†These authors contributed equally.
Academic Editor: Wayne L. Miller
Background: Elevated left ventricular mass index contributes to
morbidity and mortality induced by heart failure and M2 macrophages play a
critical role in left ventricular remodeling. Here, our aim was to investigate
the roles of M2 macrophage-related genes in heart failure. Methods:
GSE10161 was downloaded and the abundance of immune cells were estimated utilizing the
CIBERSORT algorithm. Using the limma test and correlation analysis,
differentially expressed plasm B cells and M2 macrophages-related genes (DEBRGs
and DEMRGs) were documented. Functional pathways and the protein-protein
interaction network were analyzed and the hub DEMRGs were obtained. The hub
DEMRGs and their interactions were analyzed using NetworkAnalyst 3.0 and for
validation, the hub DEMRGs expressions were analyzed using the GSE135055,
GSE116250 and GSE74144 datasets. Results: 103 differentially expressed
genes were correlated with the abundance of M2 Macrophages and were identified as
DEMRGs (PCC
Currently, coronary artery disease (CAD) is still one of the leading causes of death in patients [1]. In addition, acute myocardial infarction (MI) mortality has increased 5.6-fold in the past 30 years [2]. Young patients with type 2 diabetes and MI have higher long-term cardiovascular and all-cause mortality and more than one-third of patients die within 10 years, which may emphasize more aggressive secondary prevention for those patients [3]. Acute heart failure (HF) contributes to the mortality above mentioned, which is characterized by an acute or subacute deterioration in cardiac function due to the underlying heart diseases and precipitating factors.
Isabelle et al. [4] reported that 23,291 patients with HF from 40 countries in 8 different world regions were investigated and there were 4460 (19%) deaths, 3885 (17%) HF hospitalizations, and 6949 (30%) instances of either adverse event, suggesting that HF has become the leading problem in CAD. Previous researches have confirmed the importance of neurohormonal systems, such as the renin-angiotensin-aldosterone axis in the pathogenesis of heart failure phenotypes [5, 6]. However, a large number of drug trials have failed, highlighting the limitations of animal models in replicating complex diseases and investigating critical regulators in HF [7]. The underlying critical genes and interactions that control the transformation of heart failure remain largely unknown.
Elevated left ventricular mass (LVM) contributes to morbidity and mortality induced by heart failure, which is partially regulated by hemodynamic indicators [8]. However, only a small portion of LVM variation is investigated by hemodynamic effects, and it has been suggested that genetic influences may also be important.
In this study, GSE10161 was downloaded and immune cells analysis was carried out to screen out the hub genes associated with immune cells. Then the expression levels of the hub genes in heart failure, dilated cardiomyopathy, ischemic cardiomyopathy and hypertension patients were demonstrated to validate the correlation to left ventricular remodeling, which will help to diagnose the adverse left ventricular remodeling at an early stage and reduce the patients’ mortality.
Using the keywords “left ventricular” in “Homo sapiens”, GSE10161 from the Gene Expression Omnibus (GEO) database was downloaded and analyzed [9]. There were 7 left ventricular biopsies from control patients and 20 cardiac biopsies from aortic stenosis (AS) patients based on the Affymetrix Human Genome U133A Array. Firstly, the raw transcriptomic data were processed with log2 transformation for normalization utilizing a robust multichip average algorithm [10].
Based on their gene expression profiles CIBERSORT is a deconvolution approach to
characterize the cell compositions in bulk tissues [11]. To obtain the abundance
of immune cells in the left ventricle, the CIBERSORT algorithm with 100
permutations was applied in the
GSE10161 dataset, utilizing the LM22 matrix as reference. CIBERSORT outputs a
deconvolution p-value for each sample to determine the reliability of
the results. In this study, we retained the samples with p
The differentially expressed genes (DEGs) between left ventricle samples from
control patients and aortic stenosis patients were investigated utilizing the
“limma” package. The genes with
To understand the functions of DEMRGs and DEBRGs, GO and KEGG pathway analysis,
as well as Gene Set Enrichment Analysis (GSEA) analysis, were applied utilizing
the “clusterProfiler” package in R [12]. A p
STRING (https://string-db.org) is a biological
resource that provides systematic screens of human protein interactions [13]. To
investigate the hub genes, DEMRGs and DEBRGs were uploaded to STRING to
investigate the protein network interaction diagram and significant PPIs were
identified with a combined score
NetworkAnalyst 3.0 (https://www.networkanalyst.ca/) is a comprehensive network visual analyzed platform for gene expression analysis [14]. The hub DEMRGs and their interactions were analyzed using NetworkAnalyst 3.0. Specifically, transcription factors (TFs)-miRNA coregulatory interactions with 5 screened hub DEMRGs were shown using the RegNetwork repository where the literature curated regulatory interaction information was collected (Applicable for human and mouse data only). Left ventricle tissue-specific PPI were shown using the DifferentialNet database (Filter is 15), which shows the differential protein-protein interactions across human tissues. The hub DEMRGs-chemical interactions were shown using the Comparative Toxicogenomics Database (CTD). Left ventricle tissue-specific co-expression with 5 screened hub DEMRGs were shown using the TCSBN database. The hub DEMRGs-drugs interactions were shown using the DrugBank database (Version 5.0) (https://go.drugbank.com/releases/5-0-11). MMP2 and its edges were validated in THP-1 cells from the Immuno-Navigator database.
