirical data rather than synthesized or secondary interpretations commonly found in review articles.
The preliminary assessment of each article’s appropriateness for inclusion was made based on a thorough review of its title and abstract. This scrutiny ensured that only articles explicitly focusing on topics that we wanted to approach in each section of the review were considered. Articles were required to have titles and abstracts that clearly indicated a direct relevance to these specific topics, thereby serving as an initial filter to maintain the thematic coherence and specialized focus of the review.
To guarantee the reliability of the findings discussed in this review, stringent methodological criteria were established for article inclusion. Articles were required to have well-designed experimental or observational protocols, clearly defined variables, and appropriate statistical analyses. Any study with evident methodological flaws, such as a lack of control groups or inadequate sample sizes, was excluded.
Articles were preferred if they provided sufficient detail to allow for the reproduction of experiments or analyses. Transparency in reporting results, including the provision of raw data or supplementary materials, was considered a mark of high quality and contributed to an article’s eligibility for inclusion.
While not a strict requirement, articles published in journals with high impact factors and those that were frequently cited were given preferential consideration. These metrics were seen as indicators of an article’s influence and the scientific community’s validation of its quality.
Ethical considerations were also integral to the article selection process. Studies were expected to comply with recognized ethical guidelines, such as obtaining informed consent from human subjects or ethical approval for animal research practices, as applicable. This ethical criterion was enforced to ensure the credibility and ethical soundness of the information presented.
The interplay between epigenetics and metabolism in ageing stem cells has been shown to have a significant impact on cellular senescence and the regulation of gene expression. Epigenetic modifications, primarily examined through cytosine guanine islands (CpG) dinucleotide methylation, play a crucial role in the ageing process of stem cells, as highlighted in a study by Brunet & Rando [7]. This challenges the prevailing belief that mutations are the primary driver of ageing. Surprisingly, seemingly healthy mammals can be cloned from aged somatic cells, and certain mice species or individuals with higher mutation rates show minimal signs of premature ageing [8, 9, 10]. Instead, research on lower organisms suggests a potential link between ageing and a loss of epigenomic information [11, 12]. Recent studies have further confirmed this hypothesis in eukaryotic cells and mice [13].
In a recently developed mouse model designed to accelerated ageing, researchers successfully demonstrated the potential for rejuvenating the cells and organs of a living animal [14]. As a result of this treatment, the treated mice experienced a notable 50% increase in their average lifespan compared with untreated mice with accelerated ageing. However, it is worth mentioning that not all ageing indicators were impacted by the partial cellular reprogramming, and if the treatment was discontinued, the ageing signs reappeared. Of particular significance, the periodic partial cellular reprogramming led to a resetting of certain epigenetic ageing markers. The age-related decline in H3K9 trimethylation (H3K9me3) can also be counteracted by cyclically inducing the Yamanaka factors. These factors encompass a combination of transcription factors, such as Oct4, Sox2, Klf4, and c-Myc, which are typically utilized for cell reprogramming [15].
In a recent study, Rodríguez-Matellán and colleagues [15] made significant strides in our comprehension of partial cellular reprogramming, shedding light on its potential clinical applications. The research demonstrated a favorable impact on memory in aged mice utilizing this process. Similar to earlier investigations, this study underscored the successful achievement of a delicate equilibrium between epigenetic rejuvenation and the resetting of ageing processes. Importantly, this reprogramming approach preserves the cells’ identities, safeguarding their distinct characteristics [15].
Furthermore, the study delved into the consequences of the ectopic expression of a combination of neural transcription factors, specifically the Oct4, Sox2, and Klf4 genes (referred to as the OSK mix), in mouse retinal ganglion cells. The findings showcased the restoration of youthful DNAm patterns and transcriptomes, the stimulation of axon regeneration following injury, and the reversal of vision loss in both a mouse model of glaucoma and aged mice [16].
