†These authors contributed equally.
Academic Editor: Zhonghua Sun
Background: Recent studies have shown that epicardial adipose tissue
(EAT) is an independent atrial fibrillation (AF) prognostic marker and has
influence on the myocardial function. In computed tomography (CT), EAT volume
(EATv) and density (EATd) are parameters that are often used to quantify EAT.
While increased EATv has been found to correlate with the prevalence and the
recurrence of AF after ablation therapy, higher EATd correlates with inflammation
due to arrest of lipid maturation and with high risk of plaque presence and
plaque progression. Automation of the quantification task diminishes the
variability in readings introduced by different observers in manual
quantification and results in high reproducibility of studies and less
time-consuming analysis. Our objective is to develop a fully automated
quantification of EATv and EATd using a deep learning (DL) framework.
Methods: We proposed a framework that consists of image classification
and segmentation DL models and performs the task of selecting images with EAT
from all the CT images acquired for a patient, and the task of segmenting the EAT
from the output images of the preceding task. EATv and EATd are estimated using
the segmentation masks to define the region of interest. For our experiments, a
300-patient dataset was divided into two subsets, each consisting of 150
patients: Dataset 1 (41,979 CT slices) for training the DL models, and Dataset 2
(36,428 CT slices) for evaluating the quantification of EATv and EATd.
Results: The classification model achieved accuracies of 98% for
precision, recall and F
Epicardial adipose tissue (EAT), the fat located between the myocardium and the visceral pericardium [1] which serves as an energy store [2], has been hypothesized as a contributor to the inflammatory burden via paracrine mechanisms [3]. EAT has been suggested as an independent marker for cardiovascular risk [2, 4, 5, 6, 7, 8] and, in particular, as an independent atrial fibrillation (AF) prognostic marker [9]. In computed tomography (CT), EAT volume (EATv) and density (EATd) are parameters that are often used to quantify EAT [8]. EATv refers to the extent of EAT accumulation and increased EATv has been found to correlate with the prevalence and the recurrence of AF after ablation therapy [1, 10, 11, 12, 13, 14, 15]. In addition to AF, increased EATv is also associated with atherosclerosis [16, 17], carotid stiffness [18], myocardial infarction [19], and coronary artery calcification [20, 21]. EATv has been associated with severity of coronary artery disease (CAD) [6, 22, 23]. A higher EATd (i.e., radiodensity of EAT) in CT images is correlated with inflammation as a result of arrest of lipid maturation [24]. Researchers have also suggested a link between EATd and high risk mortality and plaque presence and progression [24].
These health risk factors emphasize the need for direct quantification of EAT (i.e., EATv and EATd). However, manual quantification is time-consuming to accomplish in clinical practice in the light of the high workload on physicians and radiographers, and so EAT is not routinely quantified. Automation of the quantification task diminishes the variability in readings introduced by different observers and removes the high dependence of state-of-the-art methods on user interaction for EAT segmentation, resulting in high reproducibility of studies and less time-consuming analysis. In general, fully automated quantification of EATv and EATd requires advanced techniques and, in this work, we propose a deep learning (DL) framework for carrying out the estimation of these two quantities autonomously. DL techniques, a set of machine learning methods, have proved to be very effective for automated detection and segmentation of a wide range of medical images with a high degree of accuracy [25, 26, 27, 28, 29]. The proposed methodology, therefore, mainly consists of image classification and segmentation DL-based models to perform the desired quantification. We compare the performance of our approach to other DL approaches proposed in the literature for accomplishing EAT quantification.
The segmentation of EAT in cardiac CT (CCT) image slices is important for EAT quantification and various semi-automatic segmentation approaches have been developed [30, 31, 32]. An overview of these approaches can be described as follows: after initial preprocessing involving the removal of all other structures in the CT images apart from the heart, these methods require an expert to scroll through the CT slices to identify some control points along the border of the pericardium, then use some interpolation methods (such as the cubic spline function techniques) to obtain smooth pericardial contour, and then identify the pericardial fat by thresholding.
