†These authors contributed equally.
Academic Editor: Giovanni Monni
Background: This study aimed to compare the use of next-generation sequencing (NGS) and single-nucleotide polymorphism (SNP) array in preimplantation genetic testing for aneuploidy (PGT-A) in the same blastocyst. Methods: We performed a retrospective study on 67 embryos (from 23 couples), where PGT-A was carried out. A trophectoderm (TE) biopsy was performed on the blastocyst, and the 24-chromosomal ploidy status was analyzed. Initially, 28 blastocysts with unknown ploidy were analyzed using both NGS and SNP array. Thereafter, 39 blastocysts with euploidy detected via NGS were re-analyzed using SNP array. Results: In the first stage, the concordance rate was 92.9% (26/28). Among the 28 blastocysts, 16 were abnormal, and 12 were euploid when analyzed using NGS. Among the 16 abnormal blastocysts, two showed mosaicisms when analyzed using NGS but were found to be euploid using the SNP array. In the second stage, the concordance rate was 100% (39/39) when analyzing the normal blastocysts. After single blastocyst transfer in 29 frozen embryo transfer cycles, the clinical pregnancy rate was 75.9% (22/29), the ongoing pregnancy rate was 69.0% (20/29), and the live birth rate was 69.0% (20/29). Nineteen couples (20 babies) had healthy babies. Their prenatal diagnosis results and karyotype analysis after delivery were concordant with the PGT results. Two cycles miscarried, and the abortion villus exhibited euploidy. Conclusions: There was a high concordance rate between NGS and SNP array. TE biopsy combined with NGS for PGT was an efficient strategy to identify the suitability of embryos for transfer.
Chromosomal abnormalities are widespread in human embryos produced in vitro [1]. The abnormalities incidence increases dramatically in embryos with advancing maternal age and if one of the parents has balanced reciprocal chromosomal translocations [2, 3, 4, 5]. Chromosomal abnormalities are one of the main reasons for spontaneous abortions [6, 7] and repeated implantation failures [8, 9].
Selecting a normal or balanced embryo for transfer increases the chances of having a healthy and live newborn child. It also lessens the stress and difficult decisions that these couples usually have to make in the case of an unbalanced pregnancy. Preimplantation genetic testing for aneuploidy (PGT-A) and preimplantation genetic testing for chromosomal structural rearrangement (PGT-SR) are widely used for advanced maternal age couples, patients suffering from recurrent implantation failure, recurrent miscarriage, and those who are carriers of chromosomal structure abnormalities, to screen the genetic condition of embryos prior to transfer [10]. It has been found that the transfer of PGT-screened normal blastocysts could significantly increase implantation and reduce pregnancy loss [11, 12, 13]. Hence, these techniques are widely used to diagnose chromosomal diseases.
With the development of genome-wide technologies, the techniques of array comparative genomic hybridization (aCGH) and single-nucleotide polymorphism (SNP) microarrays are extensively applied. These detect the chromosomes involved in the rearrangement and complete aneuploidy of 24-chromosomal aneuploidy screening to reduce the risk of aneuploidy affecting structurally normal chromosomes [14, 15]. However, next-generation sequencing (NGS) is becoming more and more popular in clinics because approximately 1 Mb of deletions or duplications and low levels of mosaicism (down to 5%) in white blood cells can be detected [16]. Furthermore, it can decrease cost as the process is high-throughput compared with the processes used in aCGH and SNP arrays [17, 18].
However, there are few studies comparing SNP array and NGS technologies in detecting chromosome aneuploidy in the same blastocysts. Hence, this study aimed to compare both technologies in the same blastocyst in the clinic. In addition, the clinical outcomes, including clinical pregnancy and miscarriage, were also recorded to evaluate the use NGS technology used in this study.
All subjects provided informed consent for participating in the study. The study was conducted in accordance with the Declaration of Helsinki, and the study protocol was approved by the Ethics Committee of Guangzhou Women and Children’s Hospital (approval number: #2016-03).
In this study, 23 couples underwent PGT-A/PGT-SR in our center between March 2017 to June 2018. The indications of PGT are reciprocal translocation, Robertsonian translocation carriers, chromosome inversion, sex chromosome aneuploidies and advanced maternal age.
