† These authors contributed equally.
Rab3a, a subtype protein in the Rab3 family amongst the small G proteins, is closely associated with the learning and memory formation process. Various neuronal stimuli can induce the expression of Rab3a; however, how DNA modification is involved in regulating its expression is not fully understood. Ten-eleven translocation (TET) proteins can oxidate methylcytosine to hydroxymethylcytosine, which can further activate gene expression. Previous studies reported that TET-mediated regulation of 5hmC induced by learning is involved in neuronal activation. However, whether Tet protein regulates Rab3a is unknown. To understand the role of TET-mediated 5hmC on Rab3a in neuronal activation, we adopted a KCl-induced depolarization protocol in cultured primary cortical neurons to mimic neuronal activity in vitro. After KCl treatment, Rab3a and Tet3 mRNA expression were induced. Moreover, we observed a decrease in the methylation level and an increase of hydroxymethylation level surrounding the CpG island near the transcription start site of Rab3a. Furthermore, recently, Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) has proven powerful in identifying open chromatin in the genome of various eukaryotes. Using FAIRE-qPCR, we observed a euchromatin state and the increased occupancy of Tet3, H3K4me3, and H3K27ac at the promoter region of Rab3a after KCl treatment. Finally, by using shRNA to knockdown Tet3 prior KCl treatment, all changes mentioned above vanished. Thus, our findings elucidated that the neuronal activity-induced accumulation of hydroxymethylation, which Tet3 mediates, can introduce an active and permissive chromatin structure at Rab3a promoter and lead to the induction of Rab3a mRNA expression.
Rab3a protein, a subtype protein in the Rab3 family in the small G proteins of the Ras superfamily, was first identified at chromosome 19 band p13.2 in 1989 [1]. It is highly enriched in the synaptic vesicles of neuronal cells, and it is also the most abundant Rab protein in the brain. Rab3a is composed of 220 amino acids and plays a role in the release of neurotransmitters. It is generally considered one of the most prominently studied activity-dependent molecules in the Rab3 family, and it is involved in exocytosis, plasma membrane repair, nerve fiber growth, and vesicle formation [2, 3, 4, 5, 6, 7]. Dysregulation of Rab3a expression may precipitate deterioration in memory, learning impairment, and neurodegenerative diseases [8, 9].
Currently, the epigenetic machinery that is involved in regulating the Rab3a expression under neuronal activation remains unknown. Moreover, DNA methylation and demethylation are two vital epigenetic regulatory mechanisms for gene expression [10], especially in neurons. There are two ways of DNA demethylation including passive demethylation (dependent on DNA replication) and active demethylation (independent of DNA replication) [11], and generally only active demethylation happens in post-mitotic neurons. Recent studies have revealed a variety of DNA modifications in post-mitotic neurons, including 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), which are the main DNA modifications that have been found in the active DNA demethylation pathway. Intriguingly, it has been shown that these 5mC oxidative derivatives are not transit products of active DNA demethylation; instead, they can form a stable epigenetic regulating state to promote gene expression [12, 13, 14]. Interestingly, there is an CpG island at the promoter region of Rab3a, and it may suggest methylation and demethylation processes would be involved in regulating Rab3a expression in an activity-dependent manner.
The Ten-eleven translocation (TET) protein family, including Tet1, Tet2, and Tet3, plays an essential role in DNA demethylation [15]. It can oxidize 5mC into 5hmC and subsequently into 5fC and 5caC [16], which is considered as the main protein family that initiates DNA demethylation mechanism. TET1 and TET3 contain a CXXC zinc finger domain at their amino-terminus known to bind CpG sequences [17], whereas TET2 partners with IDAX, an independent CXXC-containing protein [18]. Among them, Tet3 shows the highest expression in brain regions [19]. Tet3 is involved in cell survival, angiogenesis, neurogenesis, antioxidant defense, DNA repair, and metabolism. Moreover, previous research shows that Tet3, but not Tet1, exhibits a time-dependent increase in mRNA expression in an activity-dependent manner [20]. In line with this result, Tet3 may be a critical regulator of DNA demethylation during neuronal activation. However, so far, no relevant literature regarding the mechanism of Tet3 regulating Rab3a has been reported.
To date, the majority of studies have focused on the function of Rab3a, but the molecular mechanisms of how the epigenetic machinery regulates its expression under neuronal stimulation have yet to be uncovered. As described above, we hypothesize that Tet 3 may mediate the oxidation of 5mC to 5hmC, 5fC, and 5Cac in the CpG island of the Rab3a promoter region and promote Rab3a expression in an activity-dependent manner. This study can help reveal the interrelationship between Rab3a and Tet3 and providing novel insights into their functional relevance to provide the basis for further research.