Fully automated and semi-automated non-DL based EAT segmentation approaches have also been developed [33, 34, 35, 36, 37, 38]. The time taken for obtaining the segmentation masks per patient, according to [35], could be more than 15 minutes for such fully automated approaches. The DL-based automatic EAT quantification methods that have been reported for EAT segmentation or quantification include those by Commandeur et al. [39] and Li et al. [40]. In Commandeur et al. [39], two DL-based models were developed; one is used to determine heart limits and perform heart segmentation (i.e., thoracic mask segmentation) and the other was used in combination with a statistical shape model for the detection of the pericardium. EAT was then quantified by further post-processing using thresholding [–190, –30] HU. In Li et al. [40], a DL-based model was developed for the segmentation of the pericardium across multiple adjacent slices using multiple slices as input to the model. A smoothing operation is then employed by finding a solution to a partial differential equation of the 3-dimensional gradient vector flow in order to reduce the prediction of false positive and negative regions in the segmented pericardial images. EAT is then deduced by thresholding [–175, –15] HU.
To our knowledge, there is no unified method in the literature capable of both autonomous EATv and EATd. Methods exist for the EAT segmentation and EATv estimation [39, 40, 41, 42], but our approach differs from these methods in that EAT segmentation does not require any further post-processing (e.g., thresholding or smoothening operations with filters) after the prediction with the DL-based EAT segmentation model. Also, no previous work has attempted to estimate EATd alone with DL nor combine the estimation of EATv and EATd. Although some authors, such as [41, 42], have stated their quantification of EATd as the mean attenuation of EAT segmented using DL models, we are not aware of any previous work that demonstrates or carried out practical analysis of the EATd quantification using DL. Our results in the analysis of EATd strengthens the correctness of our approach of EAT segmentation and emphasizes the correctness of the results obtained for EATv estimation.
In this paper, the presentations related to DL follow the recommendation of the Proposed Requirements for Cardiovascular Imaging-Related Machine Learning Evaluation guidance [43]. The overview of the proposed framework for estimating the volume and attenuation of EAT is shown in Fig. 1 and consists of two DL models. The classification model performs the task of selecting images containing the EAT from the set of all CT images acquired for a given patient. The image segmentation model obtains the segmentation masks marking the regions of the EAT from the selected images of the preceding process. The estimation of EATv is computed using the segmentation masks while the mean attenuation of the totality of EATv, that is EATd (i.e., mean density of EAT) [8], is quantified by extracting the intensity values of the EAT from the CT images using the masks to define the region of interest (ROI). More details on these processes are given in subsequent subsections.
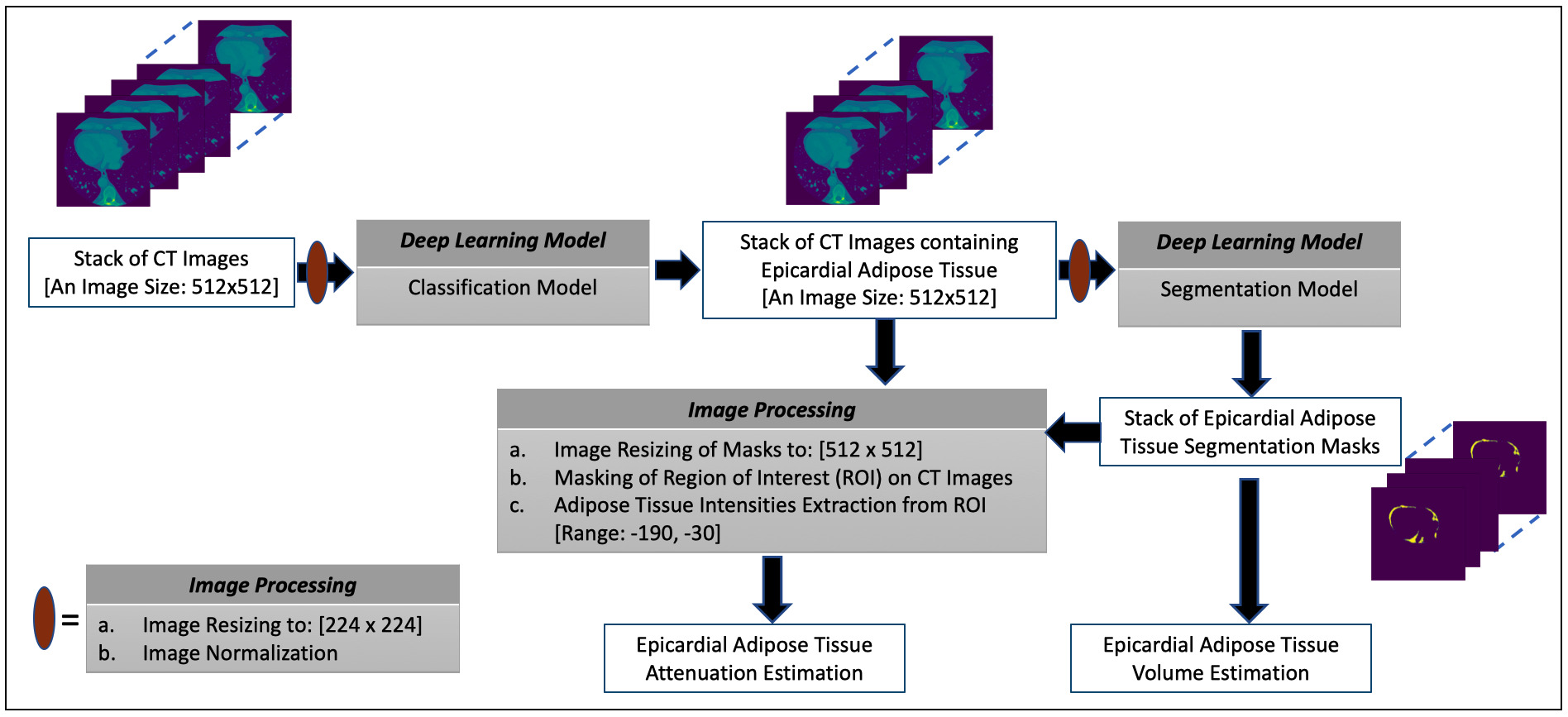 Fig. 1.
Fig. 1.The overview of the proposed framework for estimating the volume and attenuation of EAT.