In the first stage, 28 blastocysts from 5 couples were detected by both NGS and SNP array. In the second stage, 115 blastocysts from 18 couples were first screened only with NGS. Among them, 39 blastocysts were normal. These blastocysts were detected by SNP array again (Fig. 1).
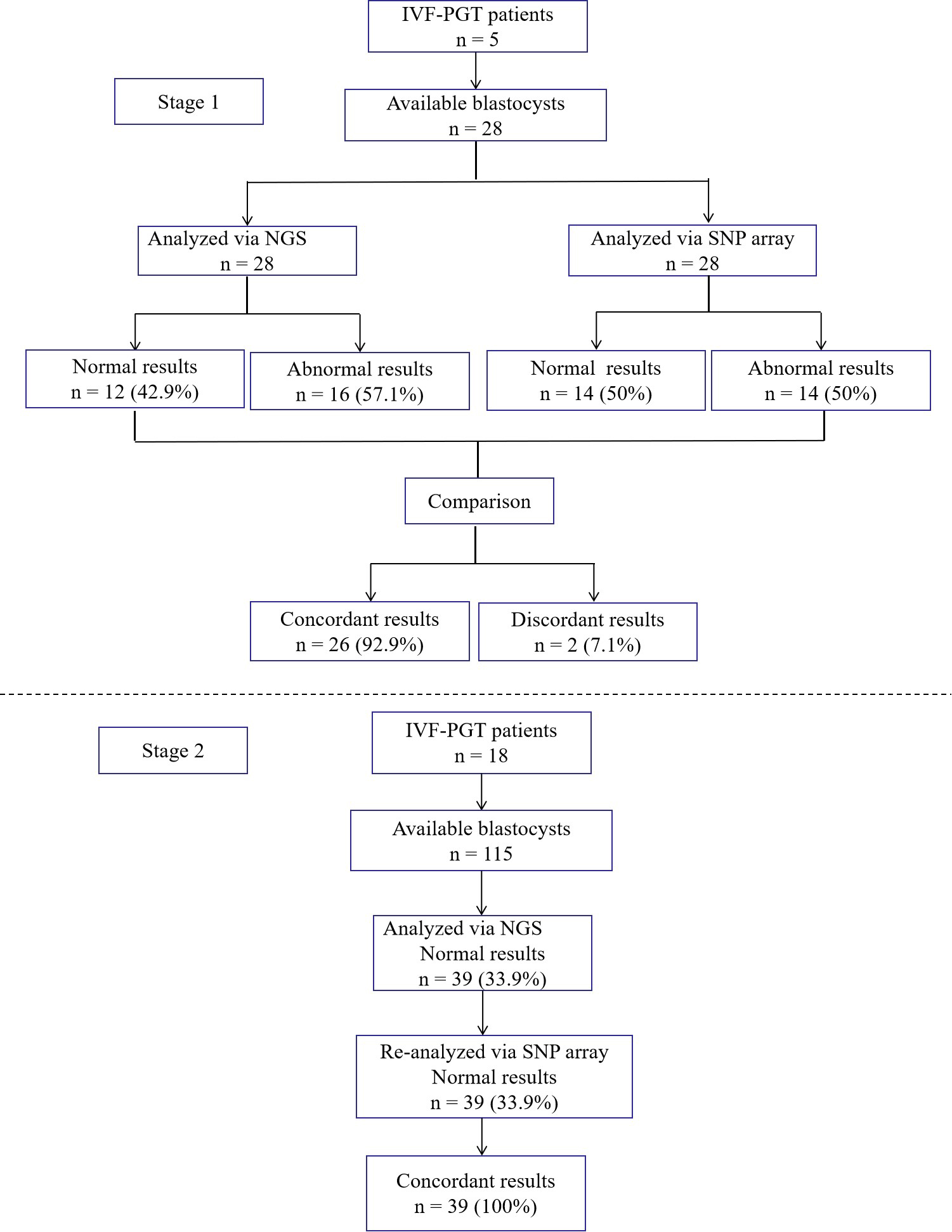 Fig. 1.