Cortical tissue was isolated from E18 mouse embryos in a sterile atmosphere. To
dissociate the tissue, it was finely chopped followed by gentle pipetting to
create single cell suspension. To prevent clumping of cells due to DNA from dead
cells, tissue was treated with 2 unit/
Chromatin immunoprecipitation (ChIP) was performed following modification of the
Invitrogen ChIP kit protocol. Cells were fixed in 1% formaldehyde and
cross-linked cell lysates were trimmed with Covaris in 1% SDS lysis buffer to
generate chromatin fragments with an average length of 300 bp by using peak
power: 75, duty factor: 2, cycle/burst: 200, duration: 900 secs and temperature:
between 5
Lentiviral plasmids were generated by inserting either Tet3 shRNA (shRNATet3: GCCTGTTAGGCAGATTGTTCT) or scrambled control (scrambled control: CCTAAGGTTAAGTCGCCCTCG) fragments immediately downstream of the human H1 promoter in a modified FG12 vector (FG12H1, derived from the FG12 vector originally provided by David Baltimore, CalTech). Lentivirus was prepared and maintained as per previously published protocols [20].
Cell pellets were collected by 1000
1 ug RNA was employed for cDNA synthesis using the PrimeScript Reverse
Transcription Kit (Takara). Quantitative PCR was performed on a RotorGeneQ
(Qiagen) cycler with SYBR-Green Master mix (Qiagen) using primers for target
genes and for beta-actin as an internal control (Rab3a Forward primer:
TCCCAGTCCCCTGGAAAAAC and Reverse primer: GCTCCTCCTTTAGGAACTCGG; Tet1 Forward
primer: ACACAGTGGTGCTAATGCAG and Reverse primer: AGCATGAACGGGAGAATCGG; Tet2
Forward primer: AGAGAAGACAATCGAGAAGTCGG and Reverse primer:
CCTTCCGTACTCCCAAACTCAT; Tet3 Forward primer: GTGGTCGGACAGTGAACACAA and Reverse
primer: GTTGGGCTGGTTGAGGTTCTT; and beta-actin Forward primer:
GGCTGTATTCCCCTCCATCG and Reverse primer: CCAGTTGGTAACAATGCCATGT (the primers were