To view the correlation between the hub DEMRGs and left ventricular remodeling, we performed unsupervised hierarchical clustering in the GSE135055 dataset utilizing the “Pheatmap” package [15]. Then, the expression patterns of the screened DEMRGs in left ventricular remodeling process induced by heart failure were validated in three independent datasets, including GSE135055 (n = 30) [15], GSE116250 (n = 64) [16, 17] and GSE74144 (n = 28). The detailed information for the datasets was shown in Supplementary Table 1.
Receiver operation characteristic (ROC) curve and joint ROC curve analysis were conducted to investigate the diagnostic performance of the DEMRGs and the area under the curve (AUC) was determined using the “pROC” package.
The CIBERSORT algorithm was conducted to obtain the immune cell compositions in
the GSE10161 dataset and Fig. 1A summarized the results investigated from the 7 left ventricular biopsies from
control patients and 20 cardiac biopsies from AS patients. Compared with control
biopsies, the biopsies from AS patients exhibited a higher infiltration of M2
Macrophages and a lower infiltration of plasm B cells (Fig. 1B; Supplementary Fig. 1). Utilizing the “limma” package, a total of
860 DEGs were obtained and of these DEGs, 103 were correlated (PCC
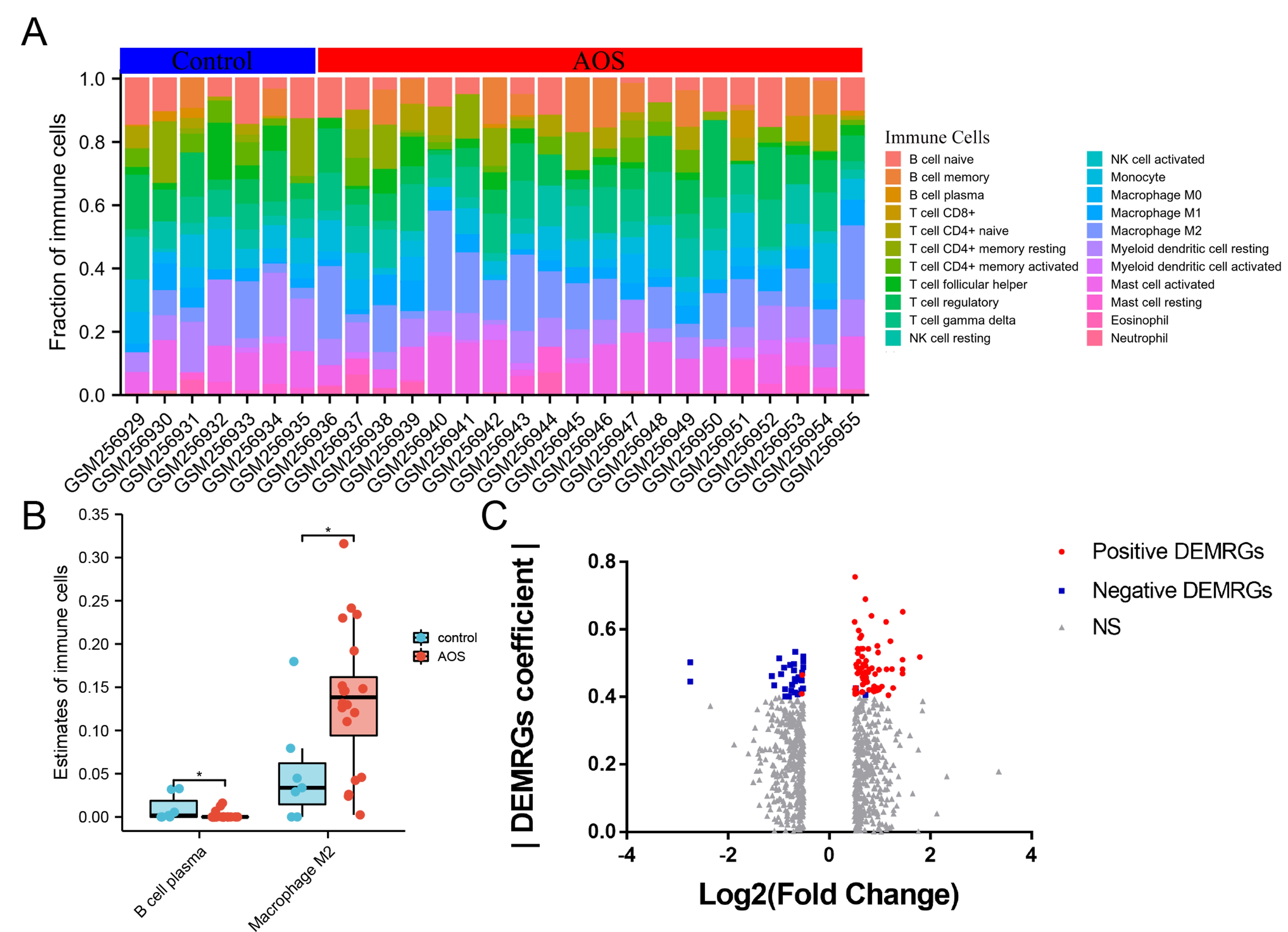 Fig. 1.
