- Academic Editor
†These authors contributed equally.
Background: The Fazekas scale is one of the most commonly used visual
grading systems for white matter hyperintensity (WMH) for brain disorders like
dementia from T2-fluid attenuated inversion recovery magnetic resonance (MR)
images (T2-FLAIRs). However, the visual grading of the Fazekas scale suffers from
low-intra and inter-rater reliability and high labor-intensive work. Therefore,
we developed a fully automated visual grading system using quantifiable
measurements. Methods: Our approach involves four stages: (1) the deep
learning-based segmentation of ventricles and WMH lesions, (2) the categorization
into periventricular white matter hyperintensity (PWMH) and deep white matter
hyperintensity (DWMH), (3) the WMH diameter measurement, and (4) automated
scoring, following the quantifiable method modified for Fazekas grading. We
compared the performances of our method and that of the modified Fazekas scale
graded by three neuroradiologists for 404 subjects with T2-FLAIR utilized from a
clinical site in Korea. Results: The Krippendorff’s alpha across our
method and raters (A) versus those only between the radiologists (R) were
comparable, showing substantial (0.694 vs. 0.732; 0.658 vs. 0.671) and moderate
(0.579 vs. 0.586) level of agreements for the modified Fazekas, the DWMH, and the
PWMH scales, respectively. Also, the average of areas under the receiver
operating characteristic curve between the radiologists (0.80
T2-weighted fluid-attenuated inversion recovery magnetic resonance imaging (T2-FLAIRs) is used to assess the severity of white matter lesions that appeared as hyperintensities (WMHs) in vivo. WMH provides important information about brain health, aging, and possible disease burden [1, 2, 3, 4]. WMH has been recognized as an important biomarker for small-vessel cerebrovascular diseases and Alzheimer’s disease [5, 6].
The Fazekas scale provides a conventional visual grading approach to quantify WMH severity into four scales and is often practiced by radiologists and in clinics worldwide [7]. The Fazekas scale classifies the severity of WMHs presented in the T2-FLAIR using the combination of the periventricular hyperintensity (PWMH) scale and the deep white matter hyperintensity (DWMH) scale [7]. Both PWMHs and DWMHs are graded from zero to three (Table 1) [7].
| The original Fazekas scale | The modified Fazekas scale | |
| Grade 0 | Absent | Absent |
| Grade 1 | PWMH: caps or pencil-thin lining | PWMH |
| DWMH: punctuate foci | ||
| Grade 2 | PWMH: smooth halo | 1. DWMH |
| DWMH: beginning confluence | OR | |
| 2. 10 mm | ||
| OR | ||
| 3. DWMH | ||
| Grade 3 | PWMH: irregular periventricular signal extending into the deep white matter | PWMH |
| DWMH: large confluent areas | ||
| DWMH, deep white matter hyperintensity; PWMH, periventricular hyperintensity. | ||
However, the use of the Fazekas scale in clinical practice or research is often limited by its labor-intensive process, as are all forms of visual grading [8], and low inter- and intra-rater reliability due to its ambiguous given criteria [9]. Over time, the age-related white matter changes (ARWMC) scale was introduced to overcome the ambiguousness of the subjectively measured Fazekas scale to provide quantifiable measurements [10]. Yet, the ARWMC scale also had limits due to not providing a detailed separation of DWMH and PWMH lesions. Hence, we had to find an advanced method that is computationally viable to implement for gratifying the original Fazekas scale. Several groups suggested a quantifiable method using the maximum diameter distance to divide DWMH and PWMH. The DWMH and PWMH scales are defined from the measured distance, which they call the modified Fazekas scale (Table 1) [11].
This study aims to provide an automated approach to the modified Fazekas scale that is efficient and easily applicable with reliable results in general clinical research and practice to assist doctors by reducing their labor-intensive process. Thus, this study shares our implementation and validation on a fully automated modified Fazekas scale using deep learning and a rule-based algorithm. Radiologists participated in this study to validate if our method is comparable to humans since this study is the first automation algorithm for the modified Fazekas scale.
The proposed approach consists of four stages (Fig. 1). First, the ventricle and WMH are segmented from the input 2D T2-FLAIR using a deep learning algorithm [12]. Second, the segmented WMHs are categorized into DWMHs and PWMHs following the rule suggested in the previous study [13]. Third, the maximum diameter is measured for both DWMHs and PWMHs according to the modified Fazekas scale. Finally, the modified Fazekas scale is calculated using the obtained maximum diameter of DWMH and PWMH. For validation, we compared the agreements of our proposed method against those of three certified radiologists.