designed based on mm10). All transcript levels were normalized to beta-actin mRNA
using the
FAIRE-qPCR was performed as previously described [21]. In details, Primary
cortical neurons were fixed with molecular grade formaldehyde (16%, Thermo
fisher) at room temperature (22–25
1
The results were expressed as mean
It has been shown that Tet-meditated 5hmC can lead to induction of
synapsis-related gene expression [22]. To extend our understanding on how Tet
family proteins and 5hmC regulate Rab3a expression, we first tested the
expression of Tet proteins and Rab3a in vitro. Neurons derived from
embryonic day 18 (E18) were maintained for 7 days in vitro (DIV). Then,
25 mM final concentration of KCl were added to culture neurons following
protocols published previously (Fig. 1A). Post-KCl treatment, compared with the
KCl-group, the mRNA expression levels of Rab3a increased 2.5 folds (P
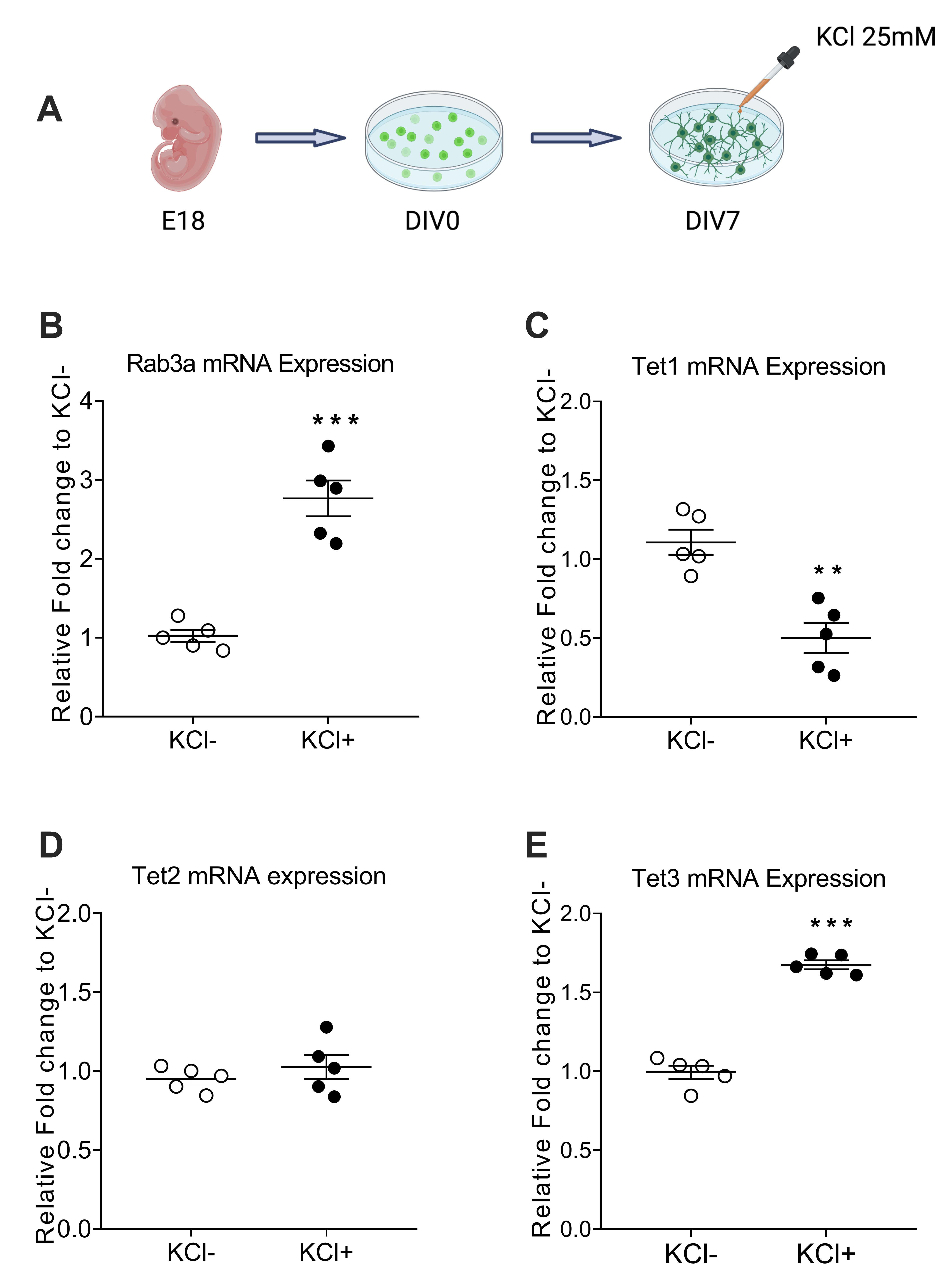 Fig. 1.
Fig. 1.The transcription levels of Rab3a and Tet3 were increased in
primary cortical neurons induced by KCl, whereas the opposite results were
observed on Tet1 expression and no obvious changes were observed in Tet2. (A)
Workflow for the path KCl-induced experiments in primary cortical neurons. (B–E) Real time quantitative PCR of reverse transcription (RT-qPCR) assay to
detect mRNA level of Rab3a (B), Tet1 (C), Tet2 (D), and Tet3 (E) in KCl- and KCl+
groups. Values represent mean
To further test our hypothesis, we examined the methylation state of the Rab3a
promoter region. By using methylated DNA immunoprecipitation (MeDIP)- and
hydroxymethylated DNA immunoprecipitation (hMeDIP)-qPCR, we accessed the 5mC and
5hmC level surrounding the transcription start site of Rab3a (Fig. 2A). To
explore the specific binding of MeDIP and hMeDIP, we add IgG control in both Tet3
and histone modification ChIP as well as DIP experiments. The result showed that