Fig. 1.Identification of DEMRGs in left ventricular biopsies from control patients and aortic stenosis patients. (A) Abundance of 22 immune cells across 27 samples estimated by the CIBERSORT algorithm. (B) Fraction of plasm B cells and M2 Macrophages between left ventricular biopsies from control patients and aortic stenosis patients. (C) Volcano plot for DEMRGs between left ventricular biopsies from control patients and aortic stenosis patients. DEMRGs, differentially expressed M2 Macrophages-related genes.
GO and KEGG pathway analysis, as well as GSEA analysis, were performed to analyze the functions of DEGs, DEMRGs and DEBRGs. These DEGs were mainly involved in extracellular matrix organization, extracellular structure organization, collagen-containing extracellular matrix, calcium-dependent protein binding and ECM-receptor interaction. Moreover, GSEA analysis showed that NABA_CORE_MATRISOME and NABA_ECM_GLYCOPROTEINS were top2 gene sets associated with DEGs (Fig. 2A,B; Table 1).
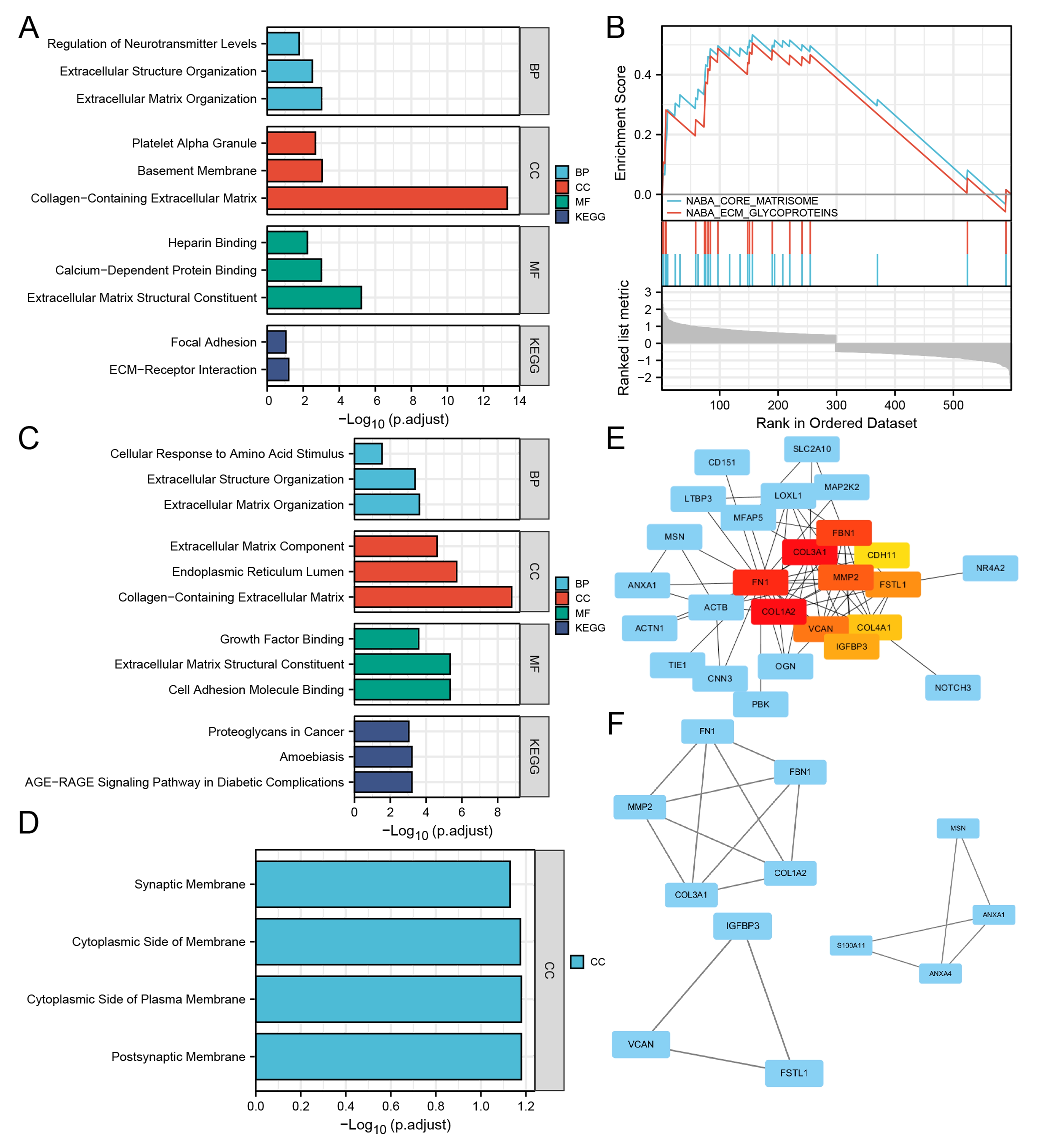 Fig. 2.