We included 300 patients for this retrospective observational study who underwent catheter ablation for symptomatic, anti-arrhythmic medication-refractory atrial fibrillation at MedStar Georgetown University Hospital in Washington, D.C. All patients gave written informed consent and underwent CCT for pre-operative assessment. The de-identification of the images was carried out prior to analysis. The approval for the study was given by the Georgetown University Institutional Review Board (STUDY-0400, approved 7/20/2017). A total of 78,407 images from the 300 patients were available for the study. This dataset is used for all the analysis, modelling and evaluation in this paper. Table 1 provides the baseline characteristics of the patients. All the 300 patients in the cohort have a history of atrial fibrillation, with 65% having paroxysmal AF, while the others have non-paroxysmal AF (i.e., either persistent or long-standing persistent AF).
| Baseline Characteristics | Total (N = 300) |
|---|---|
| Age, years | 63.2 ( |
| Men, n (%) | 200 (66) |
| BMI (kg/m |
31.1 ( |
| LAV (mL) | 135.3 ( |
| EAT Volume (mL) | 98.4 ( |
| EAT Attenuation (HU) | –84.5 ( |
| Paroxysmal AF, n (%) | 194 (65) |
| Values are numbers and percentage (%) of the variables ( Abbreviations: AF, atrial fibrillation; BMI, body mass index; EAT, epicardial adipose tissue; HU, Hounsfield Unit; LAV, left atrial volume. | |
CCT was acquired on a 256-slice Multidetector CT scanner (Brilliance iCT,
Philips Healthcare, Cleveland, OH, USA). It had a detector collimation of 128
The delivery of contrast agent was controlled by automatic bolus tracking with
defining a region of interest (ROI) in the center of descending aorta at aortic
root level. The initiation of the scan was after a post-threshold delay of 6 sec
after the signal attenuation reached a predetermined threshold of 120 HU in the
descending aorta. The intravenous contrast administration protocol included 60 mL
iohexol (Omnipaque; GE Healthcare; Chicago, IL, USA) at a rate of 5 mL/s with at
120 kV (2% were at 80 kV based on Body Mass Index (BMI)
For the reconstruction of image after scanning and evaluation of the quality of
images scanned, a dedicated workstation (Extended Brilliance Workspace [EBW]
Version V4.5.2.40007, Philips Healthcare, Cleveland, OH, USA) was used. Xres
Standard filter (XCB, Philips Healthcare, Cleveland, OH, USA) was used for the
purpose of image reconstruction with a reconstruction field of view of 500 mm and
image matrix 512
Using the semi-automated post-processing program 3D Slicer (a free, open source software – Version 4.11.0) [44], we analyzed CCT scans and carried out the manual segmentation of EAT using axial views as follows [45]: the EAT was encircled on each 2D slice of the CCT from the bifurcation of the pulmonary artery superiorly to the diaphragm inferiorly, carefully tracing the pericardium to ensure inclusion of epicardial adipose tissue only. This protocol was designed in alignment with the definition of EAT as the fat deposits inside the pericardium (i.e., adipose tissue within the pericardial sac) with the voxels between -190 and -30 Hounsfield units (HU) [8, 46]. 3D Slicer calculates the area of each corresponding EAT section using the HU range of -190 to -30 and estimates EATd and EATv taking into consideration the distance between adjacent planes. The manual segmentation task was performed for all 300 CCTs by physician MSB (with three years of experience in cardiac CT analysis and trained by JDV, a level 3 certified cardiologist with 10 years of experience) and JDV. Both inter-observer correlations (0.822 for EAT volume and 0.934 for EAT attenuation) and intra-observer correlations (0.957 for EAT volume and 0.956 for EAT attenuation) were strong for the manual segmentation task.
All experiments were conducted on a Nvidia Tesla M40 machine with Python programming language using TensorFlow 2.0 Python API machine learning framework (Version 2.7.0, Google Brain, Google Inc., Mountain View, CA, USA) [47].