As organisms undergo the ageing process, certain epigenetic markers undergo modifications [17]. DNAm-based epigenetic clocks are extremely accurate molecular indicators of age, suitable for use in diverse tissues and cell types. A meta-analysis performed on the blood of 13,089 individuals provided compelling evidence that epigenetic age predicts all-cause mortality including overall lifespan [18].
One theory proposes that the ageing process in eukaryotes may be linked to the degradation of epigenetic information and transcriptional networks caused by cellular damage. As individuals grow older, there is a decrease in the levels of Histone-3 Trimethylation at Lysine 9 (H3K9me3) [19]. Consequently, a study proposed that decreasing the activity of the methyltransferase responsible for modifying H3 leads to a reduction in H3K9me3 levels, thereby initiating the cellular ageing process [20]. Indeed, experimental evidence shows that the temporary expression of nuclear reprogramming factors in fibroblasts, endothelial cells, and muscle tissue obtained from aged mice and humans can restore H3K9me3 levels. This restoration, facilitated by mRNA expression, leads to a rapid and widespread improvement in cellular ageing. It entails a resetting of the epigenetic clock, a reduction in the inflammatory profile in chondrocytes, and the revival of youthful regenerative responses in aged mouse and human muscle stem cells, all without altering cellular identity [21].
A groundbreaking approach called the “maturation phase transient reprogramming” (MPTR) method was developed, which involves selectively expressing reprogramming factors until a specific point of rejuvenation is reached, after which they are withdrawn. When applied to dermal fibroblasts from middle-aged donors, treated with transcription factors Oct4, Sox2, Klf4, and c-Myc, the study found that the cells underwent a temporary loss and subsequent reacquisition of their fibroblast identity. Moreover, this process resulted in a partial restoration of fibroblast function, potentially due to epigenetic modifications. Notably, the transcriptome appeared to be rejuvenated by approximately 30 years, including changes in H3K9me3 levels and alterations in the DNAm ageing clock [22].
Various mood disorders, such as depression, bipolar disorder (BD), and anxiety disorders, have been associated with epigenetic changes. The development of anxiety disorders, like other psychiatric conditions, is influenced by multiple factors, including an individual’s genetic makeup and their environment. Neuroepigenetic research, focused on identifying specific patterns of epigenetic modifications affecting genes and associated protein products crucial in maintaining anxiety disorders, holds the potential to unveil novel therapeutic possibilities [23].
Depression: Studies have revealed that individuals experiencing depression often exhibit modified DNAm patterns in their brain cells, particularly in genes associated with the stress response and neurotransmission. For instance, people with depression have shown increased methylation levels of Nuclear Receptor Subfamily 3 Group C Member 1 (NR3C1) (exon 1 F) and Solute Carrier Family 6 Member 4 (SLC6A4), CpG islands, leading to reduced gene expression and dysregulation of the hypothalamic-pituitary-adrenal axis stress response. This may make these individuals more susceptible to the impact of stress, which is a significant risk factor for depression [24].
Bipolar Disorder: Likewise, individuals diagnosed with BD have shown altered epigenetic patterns in several genes linked to neurotransmission and circadian rhythms. In a specific study, it was observed that individuals with BD displayed heightened methylation in the gene encoding the dopamine receptor D2 when compared with control subjects. Such alterations could potentially influence dopamine signaling and the regulation of mood [25].
Anxiety Disorders: Epigenetic modifications have been linked to anxiety disorders as well. For example, research has revealed altered DNAm patterns in the genes responsible for the glucocorticoid receptor and brain-derived neurotrophic factor (BDNF) in individuals diagnosed with post-traumatic stress disorder (PTSD) and panic disorder. These genes play significant roles in the stress response and neuronal plasticity, suggesting that they could potentially influence the development and severity of anxiety disorders [26, 27].