Fig. 2.Enrichment pathway analysis and protein-protein interaction network for the DEMRGs. (A) The significant GO and KEGG pathways enriched by DEGs. (B) The top2 gene sets associated with DEGs using GSEA analysis. (C) The significant GO and KEGG pathways enriched by up-regulated DEMRGs compared left ventricular biopsies from aortic stenosis patients to control patients. (D) The significant GO-Cellular Component enriched by down-regulated DEMRGs. (E) Protein-protein interaction network for the DEMRGs using Cytohubba. The nodes were ranked by MCC, which were labeled successively as red, yellow and blue. (F) 3 modules were screened out using MCODE in Cytoscape.
| Gene ONTOLOGY | ID | Description | GeneRatio | BgRatio | p value | p.adjust | q value |
| BP | GO:0030198 | extracellular matrix organization | 31/555 | 368/18670 | 2.07e-07 | 9.52e-04 | 8.77e-04 |
| BP | GO:0043062 | extracellular structure organization | 32/555 | 422/18670 | 1.36e-06 | 0.003 | 0.003 |
| BP | GO:0001505 | regulation of neurotransmitter levels | 26/555 | 354/18670 | 2.31e-05 | 0.017 | 0.015 |
| BP | GO:0007596 | blood coagulation | 25/555 | 336/18670 | 2.66e-05 | 0.017 | 0.015 |
| BP | GO:0050849 | negative regulation of calcium-mediated signaling | 7/555 | 31/18670 | 2.79e-05 | 0.017 | 0.015 |
| CC | GO:0062023 | collagen-containing extracellular matrix | 48/568 | 406/19717 | 8.82e-17 | 4.82e-14 | 4.19e-14 |
| CC | GO:0005604 | basement membrane | 13/568 | 95/19717 | 3.30e-06 | 9.02e-04 | 7.85e-04 |
| CC | GO:0031091 | platelet alpha granule | 12/568 | 91/19717 | 1.16e-05 | 0.002 | 0.002 |
| CC | GO:0005925 | focal adhesion | 27/568 | 405/19717 | 5.13e-05 | 0.006 | 0.005 |
| CC | GO:0005924 | cell-substrate adherens junction | 27/568 | 408/19717 | 5.82e-05 | 0.006 | 0.005 |
| MF | GO:0005201 | extracellular matrix structural constituent | 22/552 | 163/17697 | 7.63e-09 | 5.99e-06 | 5.43e-06 |
| MF | GO:0048306 | calcium-dependent protein binding | 11/552 | 61/17697 | 2.50e-06 | 9.82e-04 | 8.90e-04 |
| MF | GO:0008201 | heparin binding | 17/552 | 169/17697 | 2.23e-05 | 0.006 | 0.005 |
| MF | GO:0005539 | glycosaminoglycan binding | 20/552 | 229/17697 | 3.48e-05 | 0.007 | 0.006 |
| MF | GO:0005178 | integrin binding | 14/552 | 132/17697 | 6.55e-05 | 0.010 | 0.009 |
| KEGG | hsa04512 | ECM-receptor interaction | 11/283 | 88/8076 | 2.34e-04 | 0.064 | 0.059 |
| KEGG | hsa04510 | Focal adhesion | 17/283 | 201/8076 | 6.68e-04 | 0.091 | 0.084 |
| DEGs, Different Expressed Genes; GO, Gene ONTOLOGY; BP, Biological Process; CC, cellular component; MF, Molecular Function; KEGG, Kyoto Encyclopedia of Genes and Genomes. | |||||||
The up-regulated DEMRGs were mainly involved in extracellular matrix organization, collagen-containing extracellular matrix, cell adhesion molecule binding and AGE-RAGE signaling pathway in diabetic complications, while the down-regulated DEMRGs were mainly involved in GO-cellular components, such as postsynaptic membrane and postsynaptic membrane (Fig. 2C,D; Tables 2,3). The up-regulated DEBRGs were mainly involved in regulation of cell growth, cell growth, external side of plasma membrane and Glycine, serine and threonine metabolism, while the down-regulated DEBRGs were mainly involved in Retrograde endocannabinoid signaling and Chemokine signaling pathway (Supplementary Fig. 3; Supplementary Tables 2,3).
