1 Department of Obstetrics and Gynecology, Peking University First Hospital, 100034 Beijing, China
Abstract
Background: Endometriosis (EM), a gynecological disorder that is dependent on estrogen and causes inflammation, is prevalent among women of reproductive age and is considered a chronic condition. The involvement of noncoding RNAs in exosomes is crucial for the progression of EM. This study aimed to determine exosomal microRNA (miRNA) biomarkers in EM. Methods: Exosomes were isolated and characterized from the plasma of patients with EM and controls. Exosomal miRNA was sequenced using microarrays. EM-related differential miRNAs (DE-miRNAs) were identified using analysis of differential miRNA expression and weighted coexpression network analysis. The common pairs of long noncoding RNA (lncRNA)-miRNA and miRNA-mRNA were determined. Cytoscape was used to establish the regulatory network of characteristic genes known as competitive endogenous RNA (ceRNA), and the hub miRNAs, hub mRNAs, and hub lncRNAs were identified. Results: We isolated plasma exosomes from 10 control and 10 EM patients. We obtained a total of 50 DE-miRNAs, consisting of 7 miRNAs that were upregulated and 43 miRNAs that were downregulated. A network of ceRNA regulation was constructed using the diagnostic miRNAs, which revealed a total of 36 lncRNAs, 20 miRNAs, and 264 mRNAs associated with EM. Additionally, 10 lncRNAs (GAS5, MALAT1, FGD5-AS1, HCG18, SNHG16, XIST, OIP5-AS1, NEAT1, KCNQ1OT1, and SNHG12), 10 miRNAs (hsa-miR-361-5p, hsa-miR-19b-3p, hsa-let-7f-5p, hsa-miR-23a-3p, hsa-miR-199a-3p, hsa-miR-18a-5p, hsa-miR-221-3p, hsa-miR-17-5, hsa-miR-27a-3, and hsa-miR-25-3p), and 10 mRNAs (GALC, ETNK1, RNF4, SOX4, ZBTB18, SPRY2, RUNX1, MYLIP, BTG2, and MAP2K4) were identified as hub molecules. Conclusions: Thirty plasma exosomal miRNA markers associated with endometriosis were identified and reported. The miRNAs were associated with the promotion of proliferation in mesenchymal cells, as well as the tumor necrosis factor (TNF) and Toll-like receptor signaling pathways, and the differentiation of T helper 1 (Th1) and Th2 cells. These biological processes and pathways could potentially play a significant role in the pathogenesis and progression of EM. The potential clinical value of these miRNAs indicates potential targets for diagnosing and treating endometriosis while also offering new insights into the molecular mechanisms of the disease.
Keywords
- endometriosis
- ceRNA
- miRNA
- enrichment analysis
- bioinformatic analysis
Endometriosis, also known as EM, is a medical condition characterized by the growth of endometrial tissue (including glands and stroma) outside the uterus and myometrium [1]. Endometriosis, a prevalent gynecological condition, frequently causes enduring pelvic pain, painful periods, and reduced fertility, impacting the well-being of approximately 10–15% of women in their reproductive years [2]. Endometriosis lesions are frequently found in the ovaries and pelvic peritoneum, although they can also develop in the fallopian tubes, cervix, vagina, and abdominal wall [3]. Identifying early endometriosis is challenging in clinical practice due to the nonspecific symptoms, despite operative laparoscopy being the gold standard for diagnosis. In addition to the requirement for surgical verification in certain instances, the typical duration from the beginning of symptoms to the identification of endometriosis is 6.7 years [4]. Currently, surgical resection combined with pharmacological therapy is considered an effective treatment for endometriosis [5]. Nevertheless, the failure to diagnose early often leads to a substantial recurrence rate and adverse effects in surgical interventions [6]. Hence, the development of diagnostic indicators and therapeutic objectives for endometriosis is crucial to offer novel treatment strategies and establish a foundation for deeper comprehension of the disease’s molecular mechanisms.
Cells secrete exosomes, which are vesicles enclosed by a lipid bilayer and
contain releasable RNA and proteins. These exosomes have a crucial function in
facilitating the exchange of information among cells [7]. The RNA in exosomes can
remain stable due to protection from degradation by RNA enzymes [8]. MicroRNAs
(miRNAs) are noncoding RNAs that naturally function as short RNAs consisting of
22–24 nucleotides. Their main role is to regulate protein expression by
primarily targeting the 3
This research utilized miRNA sequencing and bioinformatics analysis to discover blood exosomal miRNA indicators for endometriosis, aiming to offer potential targets for clinical diagnosis and treatment.
This study included patients who were diagnosed with endometriosis through laparoscopy at Peking University First Hospital. Exclusion criteria included individuals experiencing abnormal menstrual cycles, those who had taken hormonal medications or used intrauterine devices in the three months preceding the surgery, and individuals with medical complications or pelvic inflammatory disease. A group of individuals ranging in age from 27 to 40 years who were experiencing menstrual periods in the proliferative phase were chosen for inclusion in the disease group. The control group consisted of ten patients (aged 28–40 years) who had laparoscopy for ovarian teratoma, simple ovarian cyst, fallopian tube cyst, and uterine fibroids during the proliferative phase of menstruation. Supplementary Table 1 displays the characteristics of the patients. The research was carried out following the guidelines of the Declaration of Helsinki, and the Ethics Committee of Peking University First Hospital approved the protocol (approval number No. 2020KEYAN343). Following the acquisition of informed consent from every participant, 10 mL of plasma was extracted from both the patient and control groups.
As per the established procedure, plasma samples were spun at 2000
Initially, the exosome microstructure was examined using transmission electron microscopy (TEM). A copper grid was used to place a 10 µL sample of extracellular vesicles, which was then precipitated for 1 min. Afterwards, the excess liquid was eliminated using filter paper. Afterwards, phosphotungstic acid was introduced onto the copper grid and left to precipitate for a duration of 1 minute, following which the excess liquid was once more eliminated using filter paper. The specimen was permitted to air dry for a few minutes at ambient temperature. The specimen was left to dry naturally at ambient temperature for a few minutes. After the sample had dried, imaging results were obtained using a Hitachi HT7700 electron microscope (Hitachi, Ltd., Tokyo, Japan) with an acceleration voltage set between 40–120 kV (in 100 V increments). Ultimately, the instrument yielded imaging results through transmission electron microscopy.