It is crucial to recognize that while these studies suggest a connection between epigenetic changes and mood disorders, they do not establish a direct cause-and-effect relationship. It is plausible that these epigenetic changes are a consequence of the disorder rather than its root cause. Moreover, both the disorder and the epigenetic alterations may be influenced by shared environmental or genetic factors. Nonetheless, these findings open up potential avenues for treatment and prevention strategies and offer valuable insights into the biological underpinnings of these disorders.
The regulation of emotional responses in varying environmental conditions plays a vital role in maintaining mental well-being. One fundamental way of regulating emotional responses is through extinction, where the conditioned response to a stimulus weakens when the reinforcement (stimulus) is removed. Studies have indicated the significant involvement of the endocannabinoid (eCB) and glucocorticoid systems in modulating emotional states and facilitating the extinction of aversive memories in animal models [28].
Newly identified breakthroughs have shed light on the epigenetic modifications taking place within the glucocorticoid system, revealing its capacity to impact susceptibility to post-traumatic stress and anxiety disorders. These revelations suggest a potential association with future psychiatric conditions [29]. Consequently, there is considerable interest in investigating the memory-altering effects of cannabinoids and glucocorticoids, aiming to explore their potential applications in translational research [30].
Anxiety is marked by alterations in the activity of different DNA
methyltransferases and histone acetylation, which affect how the
hypothalamic-pituitary-adrenal axis functions and how the body responds to
stress. Additionally, these changes impact the signaling pathways of
Clinical studies conducted on individuals with post-traumatic stress disorder [31] and research involving animal models of acquired fear have uncovered two significant effects of glucocorticoid (GC) treatment. Firstly, it reduces the recall of distressing memories and, secondly, it enhances the process of extinction [30]. These combined actions of GCs make them particularly promising for addressing acquired fear. By hindering memory retrieval, GCs may alleviate symptoms associated with distressing memories. Furthermore, by promoting the consolidation of extinction memory, GCs may assist in retaining experiences linked to less fear. As a result, incorporating GCs into extinction-based psychotherapy along with exposure techniques shows potential as an effective approach. Indeed, initial evidence indicates that the combination of cortisol with exposure therapy has led to improved treatment outcomes in patients with phobic fear [32]. Lastly, recent research suggests that stress and GCs’ impact on memory is influenced by epigenetic changes in the gene promoter region of the glucocorticoid receptor (GR) gene [33]. These discoveries may contribute to a better understanding of why some individuals are more susceptible to the effects of stress compared with others [34].
Schizophrenia is a multifaceted and intricate condition believed to arise from the interaction of genetic and environmental elements. Epigenetic changes, such as DNAm and histone modification, represent ways through which environmental influences can modify gene function without modifying the DNA sequence itself. These alterations can affect diverse biological processes, including neuronal development and function, which have been associated with schizophrenia [35]. These alterations involve three main mechanisms:
(1) DNA Methylation: Within the brains of individuals with schizophrenia, certain genes linked to dopamine function (a crucial neurotransmitter implicated in the disorder) and brain development exhibit changes in their methylation status. Such alterations could influence the expression of these genes, potentially contributing to the pathogenesis of schizophrenia [36].
(2) Histone Modification: Histones are proteins that assist in compactly organizing DNA within the cell nucleus. Modifications to these histones can affect how accessible the DNA is to the transcription machinery, thereby influencing gene expression. Studies have identified changes in histone modification in schizophrenia, which could potentially impact the expression of genes relevant to the disorder [37].
(3) Non-coding RNAs: miRNAs and other ncRNAs play a role in regulating gene expression post-transcriptionally. Some research suggests that the levels of specific miRNAs within the brains of individuals with schizophrenia are altered, indicating the potential involvement of this epigenetic mechanism in the disorder [37].