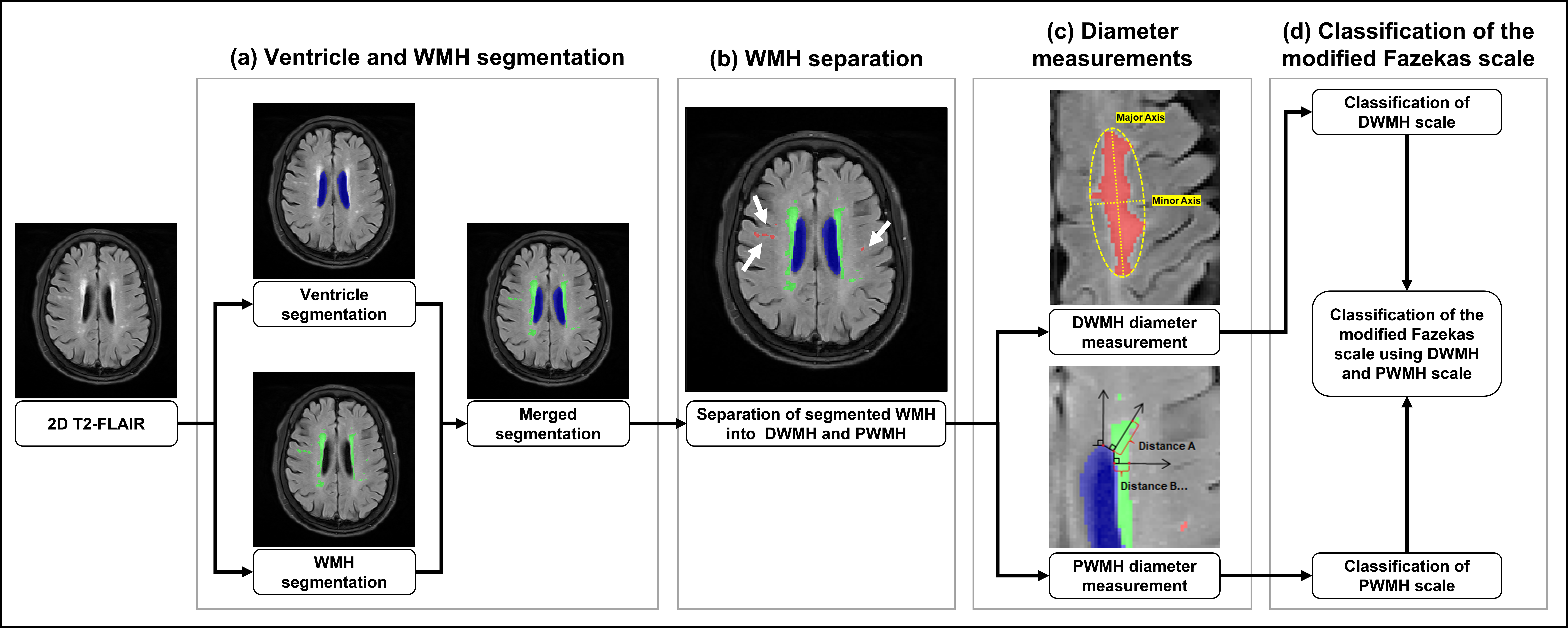 Fig. 1.
Fig. 1.The pipeline of the proposed method. Automated scoring for the Fazekas scale involves four stages and is based on T2-FLAIR MR images. (a) Brain tissue and WMH segmentation. (b) WMH separation. (c) Diameter measurement. (d) Fazekas scale prediction.
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Eunpyeon St. Mary’s Hospital, College of Medicine, The Catholic University of Korea (IRB No. PC20EISI0094 on 02 July 2020).