| Gene ONTOLOGY | ID | Description | GeneRatio | BgRatio | p value | p.adjust | q value |
| BP | GO:0030198 | extracellular matrix organization | 10/57 | 368/18670 | 1.48e-07 | 2.36e-04 | 2.01e-04 |
| BP | GO:0043062 | extracellular structure organization | 10/57 | 422/18670 | 5.23e-07 | 4.16e-04 | 3.54e-04 |
| BP | GO:0071230 | cellular response to amino acid stimulus | 4/57 | 68/18670 | 5.50e-05 | 0.029 | 0.025 |
| BP | GO:0001101 | response to acid chemical | 7/57 | 343/18670 | 7.98e-05 | 0.032 | 0.027 |
| BP | GO:0046651 | lymphocyte proliferation | 6/57 | 272/18670 | 1.76e-04 | 0.046 | 0.039 |
| CC | GO:0062023 | collagen-containing extracellular matrix | 14/58 | 406/19717 | 8.78e-12 | 1.65e-09 | 1.28e-09 |
| CC | GO:0005788 | endoplasmic reticulum lumen | 10/58 | 309/19717 | 2.07e-08 | 1.95e-06 | 1.51e-06 |
| CC | GO:0044420 | extracellular matrix component | 5/58 | 51/19717 | 3.91e-07 | 2.45e-05 | 1.89e-05 |
| CC | GO:0005604 | basement membrane | 5/58 | 95/19717 | 8.74e-06 | 4.11e-04 | 3.18e-04 |
| CC | GO:0098644 | complex of collagen trimers | 3/58 | 19/19717 | 2.26e-05 | 5.45e-04 | 4.21e-04 |
| MF | GO:0050839 | cell adhesion molecule binding | 12/56 | 499/17697 | 4.00e-08 | 4.53e-06 | 3.56e-06 |
| MF | GO:0005201 | extracellular matrix structural constituent | 8/56 | 163/17697 | 4.25e-08 | 4.53e-06 | 3.56e-06 |
| MF | GO:0019838 | growth factor binding | 6/56 | 137/17697 | 4.56e-06 | 2.59e-04 | 2.04e-04 |
| MF | GO:0048407 | platelet-derived growth factor binding | 3/56 | 11/17697 | 4.86e-06 | 2.59e-04 | 2.04e-04 |
| MF | GO:0098641 | cadherin binding involved in cell-cell adhesion | 3/56 | 19/17697 | 2.81e-05 | 0.001 | 9.39e-04 |
| KEGG | hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 5/24 | 100/8076 | 9.28e-06 | 6.19e-04 | 5.22e-04 |
| KEGG | hsa05146 | Amoebiasis | 5/24 | 102/8076 | 1.02e-05 | 6.19e-04 | 5.22e-04 |
| KEGG | hsa05205 | Proteoglycans in cancer | 6/24 | 205/8076 | 2.28e-05 | 9.21e-04 | 7.77e-04 |
| KEGG | hsa04926 | Relaxin signaling pathway | 5/24 | 129/8076 | 3.20e-05 | 9.69e-04 | 8.18e-04 |
| KEGG | hsa04510 | Focal adhesion | 5/24 | 201/8076 | 2.63e-04 | 0.006 | 0.005 |
| DEGs, Different Expressed Macrophage(M2)-Related Genes; GO, Gene ONTOLOGY; BP, Biological Process; CC, cellular component; MF, Molecular Function; KEGG, Kyoto Encyclopedia of Genes and Genomes. | |||||||
| Gene ONTOLOGY | ID | Description | GeneRatio | BgRatio | p value | p.adjust | q value |
| CC | GO:0045211 | postsynaptic membrane | 4/27 | 323/19717 | 9.21e-04 | 0.066 | 0.055 |
| CC | GO:0009898 | cytoplasmic side of plasma membrane | 3/27 | 154/19717 | 0.001 | 0.066 | 0.055 |
| CC | GO:0098562 | cytoplasmic side of membrane | 3/27 | 178/19717 | 0.002 | 0.067 | 0.056 |
| CC | GO:0097060 | synaptic membrane | 4/27 | 432/19717 | 0.003 | 0.074 | 0.062 |
| DEGs, Different Expressed Macrophage(M2)-Related Genes; GO, Gene ONTOLOGY; CC, cellular component. | |||||||
The PPI network of DEMRGs and DEBRGs constructed utilizing the STRING database
was a scale-free network and the genes with connective degrees
The hub DEMRGs and their interactions were demonstrated (Fig. 3). TFs-miRNA- hub DEMRGs coregulatory network, left ventricle tissue-specific co-expression and left ventricle tissue-specific PPI were constructed. Besides, the hub DEMRGs-chemical interactions were shown and 7 chemicals, including 4-(5-benzo(1,3)dioxol-5-yl-4-pyridin-2-yl-1H-imidazol-2-yl)benzamide, (6-(4-(2-piperidin-1-ylethoxy)phenyl))-3-pyridin-4-ylpyrazolo(1,5-a)pyrimidine, Copper, (+)-JQ1 compound, Paclitaxel, Phenylmercuric Acetate, Tretinoin and Valproic Acid, can all be interacted with 5 screened hub DEMRGs. The hub DEMRGs-drugs interactions were also shown utilizing the DrugBank database and 6 drugs seemed to have antagonistic effects on adverse left ventricular remodeling. Besides, MMP2 and its edges were validated in THP-1 cells, which can provide further evidence that MMP2 was high-expressed in M2 Macrophages for left ventricular remodeling.