In recent years, significant advancements have been made in the field of
epidemiological and molecular genetic investigations concerning the interplay of
genetic and environmental factors in schizophrenia. A handful of studies have
delved into interactions between candidate genes and environmental factors at a
molecular level (known as G
Epigenetic modifications possess the potential to shape the impact of environmental factors known to increase the risk of schizophrenia. For instance, exposure to infections, malnutrition, and prenatal stress have been associated with a higher likelihood of developing schizophrenia, potentially due to epigenetic changes. It is crucial to highlight that our comprehension of the role of epigenetics in schizophrenia is continuously evolving. While these findings suggest a potential connection to epigenetic alterations, further research is indispensable to fully grasp their contribution. It is important to acknowledge that not all changes are necessarily causative; some may result from the disorder itself or be influenced by related factors, such as medication use or lifestyle choices. Lastly, the intricate interplay between genetics, epigenetics, and the environment likely involves numerous genes and pathways [41].
BD has also been associated with a decreased lifespan, a phenomenon that scientists attribute to increased inflammation, oxidative stress, shortened telomeres, and metabolic disturbances [42, 43, 44]. These indicators suggest that individuals diagnosed with these conditions may experience accelerated ageing effects. Methylation patterns typically alter throughout one’s lifetime; however, external factors can influence their rate of change, possibly becoming significant risk factors for the development of these disorders. Consequently, young individuals undergoing migration, those experiencing lower socio-economic statuses, and those residing in urban environments—conditions often associated with social exclusion and isolation—are also at an increased risk of developing schizophrenia [45, 46, 47].
Epigenetic changes during early life may play a crucial role in triggering severe mental disorders and shaping their clinical trajectory. Although these alterations can take place at any age, their impact may not be immediately evident or observable until later in life. This delay is attributed to the cumulative nature of numerous epigenetic changes, which accumulate gradually and only result in noticeable effects on function or health once they surpass a specific threshold. Moreover, the expression of these changes can be influenced by various factors, including an individual’s genetic composition, overall well-being, and additional environmental influences.
BD has been previously associated with signs of accelerated aging, including rapid epigenetic aging in both blood and brain tissues. Additionally, there’s a faster decline in cognitive function linked to the progression of age and premature mortality in BD patients [48]. A recent study examined accelerated epigenetic aging in people with BD by employing a combination of five epigenetic clocks (Horvath, Hannum, EN, PhenoAge, and GrimAge), as well as a DNAm-based telomere length clock. This investigation used complete blood samples from 184 BD individuals. Additionally, adjustments were made for blood cell counts using DNAm levels. The study revealed significant associations between individual epigenetic clocks and various factors influencing the aging mechanism, including body mass index, sex, smoking habits, and experiences of childhood abuse [49]. Using a comparable methodological strategy, a study involving blood samples from individuals with BD who were on a regimen of mood stabilizers has presented fresh proof indicating a slowdown in epigenetic aging among BD patients [50].
Recent research has demonstrated a correlation between childhood adversities and an accelerated pace of biological aging. In a study involving 82 adults diagnosed with major depression, an investigation was conducted into how two types of childhood adversity—maltreatment and household dysfunction—influence the process of epigenetic aging. Using two distinct clocks, GrimAge and PhenoAge, the study revealed that maltreatment is linked to a heightened rate of aging according to the PhenoAge clock. In contrast, no such association was observed with the GrimAge clock. Conversely, household dysfunction appears to be connected to a decelerated aging process, especially when accounting for the presence of maltreatment [51].
Recently, Kebir and colleagues conducted a study focusing on methylomic associations to explore blood methylation biomarkers connected to the transition to psychosis in a cohort of young individuals seeking help, who were part of a longitudinal follow-up. Their findings revealed that the conversion to psychosis did not result in a comprehensive alteration of methylation, and no individual CpG sites showed significant associations with the transition to psychosis, which is consistent with previous research findings. However, they did discover that the conversion to psychosis was linked to specific types of methylation changes in genes associated with axon guidance, as well as genes belonging to the IL-17 pathway and the glutathione-S-transferase family [52].