Two-dimensional (2D) T2-FLAIR scans from the Catholic University of Korea
Eunpyeong St. Mary’s Hospital were used in this study. The dataset was collected
with the inclusion criteria of magnetic resonance imaging (MRI) containing WMH
diagnosed with dementia. The exclusion criteria were WMHs with multiple
pathologies, such as stroke or other disorders that may cause different
components (e.g., cerebrospinal fluid, microbleeds) within the WMHs. The average
age of the 404 participants was 68.7
All images were acquired using a 3T MRI scanner (MAGNETOM Vida, Siemens Medical Solutions Inc., Malvern, PA, USA) with the following parameters: axial, time of echo (TE) = 114 ms,
time of repetition (TR) = 8 s, time of inversion (TI) = 2370 ms, field of view
(FOV) = 21 cm
The modified Fazekas scale is based on measuring the maximum diameter (mm) of DWMH and PWMH, which is quantitative (Table 1). Theoretically, our computationally implemented measuring method would be more accurate than the human raters. Yet, we compared our automated results to the human raters to demonstrate the similarity since the main goal of developing this method is to help out the intense labor of humans. For human raters, each T2-FLAIR images were assessed by three certified radiologists with a subspecialty in neuroradiology. All patient information was blinded to make no bias in rating, and also that mutual information shall not be shared between the raters. The images were visually graded independently by raters following the criteria of the modified Fazekas scale. The raters manually used a MRI measuring tool to measure the diameter (mm) of the longest axis on the PWMH and DWMH. Measurement was done on raw MRI without any provided annotations. Then, radiologists provided the modified Fazekas scale on the basis of the measurement [11]. For our proposed method, we proceed with the automated pipeline shown in the overview of the proposed method (Fig. 1), then provide the modified Fazekas scale.
We used our previously reported in-house method for simultaneous ventricle and WMH segmentation (Fig. 1a) [12]. The publication introduced two individual deep learning-based segmentation methods for T2-FLAIR. This research aimed to produce brain tissues and WMH segmentation using T2-FLAIR without its paired T1-weighted MRI (T1). We utilized the semi-supervised learning method and constructed the deep learning-based segmentation model to train FreeSurfer-generated brain tissue, including the ventricle from T1 to T2-FLAIR [14, 15]. Then, the WMH model was trained with U-Net-based architecture using manually annotated and clinically confirmed WMH labels from radiologists utilizing PyTorch (version 1.7.1, python software foundation, Wilmington, DE, USA) [16, 17]. The previous research datasets are unrelated to our automated approach. The in-house segmentations demonstrated promising results for further clinical relevance and application.
All processed segmentation labels from the models used for this study were set
to right-anterior-superior (RAS) orientation and resampled to 1
We categorized the segmented WMH region further into DWMH and PWMH regions (Fig. 2). The separation was based on the calculated distance between the DWMHs/PWMHs
and the boundaries of the segmented ventricle regions. For the X and Y axes, we
separated PWMHs and DWMHs in 2D slice-based where ventricle segmentation exists
in the axial plane: PWMHs were specified from WMHs within
 Fig. 2.
Fig. 2.DWMH and PWMH separation results in multiple planes. The blue, green, and red labels represent the ventricles, PWMH, and DWMH segmentations, respectively. (a) Axial plane. (b) Sagittal plane. (c) Coronal plane. (d) 3D view from the top.
We measured the diameters of the separated DWMH and PWMH (Fig. 3). The vertical distance was used for DWMHs, and the horizontal distance was used for PWMHs, as suggested in the modified Fazekas scale [11].
 Fig. 3.
Fig. 3.Overview of the WMH separation into DWMH and PWMH. WMH, white matter hyperintensity; DWMH, deep white matter hyperintensity; PWMH, periventricular white matter hyperintensity.
Principal Component Analysis (PCA) based on the euclidean distance was performed on DWMHs in all 2D axial planes to measure the vertical diameter [18]. Taking the irregularly shaped DWMH as an input, the PCA-based measurement generates an approximated ellipse around the DWMH (Fig. 4d). Then, the major and minor axes are suggested for the eclipse. Since the DWMH scale is measured from the maximum diameter, we utilized the distance of the major axis [18].
 Fig. 4.
Fig. 4.Measurement of DWMH performed with PCA method. (a) T2-FLAIR MRI input. (b) DWMH segmentation. (c) PCA method. (d) Calculation of the major and minor axes.
PWMH is measured by measuring the horizontal diameter between the ventricle and the PWMH. Since the horizontal diameter varies from the starting point of the ventricle, we created a 2D Danielsson distance map for all 2D axial slices containing PWMHs and ventricles (Fig. 5) [19]. We extracted the ventricle contour from the distance map. We created perpendicular rays with a length of 13 mm from each pixel coordinate of the ventricle contour, representing the cut-off distance between PWMH and DWMH [13]. For each cluster of PWMH, we measured the mean distance of every ray that intersected the PWMH.