Furthermore, the exosomes’ size distribution and quantification were examined
using nanoparticle tracking analysis (NTA). The exosome samples, which were
isolated, were diluted in 1
In the end, an analysis called western blotting was performed to investigate the presence of exosome-positive indicators (CD9, CD63, and TSG101) as well as an exosome-negative indicator (calnexin). Exosome lysates were acquired by combining radio immunoprecipitation assay (RIPA, Catalog# R0010, Solarbio, Beijing, China) solution with protease and phosphatase inhibitors, then measured using a bicinchoninic acid assay (BCA, Catalog# PC0020Solarbio, Beijing, China) Protein Assay Kit. Equivalent quantities of exosomal proteins from every group were analyzed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‒PAGE, Catalog# P1200, Solarbio, Beijing, China), followed by the transfer of proteins on the gel onto a nitrocellulose membrane. The filters were obstructed using 5% low-fat milk and subsequently placed in an incubator with the primary antibodies for an extended period at a temperature of 4 °C. Following the wash with phosphate buffered solution (PBST, Catalog# P1033, Solarbio, Beijing, China), the membranes were subjected to incubation with a secondary antibody that had been modified with horseradish peroxidase.
Afterwards, the exosome size distribution and quantification were examined using
nanoparticle tracking analysis (NTA). After being moderately diluted with
1
In the end, western blotting detected exosome-positive indicators (CD9, CD63, and TSG101) as well as exosome-negative indicator (calnexin). To acquire exosome lysates, RIPA-containing protease and phosphatase inhibitors were added and then quantified using a BCA protein analysis kit. The same quantity of exosome proteins from both groups underwent SDS‒PAGE, followed by the transfer of the proteins onto a nitrocellulose membrane. The membranes were obstructed using 5% skim milk and subsequently subjected to incubation with primary antibody for an extended period at 4 °C. Following the washing process using PBST solution, the membranes were subjected to incubation with a secondary antibody conditioned with horseradish peroxidase.
The main antibodies utilized were rabbit anti-CD63 [1:1000; System Biosciences (SBI), Catalog# EXOAB-CD63A-1; Palo Alto, CA, USA], rabbit anti-CD9 (1:1000; SBI, Catalog# EXOAB-CD9-1; Palo Alto, CA, USA), rabbit anti-TSG101 (1:1000; SBI, Catalog# EXOAB-TSG101-1; Palo Alto, CA, USA), and rabbit anti-Calnexin (1:1000; Immunoway Bio-technology , Catalog# YM0089, Plano, TX, USA). Exosome-validated goat anti-rabbit IgG (CD63, CD9; 1:5000; SBI) and BeriBlot for IP Detection Reagent (Calnexin, 1:2000; Catalog# ab10286, Abcam, Cambridge, UK) were the secondary antibodies used.
The miRNeasy Micro Kit (QIAGEN, Catalog #217084; Hilden, North Rhine-Westphalia, Germany) was utilized for the isolation of total RNA. Next, the RNA samples were utilized to produce a biotinylated cRNA that targeted the gene chip® miRNA 4.0 array. In short, after that, marked examples were added to the mixture for hybridization and then incubated as per the instructions provided by the manufacturer before being injected into the miRNA array. Following a 16-hours hybridization at a temperature of 48 °C, the array underwent washing, staining, and scanning with a GeneChip scanner (3000 7G GeneChip Scanner, Affymetrix, Santa Clara, CA, USA). Wayen Biotechnologies Inc. (Shanghai, China) carried out the chip experiment in accordance with Thermo Inc. protocol.
First, the quality was assessed by scanning the microarrays and plotting box
line plots. After annotating and counting the microarray data, the annotation
information and expression values of miRNAs were derived. To compare and screen
differentially expressed miRNAs between EM and control plasma exosomes, the R
package ‘limma’ (http://www.bioconductor.org/packages/release/bioc/html/limma.html) [15] was utilized.
The occurrence of false positive results was corrected by applying the Benjamini
and Hochberg false discovery rate (FDR) method and adjusting the p
value. To identify differential miRNAs (DE-miRNAs), the thresholds were set as an
adjusted p value
For this research, we developed a weighted gene co-expression network analysis
(WGCNA) by utilizing miRNA expression data to create a network with weighted
coexpression. To begin with, a total of 20 samples were subjected to hierarchical
clustering using the Euclidean distance of expression. The purpose was to
identify any outliers within the samples and subsequently remove them [16]. Next,
the similarity matrix was constructed by calculating the correlation degree of
each miRNA pair. To establish the correlation threshold between miRNAs, a
suitable power within the range of 1 to 20 was chosen as the soft threshold. The
relationships between the scale-free network evaluation coefficient R2 and the
soft threshold
The intersection of these DE-miRNAs and module miRNAs yielded the key DE-miRNAs.