The impact of stressors on the development of schizophrenia is crucial, as they can modify epigenetic patterns, which might serve as indicative markers of exposure to environmental risk factors. In a recent investigation, researchers assessed age-related epigenetic modifications in young individuals with familial high risk (FHR) for these disorders, comparing them with a control group, and explored their association with environmental stressors. Methylation data from blood and saliva samples were used to estimate epigenetic age using six different epigenetic clocks, which was then compared with chronological age. The study revealed notable disparities in epigenetic age among the high-risk youth, suggesting that the offspring of affected parents might experience a slower biological ageing process compared with the control group. Nonetheless, the precise role of environmental stressors in orchestrating changes in methylation patterns remains unclear [53].
Another investigation examined whole-genome DNAm profiling in cases of psychotic
BD, non-psychotic BD, and matched controls. This study found a connection between
the loss or reversal of brain laterality in schizophrenia and BD, a consistently
observed phenomenon in these disorders, and aberrant epigenetic regulation of
Transforming Growth Factor Beta 2 (TGFB2), along with changes in Transforming Growth Factor Beta 2 (TGF
In 2017, Viana and colleagues [53] investigated epigenetic modifications associated with schizophrenia in multiple brain regions. Their study demonstrated the effectiveness of polygenic risk scores in identifying molecular pathways related to the diverse origins of complex diseases. The researchers examined the DNAm patterns across the entire genome in postmortem brain samples from two distinct groups: individuals with schizophrenia and control subjects. These samples were gathered from four brain regions: the prefrontal cortex, striatum, hippocampus, and cerebellum. The findings of their study revealed that schizophrenia did not show widespread differences in methylation patterns across the probes included in the Illumina 450K array within the tested brain regions. Nevertheless, they did find significant evidence of schizophrenia-associated variation at specific locations throughout the genome in each brain region. In a different study, microarray data from the Illumina HumanMethylation450 platform was used to investigate the epigenetic age of individuals with schizophrenia, BD, and healthy controls. This investigation employed five different epigenetic clocks and various statistical metrics to detect differences between epigenetic and chronological age among the three groups. The study revealed a noteworthy increase in epigenetic age compared with chronological age within the BD group. Additionally, individuals with BD exhibited a higher mean epigenetic age acceleration compared with healthy controls across four of the five epigenetic clocks. Despite the study’s relatively limited sample size, these findings imply that both BD and schizophrenia may be linked to epigenetic markers associated with a premature ageing phenotype. Such markers could potentially indicate negative outcomes linked to these conditions [55, 56].
Individuals with schizophrenia exhibit signs of accelerated aging in both cognitive and physiological domains. To investigate this, a comprehensive study combined brain magnetic resonance imaging (MRI) scans with the use of two epigenetic clocks, which were developed based on blood tests from both healthy individuals and those with schizophrenia. The aim was to determine a potential correlation with the polygenic risk score for schizophrenia. Notably, the results highlighted a discrepancy between the aging mechanisms observed in the brain and the chronological age as determined from blood analysis [57].
Another study examined the connection between various epigenetic clocks and the recurrence of schizophrenia over a span of 3 years in a group of 91 individuals experiencing their first episode of the disorder. Through a comprehensive analysis of DNAm across the genome, four different clocks were evaluated, encompassing measures such as chronological age, mortality prediction, and telomere length. Individuals who experienced a relapse displayed notable indications of expedited aging in terms of telomere length. Although shorter telomere lengths were associated with cognitive performance, they did not exhibit a direct correlation with relapse based on cognitive function or the severity of symptoms. These findings suggest that the acceleration of epigenetic aging might play a role in the clinical progression of schizophrenia and could potentially serve as an indicator of relapse during periods of remission [58].