MeDIP and hMeDIP can have specific binding in our experiment (P
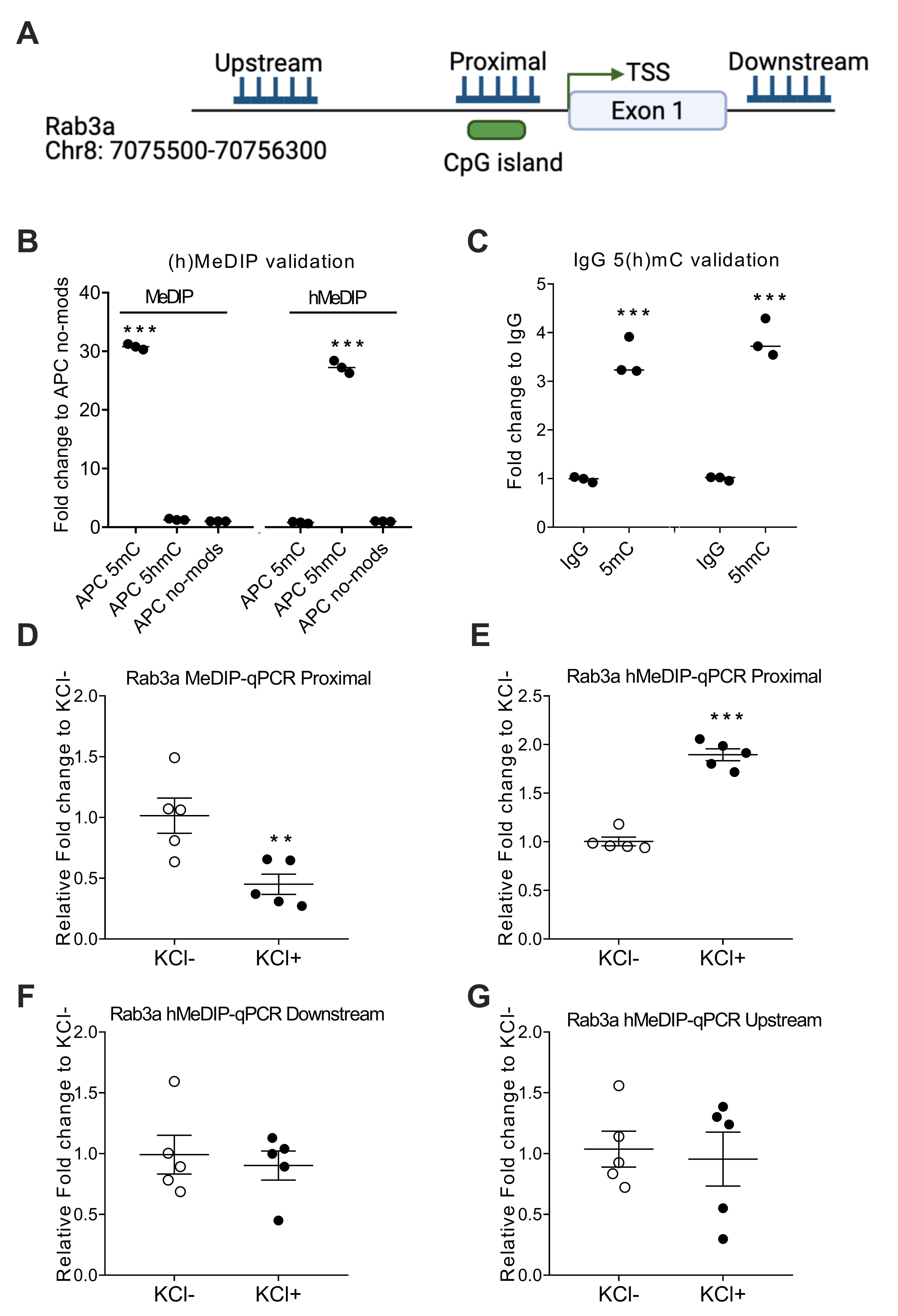 Fig. 2.
Fig. 2.CpG island demethylation levels increased at the Rab3a promoter
for KCl treatment, but not downstream or upstream. (A) Schematic illustration of
the Rab3a gene region. The gene is located on chromosome 8 from the 7,075,500 bp
to 70,756,300 bp regions. The target sequence was located in the CpG islands
region of the Rab3a promoter. TSS: transcriptional start site. (B) Validation of
(h)MeDIP-seq data using ChIP-qPCR for adenomatous polyposis coli (APC) no-mods.
(C) Validation of IgG 5(h)mC at CpG sites compared with corresponding normal IgG.
(D) MeDIP results showing the DNA methylation status of the Rab3a promoter. Five
independent repeats were performed. (E–G) hMeDIP results showing the DNA
demethylation status of the Rab3a promoter, downstream, and upstream. Values
represent mean
Previous studies have shown 5hmC accumulation is associated with euchromatin
structure. By using formaldehyde-assisted isolation of regulatory elements
(FAIRE)–qPCR, we examined chromatin accessibility in the Rab3a promoter region.
We observed a significant increase of Tet3 occupancy (P
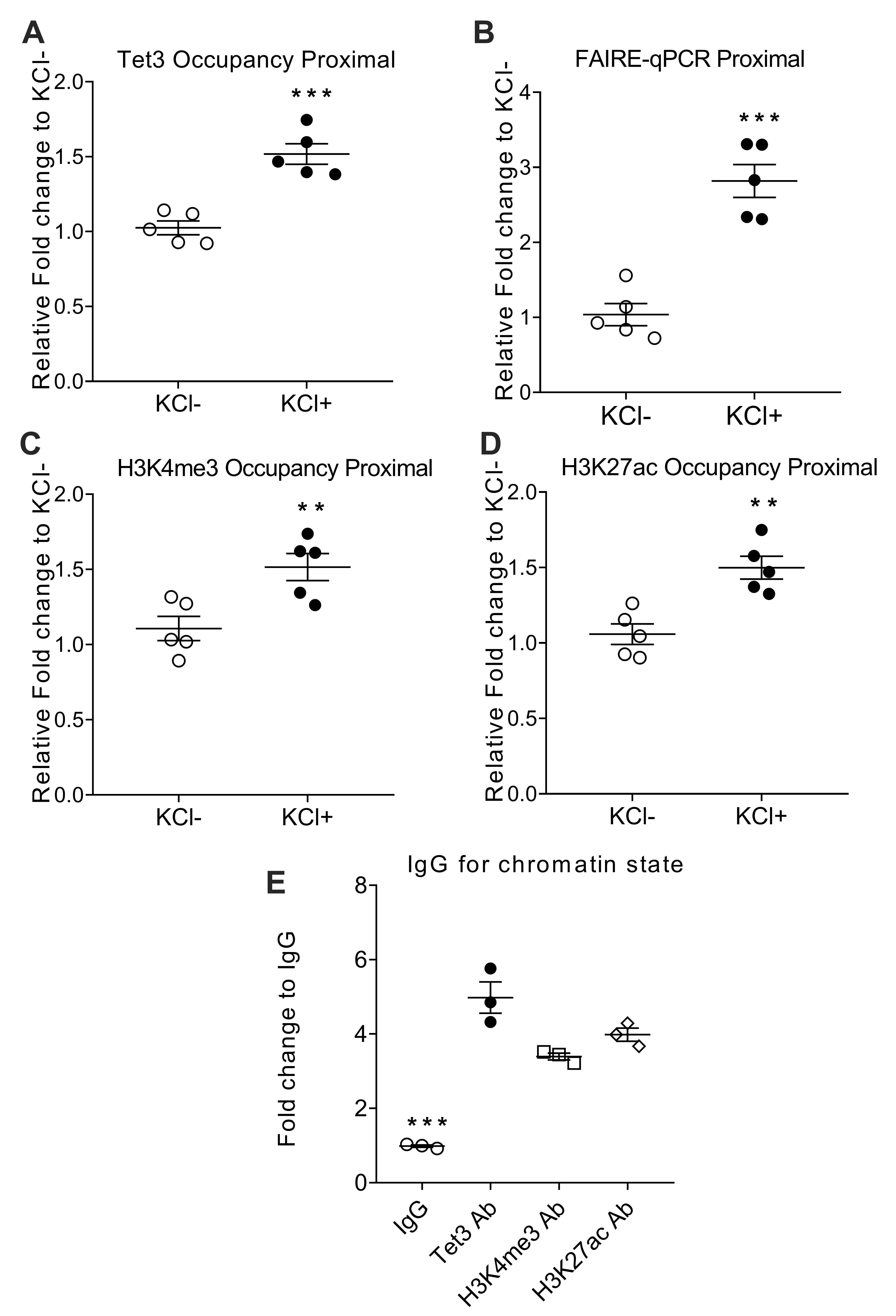 Fig. 3.
Fig. 3.Tet3-mediated 5hmC accumulation resulted in an active open
chromatin state at Rab3a promoter. (A) ChIP-qPCR occupancy of Tet3 in Rab3a
proximal region. (B) FAIRE enrichment in Rab3a promoter was quantified by
real-time PCR on the basis of the threshold cycle value (CT). (C,D) ChIP-qPCR
occupancy profiles of H3K4me3 and H3K27ac in the Rab3a proximal region. (E) The
same chromatin was used for control ChIP-qPCR experiments with control IgG.
Values for nonspecific binding (as determined by using control IgG) were
subtracted. Values represent mean
To further verify our hypothesis, we designed the shRNA specifically targeted to