We examined the significant DE-miRNAs to determine their enrichment in signaling
pathways using the miRNA Enrichment Analysis and Annotation tool (miEAA) [19]. We
searched two databases: Gene Ontology (GO) Pathway and Kyoto Encyclopedia of
Genes and Genomes (KEGG) Pathway. Retained were ontology terms and KEGG pathways
that had a significant adjusted p value (
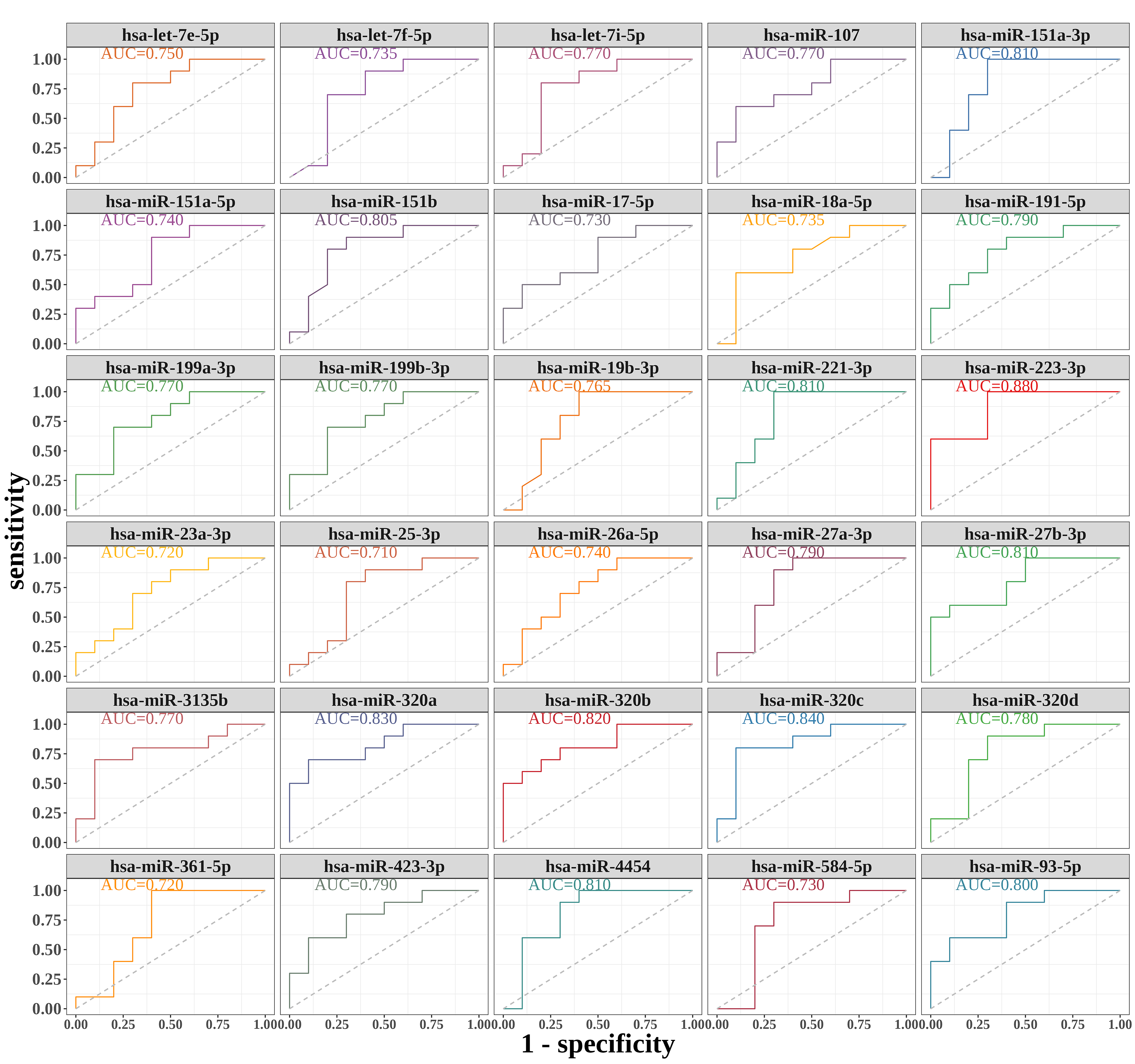
Evaluating the quality or performance of diagnostic tests was effectively done using the receiver operating characteristic (ROC) curve, which plotted the y coordinate as test sensitivity and the x coordinate as 1-specificity or false positive rate (FPR). To obtain the ROC curve and the area under the curve (AUC) value, the ‘pROC’ package was utilized [20]. The closer the AUC was to 1, the more accurate the diagnosis. The diagnostic effect was low when the AUC was between 0.5 and 0.7; there were some reference values in the diagnostic effect when the AUC was between 0.7 and 0.9; the diagnostic accuracy was high when the AUC was greater than 0.9; and there was no diagnostic value when the AUC was less than or equal to 0.5.
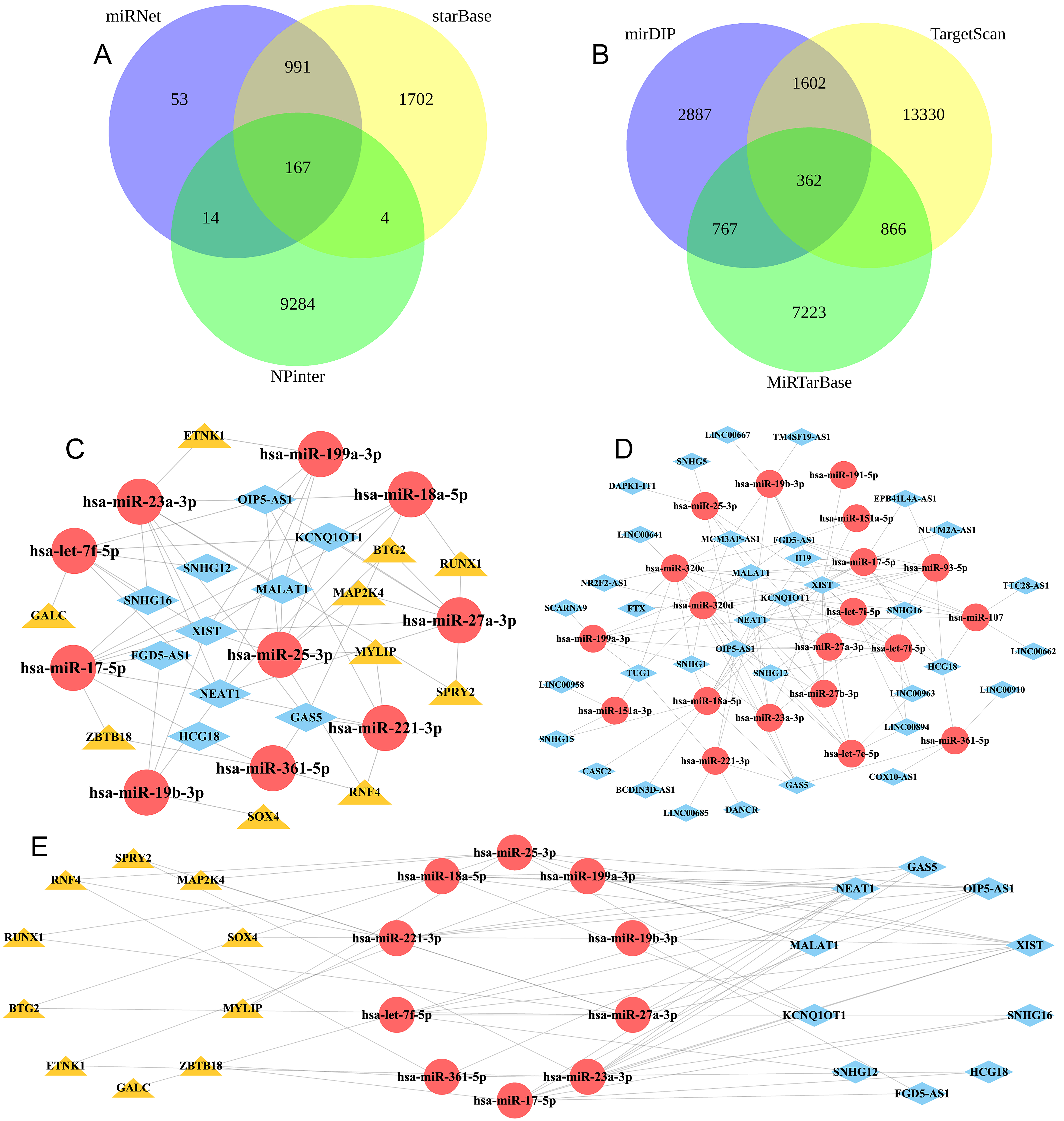
The mirDIP, TargetScan, and MiRTarBase databases can be accessed through the following links: mirDIP: http://ophid.utoronto.ca/mirDIP/; TargetScan: https://www.targetscan.org/vert_72/; MiRTarBase: https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php. The prediction of target mRNAs for diagnostic miRNAs was done using miRNA‒mRNA pairs screened from these three databases were then crossed. The Venn Diagram R package was utilized to create a Venn Diagram illustrating the intersection. To predict the diagnostic miRNAs’ target long noncoding RNAs (lncRNAs), the miRNet database (https://www.mirnet.ca/miRNet/upload/MirUploadView.xhtml), starBase database (https://rnasysu.com/encori/), and NPInter database (http://bigdata.ibp.ac.cn/npinter4/) were utilized. After that, the miRNA‒lncRNA relationship screened from these three databases was then crossed. The Venn Diagram R package was utilized to create a Venn Diagram illustrating the intersection.