 Fig. 5.
Fig. 5.Measurement of PWMH with four stages. (a) T2-FLAIR MRI input. (b) Combined segmentation results with ventricles and WMHs. (c) Distance map from ventricle segmentation. (d) PWMH measurement using ventricle segmentation and a distance map.
At this stage (Fig. 1d), we finalized the automation process by classifying the
modified Fazekas scale. Using the measured maximum diameters of the DWMHs and
PWMHs, we assigned scales ranging from 1 to 3 (Table 1) as suggested by the
modified Fazekas scale [11]. For the PWMHs, 1 represented maximum diameters
We investigated the agreements of the modified Fazekas scale from our proposed method and the experts with different years of experience. The multiple-rater agreement was assessed using Krippendorff’s alpha [20]. Krippendorff’s alpha was utilized to provide the level of agreement between the visual gradings performed by the radiologists and our proposed method. The inter-rater agreement was assessed using the areas under the receiver operating characteristic curves (AUROCs) [21] for the proposed method and each radiologist assessment. The AUROC was utilized to present the correspondence between our proposed method and the radiologists. The AUROC was used to determine the decision threshold for the classification performance of the two raters related to the true-positive rate (TPR) and false-positive rate (FPR) within the range of 0 to 1. Higher AUROCs are associated with higher performance than the gold standard [21]. All the performance evaluation was conducted either using R package software version 3.4.3 (The R Foundation for Statistical Computing, Vienna, Austria) or Python version 3.7 (Python Software Foundation) with the scikit-learn library [22, 23, 24].
To investigate the level of agreement between the different ratings, we assessed
the multiple-rater agreement using Krippendorff’s alpha (
| Multiple-rater agreements ( | ||
| ROI | (R) without proposed method | (A) with proposed method |
| R1, R2, and R3 | R1, R2, R3, and P | |
| The modified Fazekas scale | 0.732* | 0.694* |
| DWMH scale | 0.671* | 0.658 |
| PWMH scale | 0.586 | 0.579 |
| ROI, region of interest; DWMH, deep white matter hyperintensity; PWMH,
periventricular hyperintensity; P, proposed method; R1/2/3, raters 1, 2, and 3;
*, Krippendorff’s alpha ( | ||
We determined the performance agreement using AUROCs. The agreements of the
modified Fazekas scales determined by the radiologists and the proposed method
are summarized in Table 3: G shows the evaluations by the radiologists (R1 vs.
R2, R1 vs. R3, and R2 vs. R3), and M shows the evaluations by the raters and the
proposed method (R1 vs. P, R2 vs. P, R3 vs. P). The interpretations of the area
under the curve (AUROC) coefficients are as follows: 0.5, no discrimination; 0.6
to 0.7, poor discrimination; 0.7 to 0.8, acceptable discrimination; 0.8 to 0.9,
excellent discrimination; 0.9 to 1.0, outstanding discrimination [26]. The
average AUROC scores for the modified Fazekas scale determined by the
radiologists showed excellent discrimination (G 0.87
| (G) between radiologists | (M) against our proposed method | ||||||
| Modified Fazekas scale | Modified Fazekas scale | ||||||
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| R1 vs. R2 | 0.89 | 0.85 | 0.63 | R1 vs. P | 0.81 | 0.74 | 0.71 |
| R1 vs. R3 | 0.80 | 0.74 | 0.81 | R2 vs. P | 0.79 | 0.75 | 0.89 |
| R2 vs. R3 | 0.93 | 0.91 | 0.64 | R3 vs. P | 0.88 | 0.83 | 0.78 |
| Average | 0.87 |
0.83 |
0.70 |
Average | 0.83 |
0.77 |
0.79 |
| Combined | 0.80 |
Combined | 0.80 | ||||
| The inter-rater agreements of raters only (G, left part of the table) and raters vs. proposed method (M, right part of the table); R1/2/3, raters 1, 2, and 3; P, proposed method. | |||||||
In this study, we demonstrated a fully automated visual grading system for WMH using the modified Fazekas scale on T2-FLAIRs. Our approach aimed to automate the visual grading of the modified Fazekas scale utilizing deep learning and rule-based algorithms with quantifiable imaging-driven measurements using T2-FLAIR exclusively. This study was the first attempt to automate the WMH visual grading using the modified Fazekas scale [11].