Tet3, and packaged it into lenti-virus. This lenti-virus Tet3 shRNA could
sufficiently carry the shRNA into the primary cortical neuron, as shown in Fig. 4A
and this shRNA can significantly reduce the expression of Tet3 mRNA (Fig. 4B).
We then sought to examine if Tet3 is necessary for Rab3a expression under
KCl-induced conditions. The Tet3 and Rab3a mRNA expression levels after
KCl-induced depolarization significantly decreased with Tet3 shRNA included when
compared with scramble shRNA (P
Moreover, we also measured the effect of Tet3 shRNA on 5hmC levels. The result
confirmed reduction of Tet3 level can inhibit activity-induced 5hmC enrichment at
the Rab3a promoter region (P
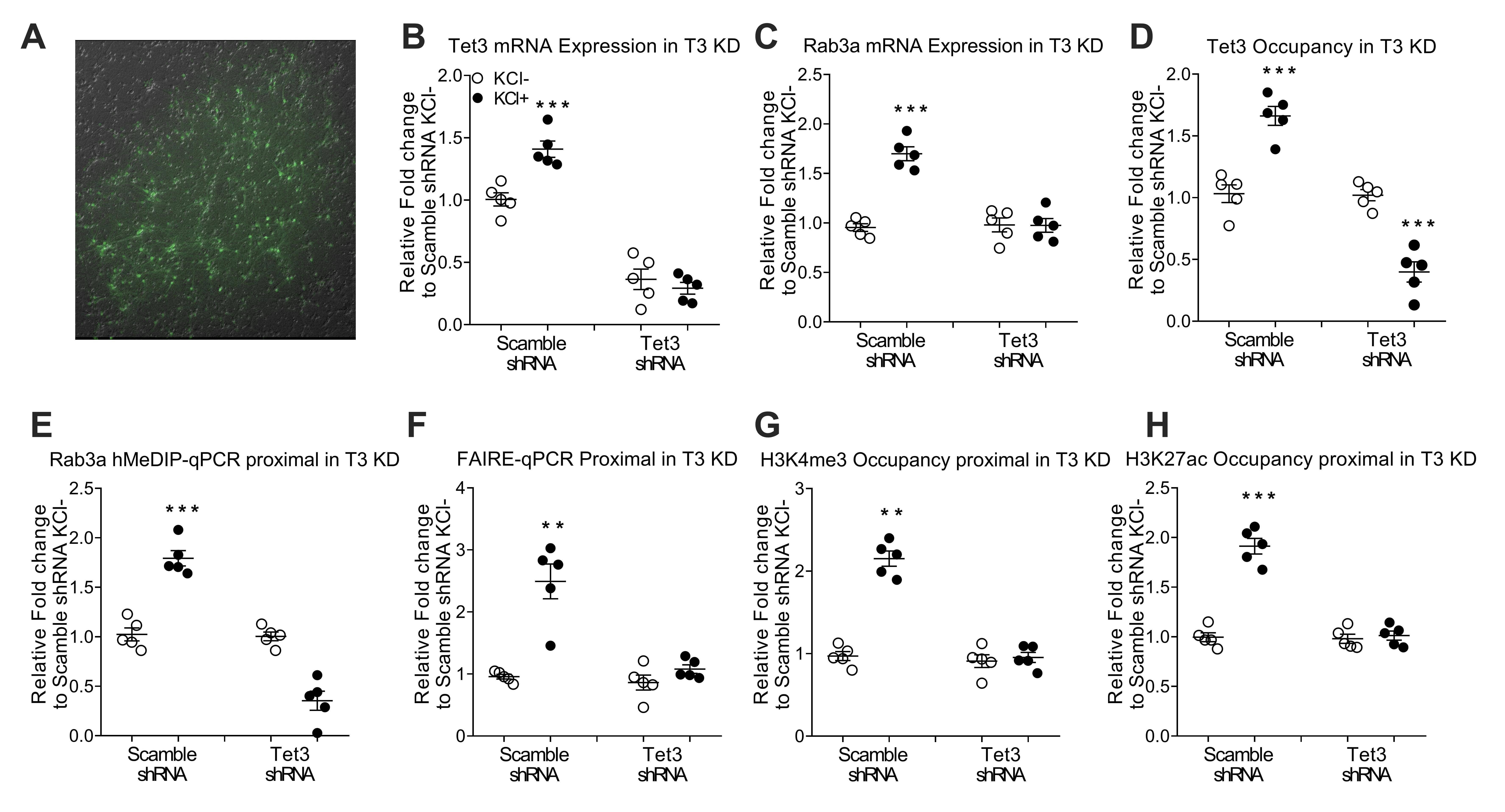 Fig. 4.
Fig. 4.Tet3-induced promoter demethylation leads to an active and
permissive chromatin structure at the Rab3a promoter accompanied by increments of
Rab3A expression. (A) Transfection efficiency of lentivirus in primary cortical
neurons. Primary neurons were transfected with plasmids expressing shRNA and
green fluorescent protein. Green fluorescent protein was detected as a marker of
transfection efficiency by fluorescence microscopy. Control: transfection vector;
Tet3 shRNA, shRNA transfection. (B,C) RT–PCR analysis of knockdown of Tet3
in primary cortical neurons transduced with lentivirus expressing Tet3 shRNA. (D)
Tet3 occupancy after knockdown of Tet3 in primary cortical neurons. (E)
hMeDIP-qPCR analysis for 5hmC enrichment after knockdown of Tet3. (F) FAIRE
enrichment in the Rab3a promoter after knockdown of Tet3. (G,H) Occupancy of
H3K4me3 and H3K27ac after knockdown of Tet3. Data are presented as the
mean
This study produced the following findings: (1) The expression of Rab3a and Tet3 increased by KCl-mediated depolarization, while the expression of Tet1 decreased. (2) Tet3 can promote 5hmC accumulation at the Rab3a promoter, which leads to an active open chromatin state at the promoter region of the Rab3a gene. (3) Accumulation of 5hmC and an open chromatin state can further facilitate the expression of Rab3a.