A competitive endogenous RNA (ceRNA) network was constructed by utilizing the interplay among lncRNAs, miRNAs, and mRNAs. Next, the data were visualized using Cytoscape software (The Cytoscape Consortium, San Diego, CA, USA), and the analyze network tool was used to calculate the nodes’ degree. LncRNAs, miRNAs, and mRNAs with degrees in the ceRNA network were considered hub lncRNAs, miRNAs, and mRNAs.
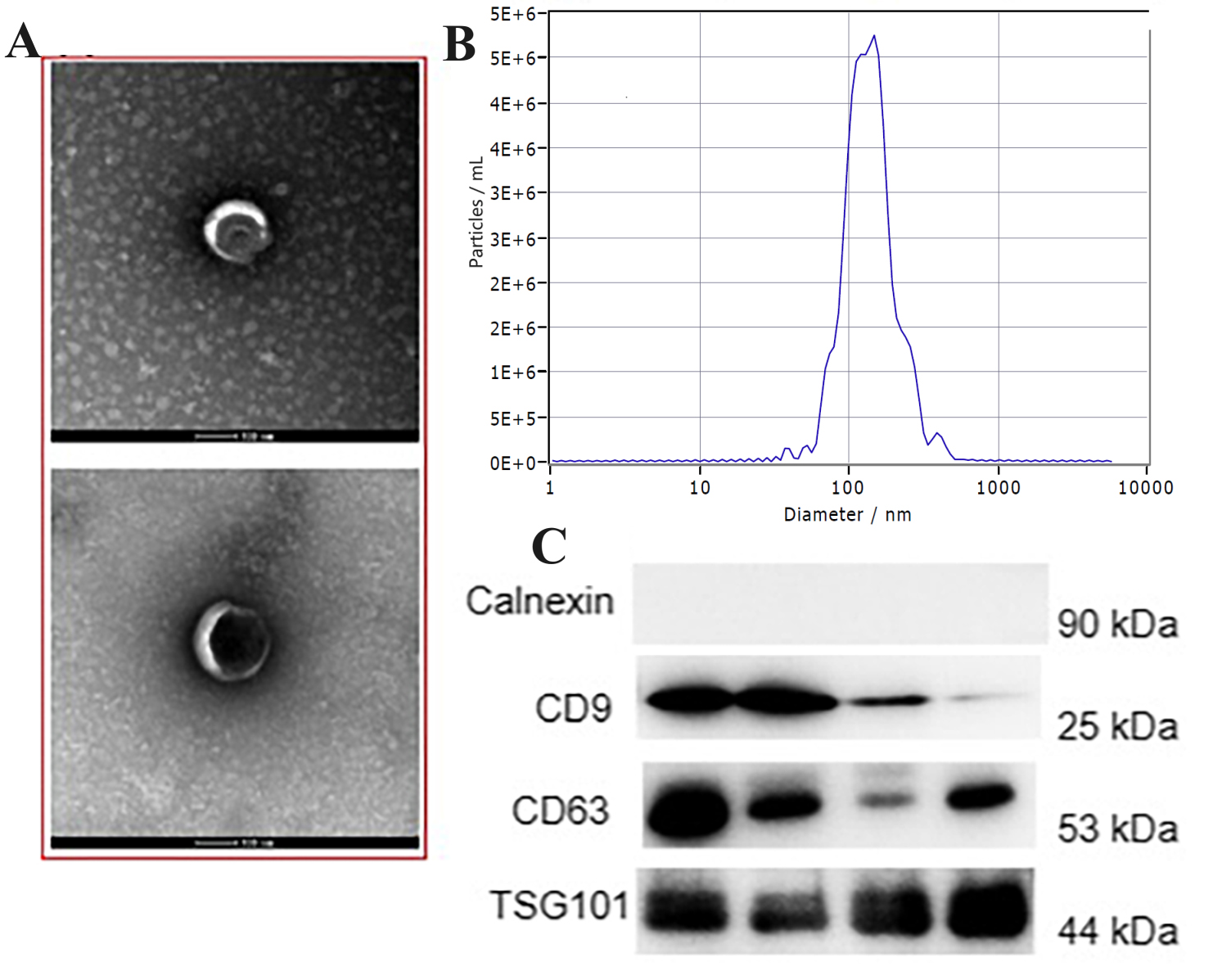
First, we isolated plasma exosomes from 10 control and 10 EM patients. Exosomes
were separated from the serum using ultracentrifugation. We employed TEM, NTA,
and western blotting techniques to ascertain the successful isolation of the
exosomes. Clear vesicle structures were visible under TEM (Fig. 1A). According to
the NTA findings, the exosomes fulfilled the requirements in terms of their
concentration (ranging from 3.5
 Fig. 1.