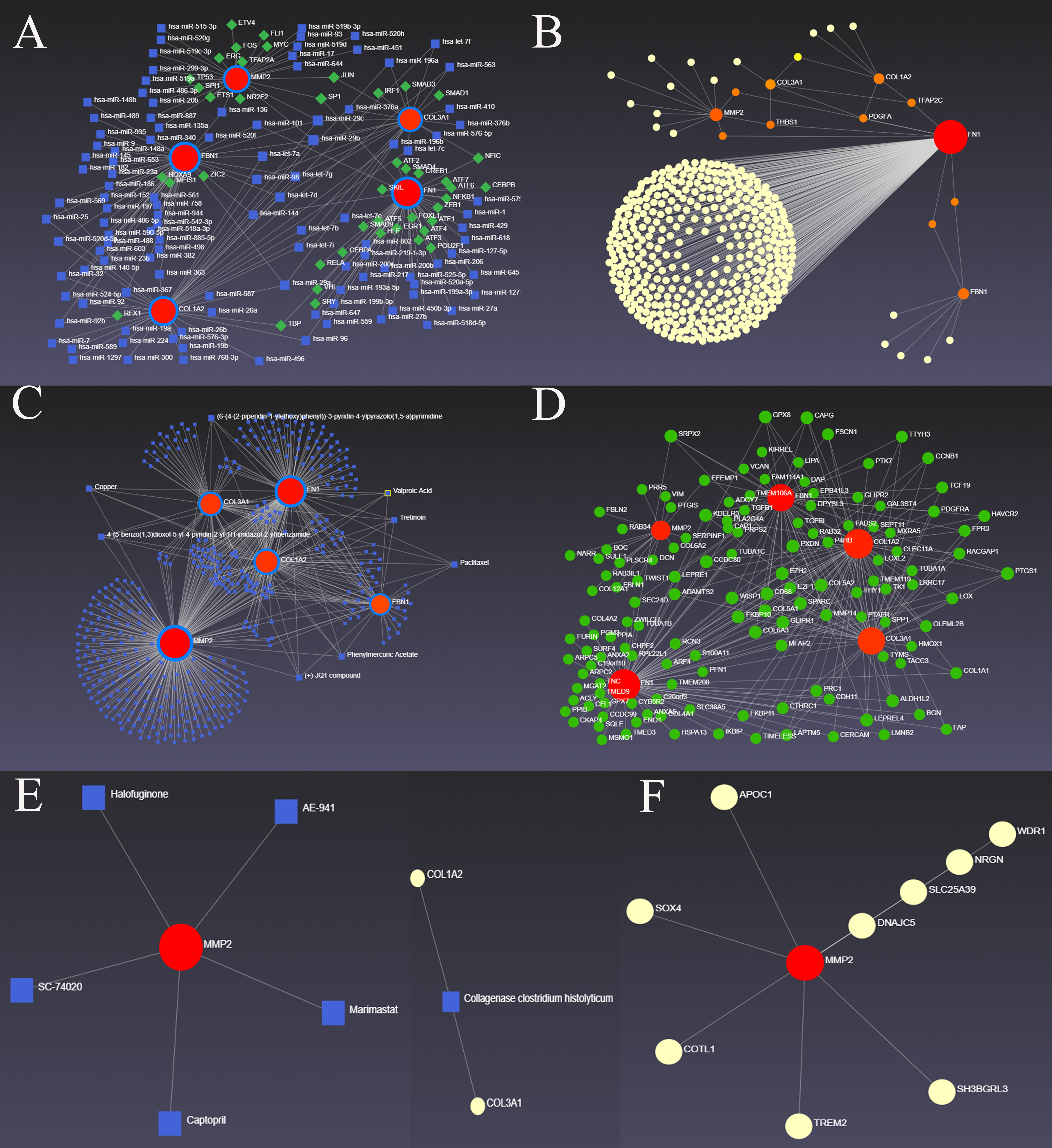 Fig. 3.
Fig. 3.The hub DEMRGs and their interactions. (A) TF-miRNA coregulatory interactions with 5 screened hub DEMRGs. (B) Left ventricle tissue-specific PPI, including 5 screened hub DEMRGs and their edges. (C) The hub DEMRGs-chemical interactions. (D) Left ventricle tissue-specific co-expression with 5 screened hub DEMRGs. (E) The hub DEMRGs-drugs interactions. (F) MMP2 and its edges validated in THP-1 cells.
Unsupervised hierarchical clustering was shown based on the expressions of hub DEMRGs in the GSE135055 dataset (Fig. 4A). Next, the DEMRGs expression patterns were validated in three independent datasets (GSE135055 (n = 30), GSE116250 (n = 64) and GSE74144 (n = 28)). In the GSE135055 dataset, MMP2, FN1, COL1A2 and COL3A1 were highly expressed and the AUC of 4 DEMRGs were 0.905 (Fig. 4B–D). Interestingly, although the AUC of FBN1 expression was 0.556, the AUC of all 5 DEMRGs was 1 (Supplementary Fig. 6A). In the GSE116250 dataset, MMP2, COL1A2 and COL3A1 were highly expressed in patients with dilated cardiomyopathy and MMP2, FN1, COL1A2 and COL3A1 were highly expressed in patients with ischemic cardiomyopathy. The ROC curves and joint ROC curves demonstrated the 5 hub DEMRGs can be diagnostic biomarkers for heart failure patients with dilated and ischemic cardiomyopathy (Fig. 4E–I; Supplementary Fig. 6B,C). In the GSE74144 dataset, FN1, FBN1, COL1A2 and COL3A1 were highly expressed in patients with hypertension and left ventricular remodeling and the AUC of the above 4 hub DEMRGs was 0.964 (Fig. 4J–L; Supplementary Fig. 6D). Taken together, MMP2, FN1, FBN1, COL1A2 and COL3A1 can be the prognostic biomarkers for the patients with left ventricular remodeling induced by heart failure and the kit of the 5 hub DEMRGs may test all the conditions of left ventricular remodeling events, which can help prevent the adverse left ventricular remodeling to decrease the mortality and morbidity.