We consider the ResNet (ResNet50) [48] DL architecture for the image classification task of selecting images containing the EAT from the set of all CT images acquired for a given patient, eliminating slices above the bifurcation of the pulmonary trunk or below the cardiac apex. That is, the model classification automatically identifies slices above the superior extent of the left main coronary artery and also those slices below the cardiac apex for elimination in further analysis. A detailed description of the ResNet50 architecture is given in Supplementary Table 1 and Supplementary Materials.
During model training, the input CCT images were resized to 224
For our experiments in this study, our 300-patient dataset is divided into two subsets, Dataset 1 (41,979 CT slices) and Dataset 2 (36,428 CT slices), with each subset consisting of 150 patients. We used Dataset 1 for training, validating and evaluating the DL models, and Dataset 2 for evaluating EATv and EATd estimations. Splitting our dataset in this way ensures that in the estimation of EATv and EATd, the DL models trained on Dataset 1 have not seen the CT slices in Dataset 2 during training, allowing us to have a proper evaluation of the proposed methods for EATv and EATd quantification.
The CT image dataset of 150 patients (Dataset 1) of which 23,771 images contain the EAT were used to train, validate and test the ResNet50 model. In particular, 15% of the images were selected; these were then divided into two equal sets representing the validation and test sets. Thus, the number of the training, validation and test (evaluation) images are 35,683, 3148 and 3148, respectively.
Of the 35,683 images in the training set, the number of images with the EAT
present and absent are 20,234 and 15449, respectively. We used weighted loss
function to address this data imbalance. Let
where
where
The image segmentation task of masking the regions of the EAT from the CT slices is performed by the UNet DL model [49]. A detailed description of the UNet architecture is given in Supplementary Table 2 and Supplementary Materials. Briefly, the architecture includes batch normalization to enhance robustness of the model [50] and ‘dropout’ operation [51] to avoid problems associated with model overfitting.
During model training, the input CCT images were resized to 224
For
where
The CT image dataset of 150 patients in Dataset 1 with the EAT present (23,771) were used to train, validate and test the EAT segmentation models. In particular, 15% of these images were selected and divided into two equal sets namely, the validation and test sets. Thus, the numbers of images in training, validation and test sets are 20,207, 1782 and 1782, respectively.
The dice score, or dice similarity coefficient (DSC), is a measure of similarity between the label and predicted segmentation masks. DSC score can be written as follows:
where
The estimation of EATv involves the integration (summation) of the interslice volumes. Each interslice volume is approximated by computing the volume between two consecutive slices using the following equation:
where
where
EATd is estimated by computing the mean attenuation of EAT across all the slices. The intensity values of the EAT for each slice, with range [–190, –30], is extracted using its segmentation mask obtained from the EAT segmentation model to mark the ROI. EATd can be expressed as follows:
where
Since the size of the input image and output segmentation mask of the
segmentation model is 224
The performance metrics of the ResNet50 classification model on the evaluation
dataset of N = 3148 are precision (0.980), recall (0.986) and F
| Predicted Label (0) | Predicted Label (1) | |
|---|---|---|
| Actual Label (0) | 1388 | 24 |
| Actual Label (1) | 34 | 1702 |
 Fig. 2.