Fig. 1.Detecting exosomes derived from the endometrium. (A) Transmission electron microscopy (TEM) was used to observe the morphologies of exosomes derived from plasma. Scale bar = 100 nm; (B) Nanoparticle tracking analysis (NTA) was used to determine the dimensions and density of exosomes; (C) Exosomes were analyzed by western blotting using antibodies against exosomal markers (Calnexin, CD63, CD9 and TSG101).
We examined the presence of exosome-associated indicators (CD9, CD63, and TSG101) as well as a marker indicating the absence of exosomes (calnexin). The findings indicated that CD9, CD63, and TSG101 were expressed positively in the exosomes, while calnexin showed negative expression. On the contrary, the serum exhibited the opposite expression when exosomes were eliminated. Thus, we obtained qualified exosomes (Fig. 1C).
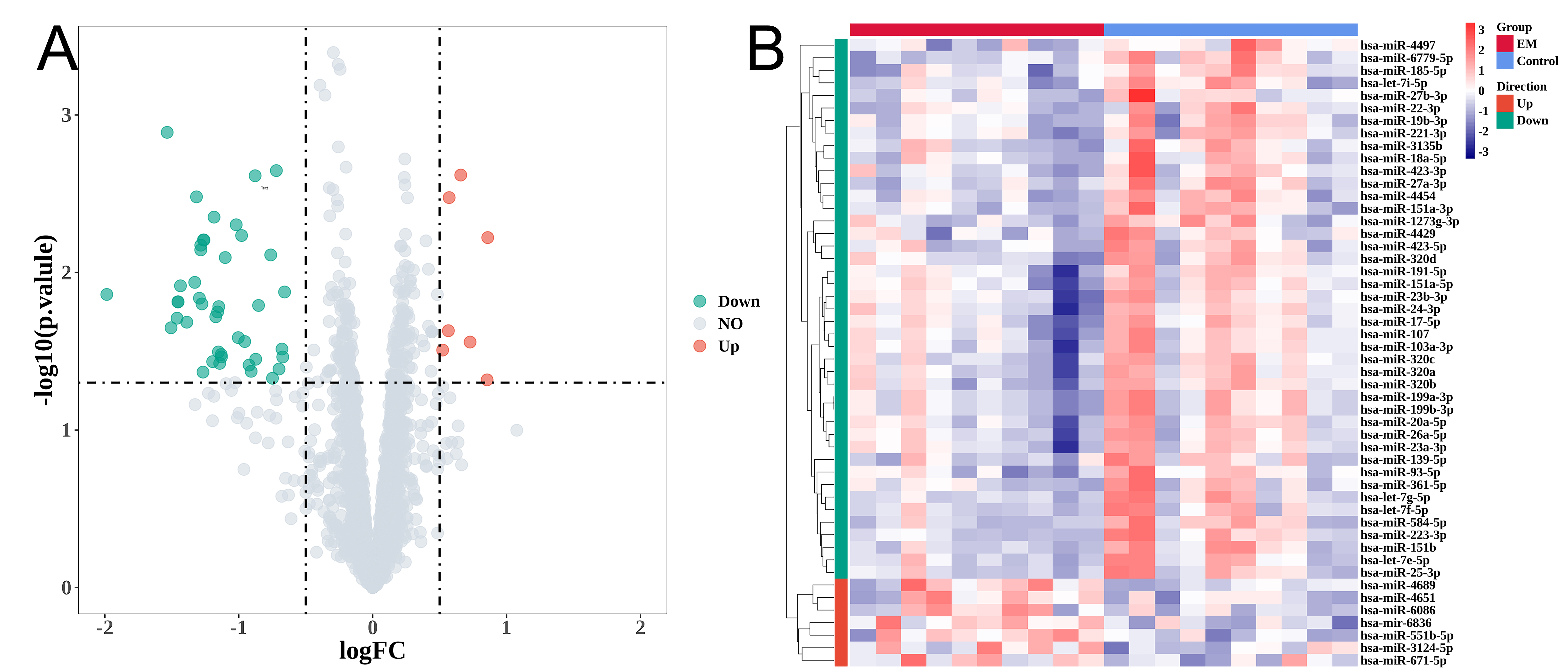
miRNA microarrays were used to extract and sequence total RNA from the eligible exosomes mentioned above. After quality control and preprocessing, we acquired the annotation information and expression values of the miRNAs. We utilized the R package ‘limma’ to conduct differential expression analysis to identify the distinctively expressed exosomal miRNAs among EM patients and control subjects. The DE-miRNAs were visualized by volcano plots (Fig. 2A). We obtained a total of 50 DE-miRNAs, consisting of 7 miRNAs that were upregulated and 43 miRNAs that were downregulated. The expression levels of the DE-miRNAs are shown in Fig. 2B.
 Fig. 2.