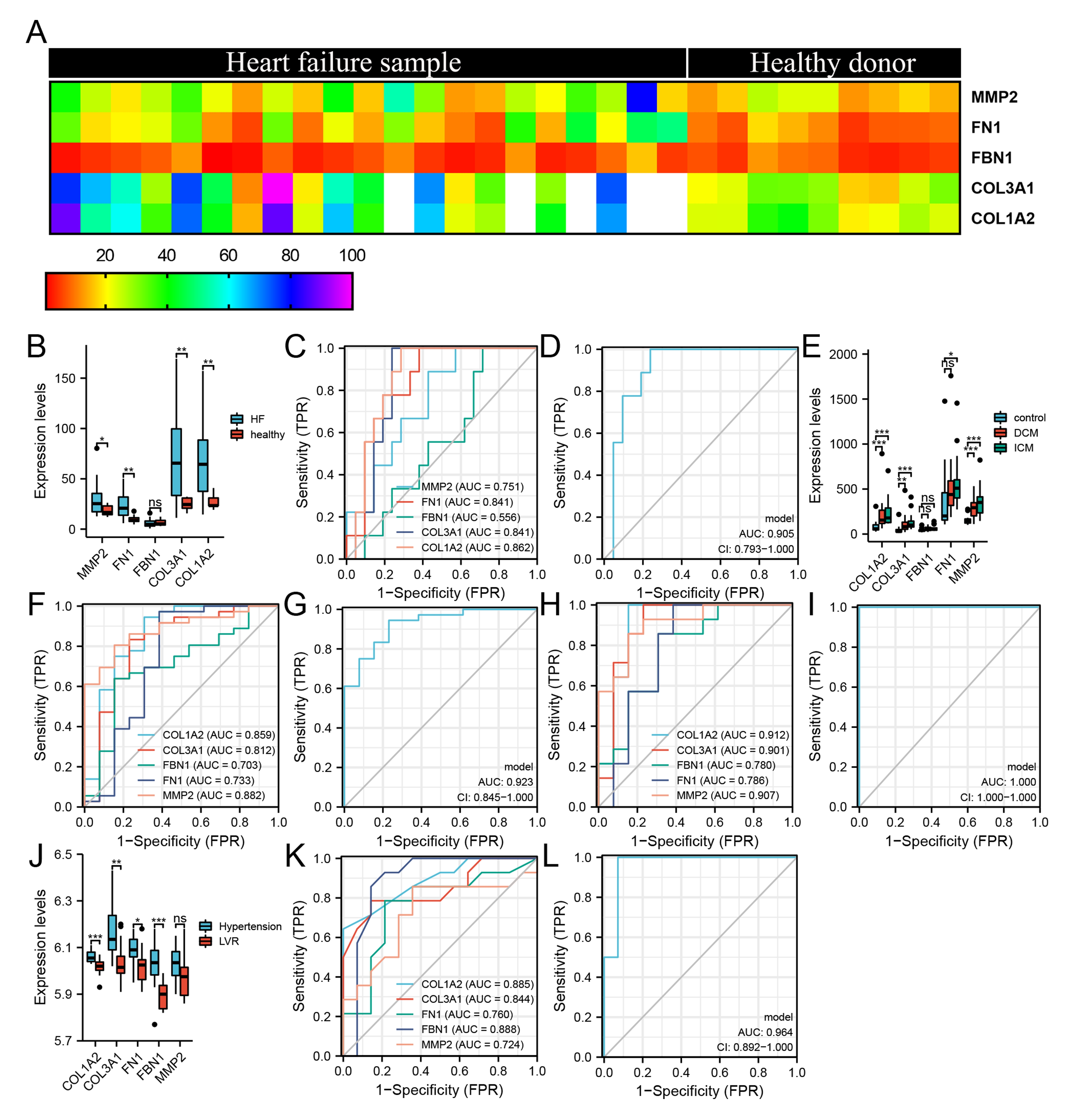 Fig. 4.
Fig. 4.Expression patterns of hub DEMRGs in left ventricular remodeling
induced by heart failure. (A) The heatmap for unsupervised hierarchical
clustering of the five hub DEMRGs in GSE135055 dataset. (B) Expression levels of
the hub DEMRGs in GSE135055 dataset. (C–D) ROC curve of 5 hub DEMRGs (C) and
joint ROC curve of MMP2, FN1, COL1A2 and COL3A1 (D) in GSE135055 dataset. (E)
Expression levels of the hub DEMRGs in GSE116250 dataset. (F–G) ROC curve of 5
hub DEMRGs (F) and joint ROC curve of MMP2, COL1A2 and COL3A1 (G) in DCM.