Fig. 2.Some examples of the prediction of the classification model. Two examples are given for each of the cases (0 – absence of EAT, 1 – presence of EAT): (0/0) (i.e., ground truth/prediction), (0/1), (1/0) and (1/1). For the (1/0) and (1/1) cases, the images to the right show the EAT in red as given by an expert human reader. The (0/1) and (1/0) cases are images which the classification model got wrong.
The performance metrics of the segmentation model on the 1782 evaluation set are
given as follows: the mean DSC is 0.844 (
| Mean (std. dev.) | Max. | Median | |
|---|---|---|---|
| Model 1 | 0.831 ( |
0.91 | 0.83 |
| Model 2 | 0.826 ( |
0.92 | 0.84 |
| Model 3 | 0.818 ( |
0.94 | 0.82 |
| Model 4 | 0.851 ( |
0.96 | 0.84 |
| Model 5 | 0.838 ( |
0.91 | 0.82 |
 Fig. 3.
Fig. 3.Some examples of the predictions of the segmentation model. The corresponding dice scores are shown at the bottom of each of the examples.
The regression, the kernel density estimates and the Bland-Altman plots of the
predicted volumes against the label volumes computed for the study population of
Dataset 1 (the ‘training’ dataset) and Dataset 2 (the ‘evaluation’ dataset), each
of which consists of 150 patients, using the proposed framework are shown in Fig. 4. Fig. 4a,c show the regression plots with volume estimated using Eqns. 5,6,7.
In our case,
 Fig. 4.
Fig. 4.The plots of the predicted volume against the label volume. Plot (a) represents the regression plot; plot (b) represents
the kernel density estimates and histogram plots of the two variables (label
volume and predicted volume) with the dashed vertical lines representing the
arithmetic mean of the distributions. The symbols
Similarly, the regression, the kernel density estimates and the Bland-Altman
plots of the predicted EATd versus the label EATd estimated for the study
population of Dataset 1 and Dataset 2 are given in Fig. 5. Fig. 5a,c show the
regression plots, and the Pearson correlation coefficient between the label and
predicted EAT mean attenuation are 0.964 (R
 Fig. 5.
Fig. 5.The plots of the predicted attenuation against the label attenuation. Plot (a) represents the regression plot and plot (b) represents
the kernel density estimates and histogram plots of the two variables (label and
predicted mean attenuations) with the dashed vertical lines representing the
arithmetic mean of the distributions. The symbols
In our task of quantifying EATv and EATd, we developed the ResNet50 DL
classification model for selecting images containing the EAT from the set of CT
images acquired for a given patient. We then used a UNet segmentation model for
obtaining the segmentation masks marking the regions of the EAT from the selected
images. The estimation of EATv and EATd are computed using the segmentation masks
and by extracting the intensity values of the EAT from the selected CT images.
Using the regression, the kernel density estimates and the Bland-Altman plots, we
have shown that the proposed framework is able to estimate EATv and EATd with a
high degree of accuracy. To summarize, we reported the performance of the
classification model in terms of precision, recall and F
The performance of the method proposed in [39] was evaluated for EAT
segmentation on 10% of the dataset of 250 subjects (i.e., 25 in the evaluation
set) and gave a median DSC of 0.823. EATv on the 250 patients gave a correlation
of (R
Also, the method proposed in [40] was trained on a dataset of 88 subjects, and reported a mean DSC score of 0.973 using an evaluation dataset of only 15 subjects; thus, the evaluation dataset is not large enough for making a fair comparison with other methods. In addition, our approach differs from the method presented in [40] in that it does not require smoothing operation by solving a differential equation nor any post-processing step via thresholding.