Fig. 2.Endometriosis is associated with differential expression of
microRNAs (miRNAs) in exosomes. (A) Volcano maps of differentially expressed
exosomal miRNAs. (B) Heatmaps displaying the miRNAs based on the magnitude of
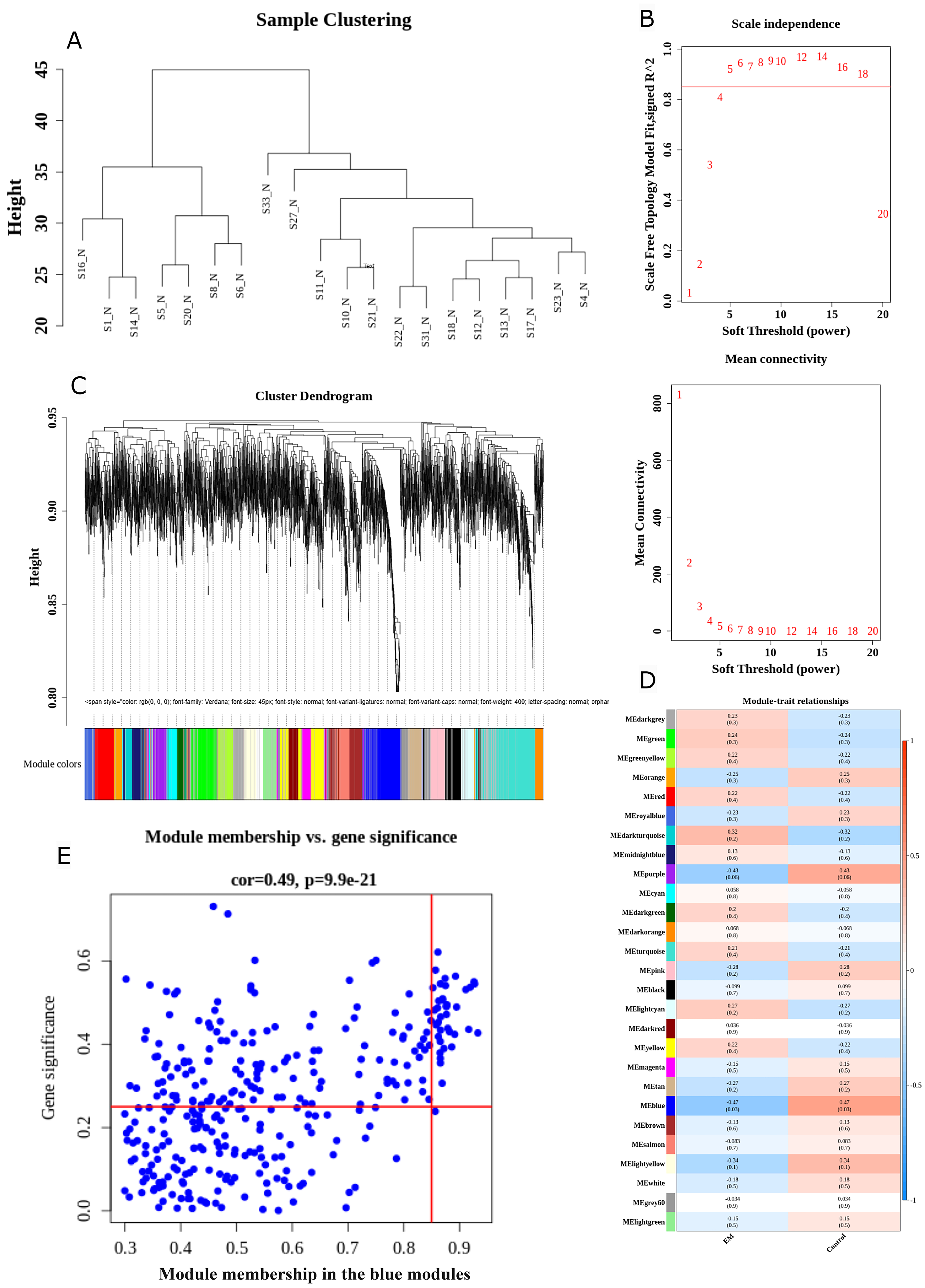
WGCNA was conducted on all miRNAs in this research utilizing the WGCNA function in the R package. To begin with first, a total of 20 samples were subjected to hierarchical clustering using the Euclidean distance of expression. The purpose was to identify any outliers within the samples and subsequently remove them [16]. As depicted in Fig. 3A, there were no apparent samples that stood out.
 Fig. 3.
Fig. 3.Analysis of dysregulated miRNAs using a weighted gene
coexpression network. (A) Sample hierarchical clustering by Euclidean distance
of expressions. (B) Examining the scale-free fit index and average connectivity,
we analyze the scale-free topology for different soft-threshold powers
(
In order to achieve a network that fulfills the scale-free topology requirement,
we computed various network structures by employing diverse soft-thresholding
powers spanning from 1 to 20. Distinct gene coexpression modules in EM were
determined by the power value (
Phenotypes and modules were correlated by utilizing samples from the EM and
control groups as phenotypes. The matrix of correlation coefficients between
module eigenvectors and phenotypic traits was computed, and heatmaps illustrating
the correlations were generated [16]. According to the module-EM relationships in
Fig. 3D, we determined that the blue module revealed the strongest correlation
(module-trait weighted correlation = –0.47, p
In order to further determine the key miRNAs, the internal module was examined to identify the genes exhibiting high GS (trait correlation with module gene, gene significance) and MM (gene correlation with module, module membership) [16]. The significant correlation between module membership (MM) in the blue module and gene significance (GS) for EM is presented in Fig. 3E. The coexpression modules that have been identified are displayed in various hues, with the blue module revealing a total of 38 core miRNAs.
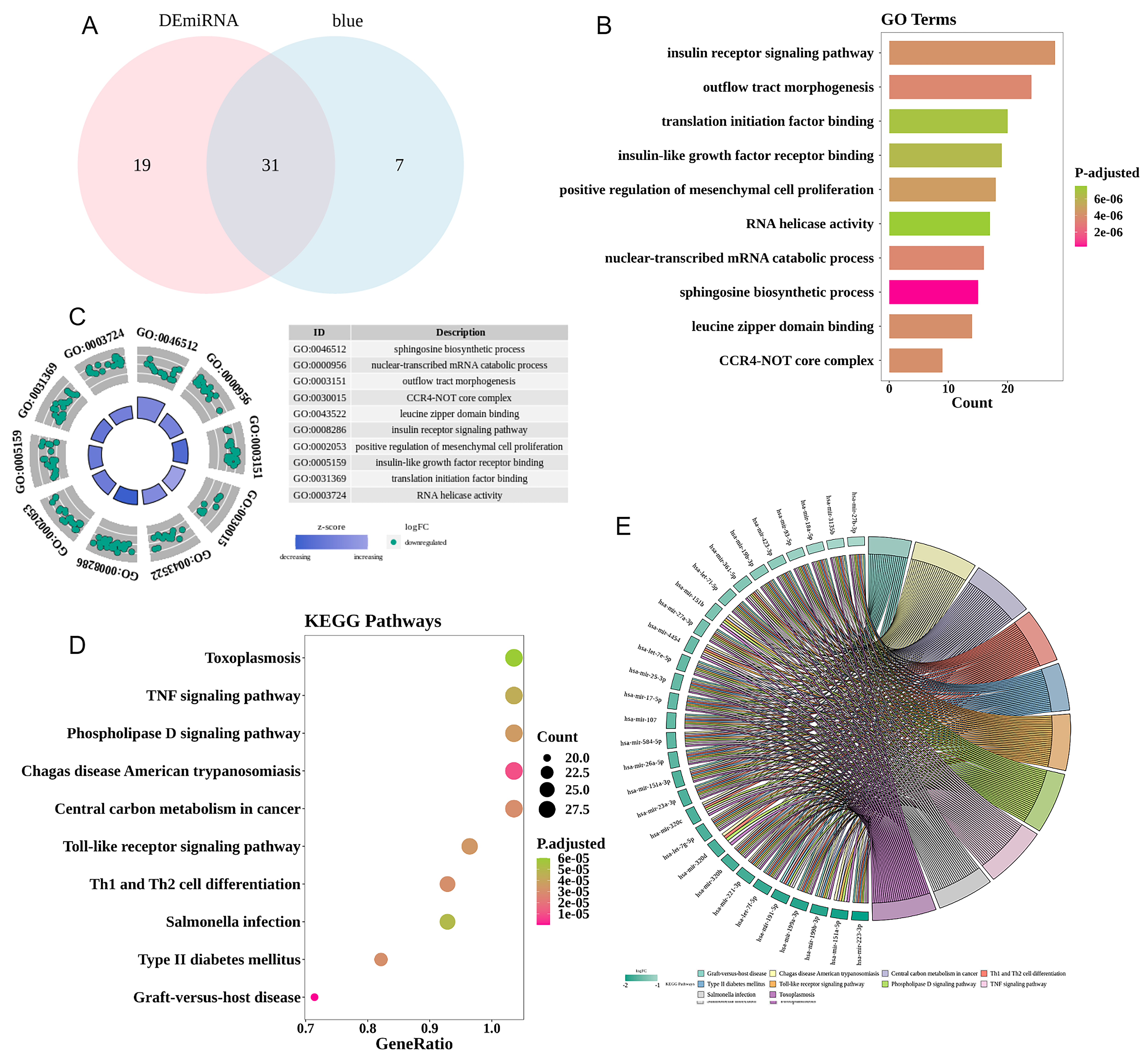
The 50 differential miRNAs (DE-miRNAs) were intersected with 38 blue module core miRNAs screened by WGCNA. Thirty-one overlapping genes from DE-miRNAs and module miRNAs were retained as key DE-miRNAs (Fig. 4A).