(H–I) ROC curve of 5 hub DEMRGs (H) and joint ROC curve of MMP2, FN1, COL1A2 and
COL3A1 (I) in ICM. (J) Expression levels of the hub DEMRGs in GSE74144
dataset. (K–L) ROC curve of 5 hub DEMRGs (K) and joint ROC curve of FN1, FBN1,
COL1A2 and COL3A1 (L) in GSE74144 dataset. *p
Heart failure contributes to increased morbidity and mortality and affects
In the event of acute inflammation, a large number of immune cells, especially monocytes, are recruited to the site of inflammation to repair the tissue to eventually restore the tissue to homeostasis [23, 24]. The monocytes differentiated into M1-type macrophages and combined with resident M1 macrophages to remove foreign bodies or necrotic tissues. In the fiber repair stage of the disease course, M1-type macrophages have been gradually replaced by M2-type macrophages, which play a role in promoting tissue repair [25, 26]. Besides, a network of macrophages can actively take up material, including mitochondria, derived from cardiomyocytes and support mitochondrial homeostasis in the heart [27]. Macrophages can also facilitate electrical conduction in the physiological and pathological heart through gap junction [28, 29, 30]. However, the specific mechanism of the differentiation of M1-type macrophages to M2-type macrophages and the role of M2-type macrophages has not been fully clarified and needs further studies. In this study, GSE10161 was downloaded and immune cells analysis was carried out to screen out the hub genes associated with immune cells. DEMRGs and DEBRGs were screened out and after further GO/KEGG enrichment analysis and PPI analysis, 5 hub DEMRGs was investigated, which may be the critical hub genes to promote left ventricular remodeling induced by heart failure. Then the expression levels of the hub genes in heart failure, dilated cardiomyopathy, ischemic cardiomyopathy and hypertension patients were demonstrated to validate the correlation to left ventricular remodeling, which will help to diagnose the adverse left ventricular remodeling at an early stage and reduce the patients’ mortality.
5 hub DEMRGs, including MMP2, FN1, FBN1, COL3A1 and COL1A2, were investigated to
be associated with M2 Macrophages phenotype. Matrix metalloproteinase 2 (MMP2),
as a fibrosis-related gene, affected extracellular matrix remodeling after an
ischemic myocardial injury and can be the detection of gelatinase expression
after MI [31, 32]. Treatment with verapamil can decline calpain-1 and MMP-2
activities and ameliorate cardiac hypertrophy [33]. MMP-2/JNK apoptotic pathway
was activated in the border zone after MI in adult sheep [34]. Transverse aortic
constriction in mice model induced cardiac hypertrophy, collagen deposition, and
the expression of transforming growth factor (TGF)-
There are some limitations which should be mentioned. Firstly, only hub DEMRGs were validated in patients with heart failure, dilated cardiomyopathy, ischemic cardiomyopathy or hypertension and left ventricular remodeling. There may be some false negatives because of the enrichment methods and validation methods. More researches are still needed to proceed with other DEGs, such as DEBRGs and neighbor DEMRGs. Secondly, the sample sizes of included datasets were not too large, however, after validation, the results are highly reliable. Lastly, further researches are still needed to confirm the functional effects of the screened hub genes in human being to improve the prognosis and decrease the adverse left ventricular remodeling mortality.
Based on our current study, our research provided a bioinformatics analysis for the patients with AS. The screened hub DEMRGs, MMP2, FN1, FBN1, COL1A2 and COL3A1, may be therapeutic targets for treatment in patients with heart failure and prevention the heart failure induced dilated cardiomyopathy, ischemic cardiomyopathy or hypertension. MMP2, FN1, FBN1, COL1A2 and COL3A1 expressions increased due to the adverse left ventricular remodeling and the kit of the 5 hub DEMRGs may test left ventricular remodeling events and help prevent the adverse left ventricular remodeling to decrease the mortality and morbidity.
CAD, Coronary artery disease; MI, Myocardial infarction; HF, Heart failure; LVM,
Left ventricular mass index; GEO, Gene Expression Omnibus; AS, aortic stenosis;
PCC, Pearson correlation coefficients; DEGs, Differentially expressed genes;
DEMRGs, Differentially expressed M2 Macrophages-related genes; DEBRGs,
ifferentially expressed B cells-related genes; GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment Analysis; PPI,
Protein-protein internet; TF, Transcription factor; CTD, Comparative
Toxicogenomics Database; ROC, Receiver operation characteristic; AUC, Area under
curve; WGCNA, Weighted gene co-expression network analysis; ECM, Extracellular
matrix protein; MMP2, Matrix metallopeptidase 2; TGF-
Conceptualization, YZ; Funding acquisition, WQG, YHL and TL; Investigation, YZ, WQG, BCQ, ZCQ and XMH; Visualization, YZ and YHL; Writing – original draft, YZ; Writing – review & editing, YZ, XMH and TL.
Not applicable.
We would like to express our gratitude to all those who helped us during the writing of this manuscript. Thanks to all the peer reviewers for their opinions and suggestions.
This work was supported by the Tianjin “Project + Team” Key Training Special Project, China (no. XC202040), the Tianjin “131” Innovative Talent Team Project (no. 201939), the Tianjin Science and Technology Support Key Project (no. 18YFZCSY01080), the Tianjin Municipal Health and Health Committee Science and Technology Project (no. ZD20001), Tianjin Health Committee traditional Chinese medicine and integrated traditional Chinese and Western medicine project (no. 2021139), and the Tianjin Municipal Health and Health Committee Science and Technology Talent Cultivation Project (no. KJ20008).
The authors declare no conflict of interest.