Extracellular vesicles (EVs) have become significant focal points in brain disorder investigation due to their role in establishing a connection between the brain and systemic circulation. For example, miRNAs enclosed within EVs have been linked to the development of conditions such as glioblastoma and Alzheimer’s disease. These miRNAs have the potential to serve as biomarkers for the early detection of these diseases, as they can be identified in peripheral fluids [59, 60].
miR-134, a type of miRNA, is linked to the processes of neurogenesis and the adaptability of synapses. A recent study explored the potential of using miR-134 as a biomarker for major depressive disorder (MDD). This was investigated by measuring the levels of miR-134 in plasma samples from rat models of depression and unpredictable mild stress. The investigation was then extended to include a group of 100 MDD patients, 50 individuals with BD, 50 with schizophrenia, and 100 healthy subjects. The analysis revealed that plasma miR-134 concentrations were significantly lower in individuals with MDD. This distinction enabled the differentiation between MDD patients and healthy controls, as well as those with BD and schizophrenia, with a high degree of accuracy. Results from the rat model further corroborated the observed decrease in miR-134 levels following stress exposure. Although the sample size was relatively limited, these findings strongly suggest the potential of miR-134 as a biomarker for MDD [61].
In a recent study, Honorato-Mauer and colleagues presented a miRNA profile extracted from serum EVs of adolescents diagnosed with mental health disorders, such as MDD, anxiety, and attention deficit hyperactivity disorder (ADHD). In the initial phase of the study, miR-328-3p displayed elevated expression in subjects with ADHD as compared with control subjects without the disorder. In the follow-up study, miR-432-5p, miR-151a-5p, and miR-584-5p demonstrated reduced expression in individuals with anxiety relative to control subjects without the disorder. Similarly, miR-4433b-5p, miR-584-5p, miR-625-3p, miR-432-5p, and miR-409-3p were observed to have decreased expression in MDD subjects when contrasted with control subjects without the disorder [62].
Although their specific roles in depression remain uncertain, ncRNAs have displayed promising potential as biomarkers for diagnosing and treating patients with depression [63].