 Fig. 4.
Fig. 4.Functional enrichment analysis of differentially expressed mRNAs. (A) A comparison of differentially expressed genes (DEGs) in the three sets using a Venn diagram. (B) Histogram of the top 10 Gene Ontology (GO) annotation terms of key differential miRNAs (DE-miRNAs). (C) Plotting a circular diagram illustrating the top 10 GO annotation terms associated with important DE-miRNAs. (D) Bubble plot of the top 10 Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation terms of key DE-miRNAs. (E) Chord plot of the top 10 KEGG annotation terms of key DE-miRNAs in the enrichment analysis of dysregulated mRNAs between the EM and control groups. TNF/Th1, the tumor necrosis factor/T helper 1; CCR4-NOT, carbon catabolite repressor 4-Negative on TATA box (Goldberg–Hogness box).
To explore the biological functions and signaling pathways in which key
DE-miRNAs were implicated, GO and KEGG enrichment analyses were conducted. A
total of 3393 biological functions had 31 DE-miRNAs enriched when using
p
A total of 223 signaling pathways had 31 DE-miRNAs enriched when using
p
The Proc data package in R was utilized to plot the ROC curves of important
DE-miRNAs in this research, and the validation of each crucial DE-miRNA’s
diagnostic significance for endometriosis was conducted. AUC
 Fig. 5.
Fig. 5.Diagnostic value of serum exosomal miRNAs. Receiver operating characteristic (ROC) curves were produced using each miRNA expression value for key DE-miRNAs. AUC, area under the curve.
We utilized three databases, namely, mirDIP, TargetScan, and MiRTarBase, to screen miRNA‒mRNA interaction pairs and forecast the target mRNAs for 30 diagnostic miRNAs. Fig. 6A shows that there were 362 pairs of miRNA-mRNA relationships, with 337 being mRNAs and 24 being miRNAs. To predict 30 diagnostic miRNAs for targeting lncRNAs, we screened miRNA‒lncRNA interaction pairs using three databases: miRNet, starBase and NPInter. Fig. 6B shows that there were 167 pairs of interactions between miRNA and lncRNA, comprising 24 different miRNAs and 37 different lncRNAs.
 Fig. 6.
Fig. 6.The networks of competitive endogenous RNA (ceRNA) regulation involving lncRNA, miRNA, and mRNA. (A) Venn diagram of miRNA‒mRNA intersections obtained from the mirDIP, TargetScan and MiRTarBase databases. (B) Venn diagram of miRNA‒lncRNA intersections obtained from the miRNet, starBase and NPInter databases. (C) mRNA‒miRNA regulatory network; red circles are miRNAs, and yellow triangles are mRNAs. (D) miRNA‒lncRNA regulatory network; red circles are miRNAs, and blue squares are lncRNAs. (E) Core mRNA‒miRNA‒lncRNA regulatory network; red circles are miRNAs, blue squares are lncRNAs, and yellow triangles are mRNAs.
Next, we utilized Cytoscape software to combine lncRNA‒miRNA and miRNA‒mRNA interaction pairs into a network consisting of mRNA‒miRNA‒lncRNA triplets. The regulatory network of lncRNA-miRNA-mRNA consists of 1796 pairs of relationships, encompassing 36 lncRNAs, 20 miRNAs, and 264 mRNAs (Fig. 6C).
The ‘Analyze Network’ tool in Cytoscape software was used to calculate the degree of each node in accordance with the ceRNA regulatory network. We screened the top 10 hub lncRNAs (GAS5, MALAT1, FGD5-AS1, HCG18, SNHG16, XIST, OIP5-AS1, NEAT1, KCNQ1OT1 and SNHG12), hub miRNAs (hsa-miR-361-5p, hsa-miR-19b-3p, hsa-let-7f-5p, hsa-miR-23a-3p, hsa-miR-199a-3p, hsa-miR-18a-5p, hsa-miR-221-3p, hsa-miR-17-5, hsa-miR-27a-3, and hsa-miR-25-3p), and hub mRNAs (GALC, ETNK1, RNF4, SOX4, ZBTB18, SPRY2, RUNX1, MYLIP, BTG2, and MAP2K4). The central network of mRNA-miRNA-lncRNA regulation was built, as depicted in Fig. 6D,E.
Numerous studies have demonstrated the crucial involvement of noncoding RNAs in exosomes in the progression of EM [21]. The challenge associated with early clinical identification has resulted in the creation of convenient peripheral indicators that can be utilized for swift diagnosis. RNA is very stable in the transcriptome and is protected from ribonuclease (RNase) in exosomes [9]. Exosomes that are released can transmit RNA data to nearby cells, potentially playing a crucial part in the development of the disease microenvironment [22]. Investigating the intricate mechanisms of EM development can be effectively achieved by identifying exosomal RNA biomarkers in EM and understanding their functions. By utilizing RNA microarray sequencing technology and bioinformatics analysis, we discovered miRNAs in EM plasma exosomes, offering novel molecular targets for the diagnosis and treatment of EM.