TET protein family consists of three members, namely Tet1, Tet2, and Tet3. They
belong to a type of dioxygenase that depends on Fe
TET proteins can catalyze the conversion of 5mC to 5hmC and subsequently into 5-fC and 5-caC. 5hmC, 5fC, and 5caC may not only be the oxidation products of 5mC but also have their specific epigenetic functions. There are significant differences in the distribution of 5hmC, 5fC, and 5caC in different tissues. For example, 5hmC is 10- if not 100- fold more prevalent than 5fC/5caC, and it is relatively enriched in neurons [26, 27, 28, 29]. Compared with 5hmC, 5fC/5caC was converted into an unmodified cytosine by the initiate base excision repair pathway [30, 31, 32]. Thus, rather than the intermediate product of the active demethylation process of 5mC, 5hmC is considered a stable epigenetic marker in mammalian cells. It is widely distributed in the nervous system and participates in memory formation, cognition, differentiation and development of neurons, as well as other multifunctional activities [33]. Thus, in our study, we only detected the accumulation of 5mC and 5hmC at the Rab3a promoter region. Nevertheless, the 5hmC level is higher than 5fC and 5caC, and the 5hmC is likely to be localized at the transcription start site [34, 35]. Also, Tet3 could directly bind to the Rab3a promoter and mediate the accumulation of 5hmC. To confirm this, we established Tet3 shRNA and found the reduction of Tet3 level can block the activity-induced 5hmC enrichment at the Rab3a promoter region. Meanwhile, the lack of available antibodies that can detect 5-fC and 5-caC prevented us from testing. Further studies are, therefore, needed to reliably check the 5fC and 5caC level at the Rab3a promoter region.