In the near future, we might be able to replace chronological age with biological age, based on the epigenetic clock. Additionally, we could potentially predict the odds of acquiring a psychiatric disease as one ages (Fig. 1).
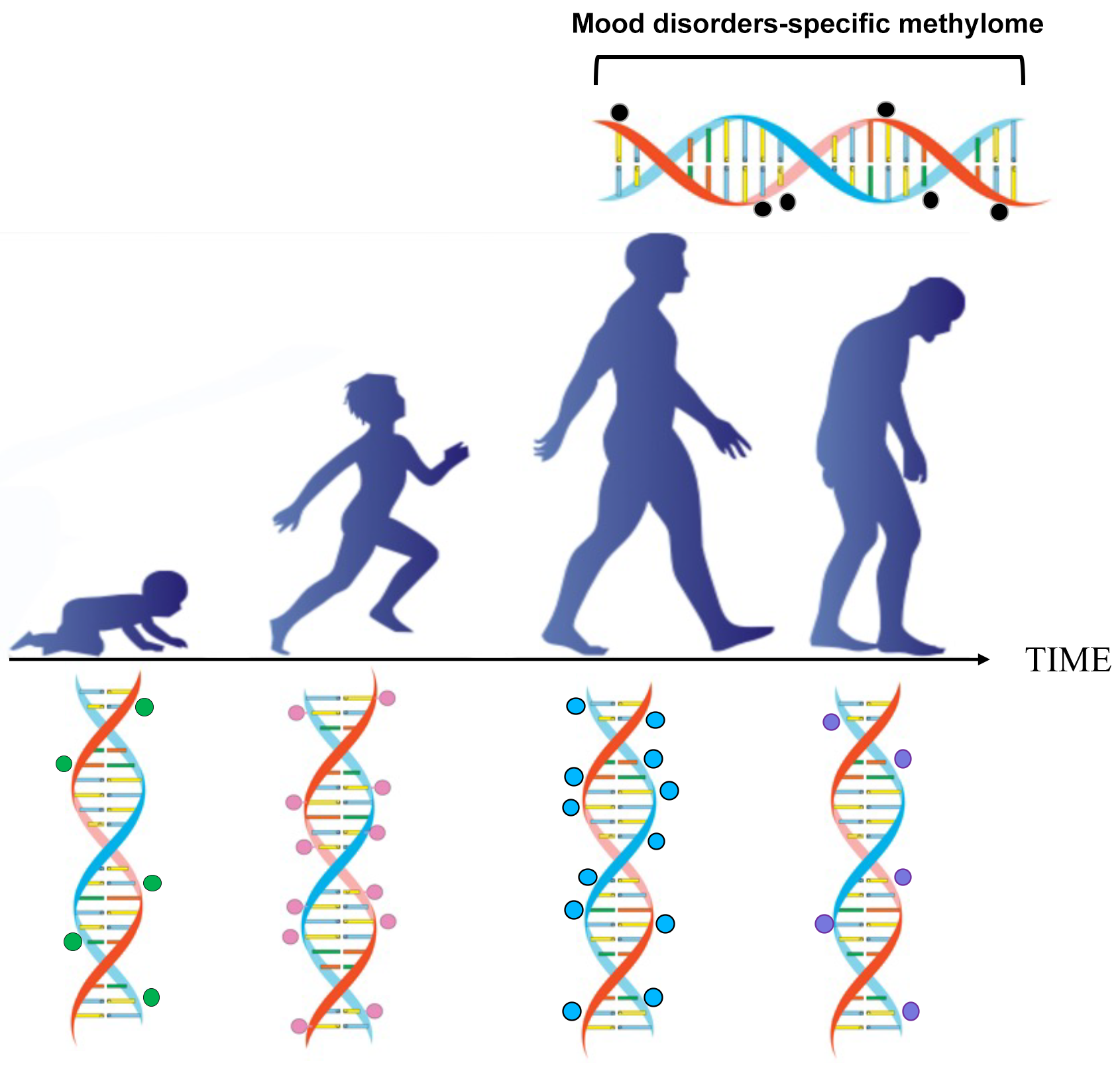 Fig. 1.
Fig. 1.Epigenetic changes occur during postnatal development, adulthood, and ageing. The methylation of cytosine-guanine islands (CpG) by DNA methyltransferase is a covalent modification of DNA, which typically results in gene inactivation. The methylation and demethylation of CpG islands are modulated during development and ageing. This process may malfunction in conditions such as ageing, schizophrenia, and mood disorders.
From a therapeutic perspective, epigenetic drugs offer a new and potentially more effective approach compared with current symptomatic treatments and hold the promise of providing greater therapeutic benefits while reducing the likelihood of adverse effects. A wide range of epigenetic drugs are currently under development at various stages. Although their full effectiveness is yet to be realized, they show great potential in the treatment of cancer, psychiatric disorders, and other complex diseases.
Another important consideration is whether targeting the persistent epigenetic mechanisms, which are responsible for lifelong changes in addiction vulnerability could open up new possibilities for medication development. As mentioned before, there are promising findings from studies using histone deacetylase and DNA methyltransferase (DNMT) inhibitors that target chromatin remodeling in animal models. The ability of DNMT inhibitors to reverse epimutations underscores their utilization in innovative approaches for cancer therapy.
Currently, ongoing clinical trials are exploring the implementation of combination therapy, employing DNMT inhibitors alongside hypomethylation agents and other chemotherapeutic drugs.
CIC, IU, DG, DLA, APW had substantial contributions to conception and drafted the manuscript. DMH, DC, and EC searched for relevant literature. APW contributed Fig. 1. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
We acknowledge the generous contribution of Dr. Roxana Surugiu.
This work was supported by National Recovery and Resilience Plan (PNRR) (Project No 760058 /11.05.2023).
The authors declare no conflict of interest. Aurel Popa-Wagner is serving as one of the Guest editors of this journal. We declare that Aurel Popa-Wagner had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Gernot Riedel.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