Theoretically, since our proposed method is a computational implementation, it is more accurate than the manually calculated results from the human raters when it comes to measuring the diameter of WMHs. Nevertheless, performance evaluations were done on comparing our results to the radiologists’ assessments, mainly due to two big reasons. First, the main goal of this method is to help doctors on reducing labor time and cost on daily basis. Second, since we are the first software to implement the modified Fazekas scale, comparison with other software was impossible. Hence, we compared our proposed method to human raters with multiple-rater and inter-rater agreements, which showed a high correspondence. Further investigation of the intra correlation coefficient (ICC) between software is preferred [27].
The multiple-rater agreement investigation (rating agreements with and without
our proposed method suggested that the level of agreement from our approach was
comparable to those among the radiologists. We used Krippendorff’s alpha
(
The inter-rater agreement between the radiologists and our proposed method demonstrated an equivalent performance on AUROC as well, which indicates the classification performance of the modified Fazekas scale between the two raters. The average AUROC showed minimal differences in the comparisons within radiologists (G) and between the radiologists and our proposed method (M) for the modified Fazekas scale 1 (G 0.87 vs. M 0.83), the modified Fazekas scale 2 (G 0.83 vs. M 0.77), and the modified Fazekas scale 3 (G 0.70 vs. M 0.79).
The average AUROC coefficient being higher in lower modified Fazekas scale means
that the radiologists performed better for small WMH burdens than our proposed
method. In contrast, our proposed method performed better than all of each
radiologist and also the average AUROC coefficient for grade 3 for the modified
Fazekas scale. This indicates out method may be clinically useful for objective
disease severity evaluation in large WMH burdens. Regardless, the combined AUROC
of the modified Fazekas scales demonstrated that the performance value between G
and P was comparable (G 0.80
Our study has a few limitations. The implemented modified Fazekas scale may not be widely used more than the original version. However, since the original Fazekas scale is not quantifiable and is based on a qualitative and subjective grading, we had to implement a scale which is applicable to automatic analysis. Additionally, our proposed system is currently being developed, and it has been mostly tested using 2D T2-FLAIRs. While this approach can be extended to any T2-FLAIR protocol, its performance may vary depending on the protocol. Future validation studies are needed to generalize our approach. Another limitation is the lack of ground-truth data, which is grand-scale collected data, on the modified Fazekas scale. We validated our approach against the three radiologists, whose results were used as the standard for comparison. As we have observed from our results, the three radiologists did not agree perfectly, and the ground-truth for the modified Fazekas scale has not been established at this point. To overcome the lack of ground-truth, further studies involving more experienced experts are needed to establish the gold standard for the modified Fazekas scale.
This study presented an automated modified Fazekas scoring approach using the objective measurements driven from T2-FLAIR and showed its performance against certified neuroradiologists. More work is needed to show our approach’s applicability to the research and clinical setting in the near future. Even so, we believe the present work could also contribute to both scientific society and clinical environments by suggesting automated analysis for the modified Fazekas scoring, especially for research related to large-scale or multi-site of WMH.
We introduced a fully automated visual grading system for WMH of T2-FLAIRs based on deep learning and rule-based algorithms utilizing the modified Fazekas scale. As we aimed, the results of our method were comparable to those of the three certified radiologists who used the visual grading method. We believe that our proposed method may assist clinic works and radiologists’ reading with its fully automated and quantifiable Fazekas scale with consistent measurement.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
ZR, REK, ML, DK, and JYK designed the research study. ZR, REK, ML, JYK, JMK, MKL, HL, and JY performed the research. ZR, REK, and HWK contributed to the interpretation of the results. ZR, REK, and JYK analyzed the data. ZR, REK, and JYK wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Eunpyeong St. Mary’s Hospital, College of Medicine, The Catholic University of Korea (IRB No. PC20EISI0094 on 02 July 2020). The authors confirm that all subjects or legally authorized representatives signed written informed consent forms.
Not applicable.
This work was supported by the Korea Medical Device Development Fund grant funded by the Korea government (Project Number: 202015X34, KMDF-PR-20200901-0306).
The authors declare no conflict of interest. ZunHyan Rieu, Regina EY Kim, Minho Lee, Hye Weon Kim, Donghyeon Kim and JeongHyun Yong belong to Research Institute, NEUROPHET Inc. The authors declare that there have no conflicts of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