5hmC has been proven to be an important epigenetic modification for euchromatin structure. DNA methylation can change chromatin conformation. DNA methylation is also related to histone modification, which may control the accessibility of transcription factors to promoters, followed by increasing transcription of specific genes. Common histone modifications mainly include H3K4me1, H3K4me3, H3K27me1, H3K27me3, and H3K27ac [36]. Among them, H3K4me3 and H3K27ac are two primary histone-modification markers and represent an active euchromatin state that is associated with gene expression [37, 38]. In the present study, we observed the elevated occupancy of H3K4me3 and H3K27ac in the KCl group relative to the control group. Moreover, shRNA-targeted Tet3 was designed to knock down Tet3 expression, which inhibits the occupancy of H3K4me3 and H3K27ac. Thus, KCl depolarization triggers the expression of Tet3, and then Tet3 converts 5mC to 5hmC at Rab3a promoter. H3K4me3 and H3K27ac at the promoter were activated at a later stage, and the structure of chromatin changes from heterochromatin (closed chromatin) to euchromatin (open chromatin). Finally, the transcription factor can directly bind to promoters to upregulate the transcription of Rab3a (Fig. 5).
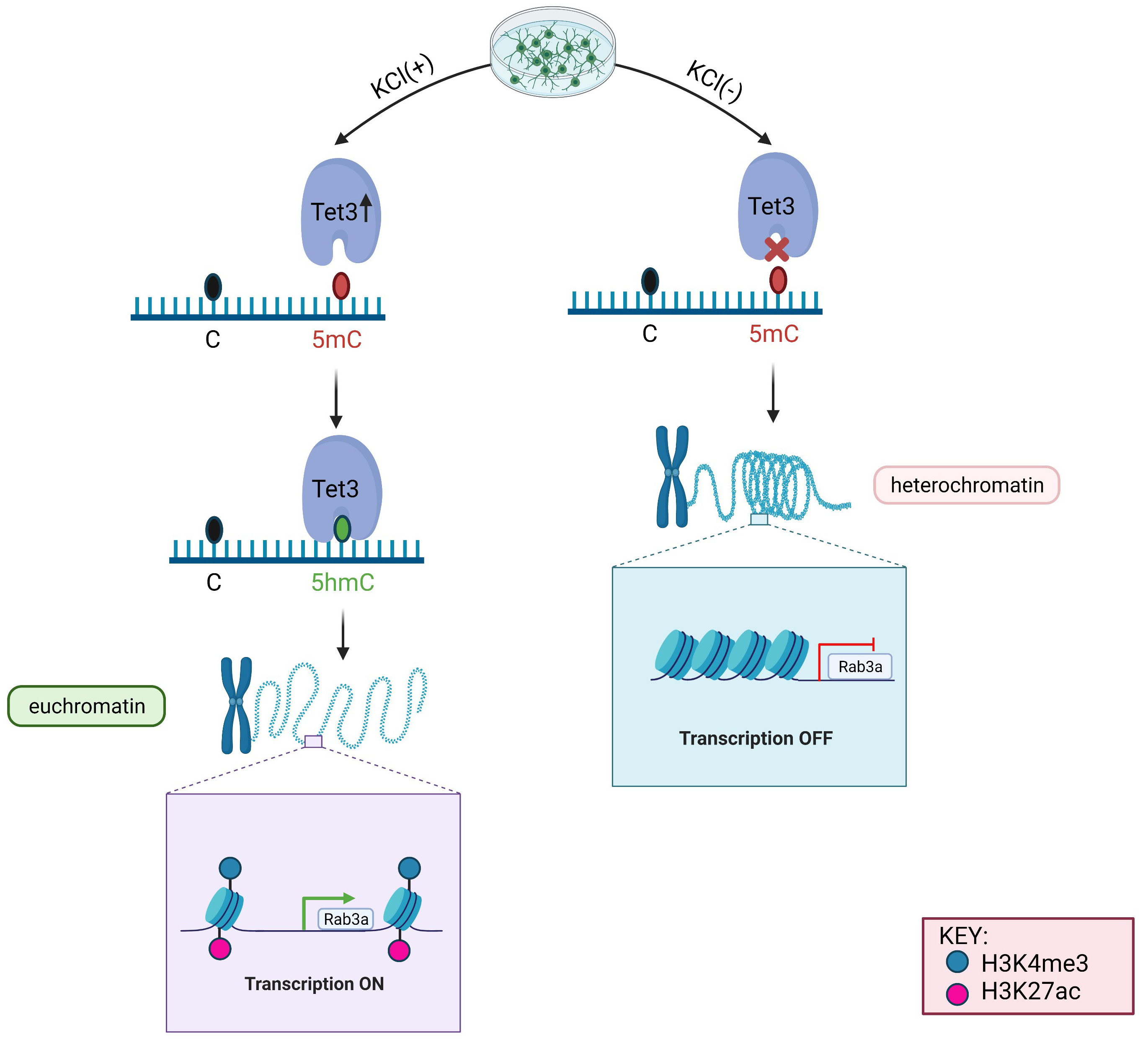 Fig. 5.