Our models were trained on a dataset from a single hospital system. The data augmentation techniques, the batch normalization and dropout operations and the weighted loss function for addressing data imbalance used during model training may have improved the chance that performance may not deteriorate significantly from datasets from elsewhere but it would be useful assessing the performance of the models using an external dataset. If needed, the performance of the models may then be improved with training on multi-centre datasets to enhance generalizability with little or no modification to the proposed methods. Approaches that may be explored to address model generalization issues include transfer learning and federated-learning [53].
In relation to gender, men constitute the majority (66%) of the dataset we have used for model training. In the evaluation of our models trained with this dataset using 5-fold cross-validation (Table 3), there is no indication of biasness of the models at slice level towards a particular gender. Biases in the training datasets can affect performance of DL models. As such, future work on this research would focus on investigating the estimation of EATv and EATd at patient-level for biasness. A possible approach for addressing biasness in CT images is by balancing the dataset using generative models in the form of data augmentation. An in-depth discussion on generative models is beyond the scope of this paper and we refer readers to [54] for more details on this technique.
Our framework focused on EATv and EATd quantification for CT images, future work will focus on training using heterogenous multi-centre dataset as well as on analysis of EAT for cardiovascular risk and outcome prediction. Future direction of this work will also include using machine learning methods for quantifying the distribution of EAT given that the location of EAT is a disease-specific risk factor (e.g., thickness of peri-atrial EAT being a predictor of AF recurrence [55]).
We proposed a novel and clinically useful framework that consists of DL models for EAT quantification. The framework provides a fast and robust strategy for accurate EAT segmentation, and volume (EATv) and attenuation (EATd) quantification tasks. It fully automates the process of computing EATv and EATd. The framework we have proposed in this paper will be useful to clinicians and other practitioners as a first step which they can build upon in order to develop DL models for carrying out reproducible EAT quantification at patient level or for large cohorts and high-throughput projects, creating prognostic EAT data for better further analyses.
AF, atrial fibrillation; CCT, cardiac CT; CT, computed tomography; DL, deep learning; DSC, dice similarity coefficient; EAT, epicardial adipose tissue; EATv, EAT volume; EATd, EAT density (mean EAT attenuation).
The datasets presented in this article are not publicly available because restrictions apply to the availability of these raw data, which were used under license for the current study from Georgetown University Institutional Review Board. Generated anonymized dataset are however available from the authors upon reasonable request and with permission of Georgetown University Institutional Review Board. Requests to access the datasets should be directed to Jose D. Vargas, jose.vargas@nih.gov.
MA, MSB, JDV, and SEP conceived the idea and contributed to the analysis; MSB, JDV, FZ, ATaylor, AThomides, PJB, and MBS developed the contouring method; MA led on the machine learning methodology and the main mathematical and statistical analysis of CT data and images; MSB and JDV advised on cardiac CT analysis and validation; MA, AML, and SEP advised on data governance and computing infrastructure; MA drafted the first version of the manuscript; JDV and SEP provided overall supervision; ER and IU provided critical feedback on the initial draft of the manuscript; all authors contributed to the content, the writing of the final version, and provided critical feedback. All authors read and approved the final manuscript.
The studies involving human participants were reviewed and approved by Georgetown University Institutional Review Board (STUDY-0400, approved 7/20/2017). Dataset was fully anonymized. The patients/participants provided their written informed consent to participate in this study.
Not applicable.
MA and SEP acknowledge support from the CAP-AI programme (led by Capital Enterprise in partnership with Barts Health NHS Trust and Digital Catapult and funded by the European Regional Development Fund and Barts Charity) and Health Data Research UK (HDR UK—an initiative funded by UK Research and Innovation (UKRI), Department of Health and Social Care (England) and the devolved administrations, and leading medical research charities; www.hdruk.ac.uk). SEP and AML acknowledge support from the National Institute for Health Research (NIHR) Biomedical Research Centre at Barts, from the SmartHeart EPSRC programme grant (www.nihr.ac.uk; EP/P001009/1) and the London Medical Imaging and AI Center for Value-Based Healthcare (AI4VBH). SEP has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 825903 (euCanSHare project). ER acknowledge support by the London Medical Imaging and AI4VBH, which is funded from the Data to Early Diagnosis and Precision Medicine strand of the government’s Industrial Strategy Challenge Fund, managed and delivered by Innovate UK on behalf of UKRI.
The authors declare no conflict of interest.