First, we isolated and characterized exosomes in serum. TEM, NTA, and protein blot analysis demonstrated that the exosomes we acquired fulfilled the criteria for exosome identification in terms of morphology, size, distribution, and specific markers. Furthermore, the obtained exosomes align with the findings reported by Ronsini et al. [23]. This result indicates that we successfully isolated exosomes from serum. Next, the RNA microarray was used to analyze the serum exosome miRNA expression profiles of 10 pairs of endometriosis and negative controls. The findings indicated that there was significant dysregulation in 50 serum exosomal miRNAs, with 7 being upregulated and 43 being downregulated.
The purpose of WGCNA is to discover gene modules that are coexpressed and investigate the relationship between gene networks and clinical traits of interest. This helps identify gene modules that are relevant to clinical matters and identify key genes in disease pathways through the use of intramodule connectivity and gene significance. WGCNA yields more dependable and biologically meaningful outcomes in comparison to alternative bioinformatics approaches. The WGCNA analysis identified 27 coexpression modules in this study. Among them, the blue module, which was most relevant to the phenotype, consisted of 320 miRNAs, and a total of 38 core miRNAs were obtained. Obtained were 31 key miRNAs (DE-miRNAs) with dysregulated expression in EM plasma exosomes, which intersected with 50 dysregulated serum exosomal miRNAs and these 38 core miRNAs.
After analyzing functional enrichment, we discovered that the primary miRNAs
were linked to the promotion of mesenchymal cell growth, the TNF signaling
pathway, the Toll-like receptor signaling pathway, and the differentiation of Th1
and Th2 cells. The study conducted by Huang et al. [24] indicated that
estrogen has the ability to control the process of epithelial-mesenchymal
transition and cell growth in endometriosis by means of the Ras homolog gene
family, member A (RhoA) /Rho-associated protein kinase (ROCK) (RhoA/ROCK)
pathway. The TNF signaling pathway is linked to cell death [25]. The study showed
that miR-191 suppressed TNF-
In order to explore plasma exosomal signature miRNAs in endometriosis more
extensively, we assessed the discriminatory potential of crucial differential
miRNAs by analyzing ROC curves. After careful analysis, we determined that a set
of 30 miRNAs has the ability to differentiate between the disease and control
groups, showing promise as a diagnostic tool. In a prior investigation, it was
demonstrated that let-7f has the ability to control he migration of endometrial
cells in cases of endometriosis [28]. Jia et al. [29] discovered that
miR-17-5p exhibited decreased expression in the plasma of individuals diagnosed
with endometriosis. Lin et al. [30] reported that serum miR-17-5p
combined with miR-424-5p, and Pang et al. [31] further demonstrated that
miR-17-5p alleviated endometriosis by directly regulating vascular endothelial
growth factor A (VEGFA). In a study conducted by Tian et al. [26], it
was discovered that miR-191 hindered the apoptosis induced by TNF-
Furthermore, we explored the governing mechanisms of diagnostic miRNAs and established a ceRNA regulatory network, offering guidance and a point of reference for future investigations into the functionality of these miRNAs. We examined the hub miRNAs, hub mRNAs and hub lncRNAs in the network and illustrated the hub ceRNA network. The hub regulatory axes could have a significant impact on the growth and advancement of endometriosis. Several studies have demonstrated the significant regulatory role of the lncRNA metastasis associated lung adenocarcinoma transcript 1(MALAT1) in endometriosis development [40, 41]. The binding of some hub lncRNAs and hub miRNAs has also been demonstrated in previous studies [42, 43]. Sun et al. [44] found that myeloma cell proliferation and adhesion are tightly regulated by the lncRNA MALAT1/miR-181a-5p axis, and silencing MALAT1 stimulates apoptosis and interferes with myeloma cell proliferation. Malat1 decreases the expression of miR-181-5p, which induces the Hippo/YAP axis to promote the progression of myeloma. Wu et al. [45] also obtained exosomal lncRNA, miRNA and mRNA expression profiles and ceRNA network construction in endometriosis. Sun et al. [46] reported that lncRNA Small nucleolar host gene 12 (SNHG12) accelerates the proliferative and migratory abilities of ovarian cancer (OC) cells by sponging miRNA-129 to upregulate SRY-Related HMG-Box Gene 4 (SOX4), which was consistent with our regulatory axes. These results bolster our assurance in delving deeper into the involvement of regulatory axes in the network in endometriosis.
A significant constraint of this research is the limited number of participants. Further in-depth study is required to support the diagnostic value of these plasma exosomal miRNA markers with increased clinical sample data, as well as to investigate the mechanism of action.
This research focused on the isolation and characterization of serum exosomes obtained from individuals diagnosed with endometriosis. Reported for the first time, a total of 30 exosomal miRNA markers linked to endometriosis were identified and documented. The miRNAs were associated with the promotion of proliferation in mesenchymal cells, as well as the TNF and Toll-like receptor signaling pathways, and the differentiation of Th1 and Th2 cells. These biological processes and pathways could potentially play a significant role in the pathogenesis and progression of EM. The potential clinical significance of these miRNAs indicates possible targets for diagnosing and treating endometriosis while also offering fresh insights into the molecular mechanisms underlying this condition.
The datasets used and/or analyzed during the current study are not publicly available for ethical and privacy reasons but are available from the corresponding author upon reasonable request.
YH, CP and YZ designed the research study. YH and DZ performed the research. YH and DZ provided help and advice on the exosomal miRNA GeneChip assay. YZ analyzed the data. All authors contributed to editorial changes in the manuscript. All the authors have read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Peking University First Hospital (No. 2020KEYAN343).
The authors wish to acknowledge their patients and the families who agreed to participate in the study.
This work was supported by the National Key Research and Development Program on Reproductive Health and Women’s and Children’s Health Protection-Study on the Etiology and Prevention of Fertility Loss in Endometriosis (NO. 2022YFC2704004) and the National Key R&D Program on Reproductive Health and Prevention and Control of Major Birth Defects-Study on Etiology and Clinical Prevention of Endometriosis (NO. 2017YFC1001200).
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/j.ceog5102051.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.