Fig. 5.Diagram of the differences between the KCl (+) and KCl (-) group involving the mechanism of Tet3-mediated DNA demethylation leading an active and permissive chromatin structure at the Rab3a promoter.
Rab3a, located on the synaptic vesicles of brain neuron cells, belongs to the small guanosine triphosphate (GTP) binding protein, which is thought to plays a key role in the process of neurotransmitter release and membrane transport [39]. Rab3A, as an active GTP hydrolyzing enzyme, can bind to GTP. When Rab3A binds to GTP, GTP/Rab3A forms an active protein conformation with lowered proteolytic activity, which triggers a series of processes by interacting with downstream proteins to regulate the transport of neuronal synaptic vesicles [40, 41]. Moreover, GTP hydrolyzed to guanosine diphosphate (GDP) by Rab3a, leads to structural conformation changes, and the protein loses its activity [42]. At the same time, this combination of Rab3A and GDP or GTP is also regulated by a variety of proteins, including guanine nucleotide exchange proteins (GEPs), GTPase-activating proteins (GAPs), and GDP dissociation inhibitors (GDIs) [43, 44, 45]. Furthermore, Rab3a is a requirement for brain-derived neurotrophic factor-induced (BDNF) synaptic plasticity associated with learning and memory [46]. Also, a previous study demonstrated that the homeostatic increase in miniature endplate current amplitude after activity blockade is diminished in the Rab3A deletion mouse [47]. This implicates that Rab3a is closely associated with neuronal activation. Generally, environmental factors engage specific receptors on nerve cells and activate ion fluxes, leading to nerve cell activation. Additionally, Rab3a regulates Ca2+-dependent exocytosis of neurotransmitter in the state of neuronal activation. Therefore, the increment of Rab3a expression can be induced by neuronal activation. However, how the expression of Rab3a is regulated remains inconclusive. In our study, we concluded that Tet3 drives KCl-induced expression of Rab3a by increasing DNA hydroxymethylation, chromatin accessibility and active histone modifications at the gene promoter from the perspective of DNA modification. These data therefore point to a critical role of Tet3-mediated oxidation of 5mC at CpG islands of the Rab3a promoter in regulating Rab3a gene expression under neuronal activation.
Admittedly, there are some limitations. Firstly, since the IgG controls should have been performed in parallel with the antibody-specific ChIP, it would be more appropriate to calculate the ddCt by normalizing first to the IgG pull-down over the same locus and then to the input DNA. IgG validation in our experiment were run by different cell batch and not same experimental run with previous ChIP sample. The changes would also not affect the conclusion. Li et al. [48], also published ChIP-qPCR and IgG validation separately. Secondly, the Rab3a gene may produce several different transcripts, which can be regulated by activation of the corresponding alternative promoter’s usage under neuronal activation. In the current study, we only focused on one promoter in the Rab3a genome. Future studies may require to fully elucidate the role of Tet3-mediated 5hmC in regulating activation of alternative promoters of Rab3a.
In summary, in this study we have confirmed that the expression of Rab3a mRNA in primary cortical neurons is activity-dependent and related to DNA demethylation mediated by Tet3. Moreover, we found accumulation of 5hmC was associated with induction of H3K4me3 and H3K27ac occupancy, which suggests an active euchromatin structure at the Rab3a promoter. Lentivirus-mediated Tet3 knockdown blocked the effect of DNA demethylation at the Rab3a promoter and reduced the expression of Rab3a mRNA. Therefore, these findings emphasize the critical role of Tet3 in promoting the active epigenetic state and lead to an induction of Rab3a expression in post-mitotic neurons. However, future work will also be needed to translate our in vitro findings into in vivo experiments with the goal of assessing the comprehensive roles of Tet3 in regulating Rab3a during learning and memory formation processes.
TET, Ten-eleven translocation; 5hmC, 5-hydroxymethylcytosine; 5fC,
5-formylcytosine, 5caC, 5-carboxylcytosine; ChIP, Chromatin immunoprecipitation;
E18, embryonic day 18; DIV, days in vitro; MeDIP, methylated DNA
immunoprecipitation; hMeDIP, hydroxymethylated DNA immunoprecipitation; FAIRE,
Formaldehyde Assisted Isolation of Regulatory Elements; APC, Adenomatous
polyposis coli. DSBH, double-stranded
KL, LSW carried out the experiments and wrote the manuscripts. YC wrote and edited manuscript and performed RT-qPCR experiments. KL conceived the study, designed experiments and wrote the manuscript. YLS, SHS performed the acquisition, analysis and interpretation of data.
Not applicable.
The authors gratefully acknowledge grant support from Zhongnan Hospital Wuhan University Start-up grant to KL, The Huaihua Hospital Young Researcher Grant to KL. LSW has been supported by postgraduate scholarship from Wuhan University. The authors would also like to thank Jianjian Zhang and Jincao Chen for helpful comments and lively discussion.
This study was supported by grants from Zhongnan Hospital of Wuhan University Start-up Grant (PTXM2021006).
The authors declare no conflict of interest.