Fig. 1.Flow diagram of this study.
Intracytoplasmic sperm injections were administered after the oocytes were
retrieved. Injected oocytes were sequentially cultured in G1-PLUS/G2-PLUS
All blastocysts were subjected to trophectoderm (TE)-cell biopsy by laser and 5–10 TE cells were biopsied. After biopsy, blastocysts were cryopreserved using vitrification technique according to the manufacturer’s protocol (ARSCI Inc, Longueuil, Canada) and stored in liquid nitrogen.
The multiple displacement amplification DNA amplification system was used for WGA. Briefly, the REPLI-g Single Cell Kit (QIAGEN, Hilden, Germany) was used for single-cell amplification following the manufacturers’ instructions.
The Illumina MiSeq platform (Illumina, San Diego, California, USA) was used, and
the amplified genome of each blastocyst’s TE cells were sequenced at an
approximate 0.01
All procedures were performed according to the manufacturer’s protocol for the Illumina human SNP array. Briefly, DNA was hybridized with the Human Cyto-12 BeadChip (Illumina, San Diego, California, USA) (which contains approximately 300,000 SNPs with average distance of 9.7 kb). The bead chips were subjected to immunostaining, followed by stringent washes to remove unhybridized and non-specifically hybridized DNA. Subsequently, the bead chips were scanned using the iScan Bead Array Reader (Illumina, San Diego, California, USA). The scanning results were processed using the B allele frequency and log R ratio using Illumina Genome Studio Genotyping Module v2.0 software (Illumina, San Diego, California, USA) to analyze the copy number of the chromosomes according to the protocol in the Illumina human SNP array.
A serum beta-hCG test was performed 14 days after transfer. Clinical and ongoing pregnancy was confirmed via transvaginal ultrasound at 5 to 6 weeks and 12 weeks after transfer, respectively. Clinical miscarriage was defined when a pregnancy failed to progress after an intrauterine gestational sac had been detected with pelvic ultrasonography. Live birth rate was defined as the number of live births per number of frozen embryo transfer.
Table 1 summarizes patient demographic characteristics for the 23 women in the
study. Their average age was 31.1
| Clinical data | Value | |
| No. of patients | 23 | |
| OPU cycles | 26 | |
| Age (years) | 31.1 | |
| Antral follicle count (n) | 12.9 | |
| Baseline follicle-stimulating hormone (mIU/mL) | 5.4 | |
| Anti-Műllerian hormone (ng/mL) | 4.8 | |
| Type of fertility disorder (n, %) | ||
| Primary | 9 (39.1) | |
| Secondary | 14 (60.9) | |
| Etiology for PGT (n, %) | ||
| Reciprocal translocation | 11 (47.8) | |
| Robertsonian translocation | 4 (17.4) | |
| Chromosome inversion | 3 (13.0) | |
| Advanced maternal age | 3 (13.0) | |
| Sex chromosome aneuploidies | 2 (8.7) | |
In stage 1, the concordance between NGS and SNP array was 92.9% (26/28). The 28 blastocytes were then analyzed by both NGS and the SNP array. NGS analysis revealed that 16 blastocysts were abnormal and 12 had normal ploidy status, whereas the SNP array identified 14 blastocysts to be abnormal and the other 14 to have normal ploidy status. Two blastocysts were completely discordant between NGS and SNP. The abnormality detected by NGS showed that those two blastocysts had a low level of mosaicism, whereas the SNP array detected that they were euploid (Table 2, embryo number 1 and 2, Fig. 2). Among the other 14 abnormal blastocysts, one blastocyst had a discordant result as the mosaicism could only be detected by NGS (Table 2, embryo number 3, Fig. 3).
| Embryo number | Indications of PGT | NGS result | SNP array result |
| 1 | 46,XY,t(9;15)(q22;q22) | seq(6) × 3[0.40],(13) × 3[0.38],(14) × 3[0.31],(15) × 3[0.40],(16) × 3[0.40] | arr(X,1–22) × 2 |
| 2 | 46,XY,t(9;15)(q22;q22) | seq(13) × 3[0.44] | arr(X,Y) × 1,(1–22) × 2 |
| 3 | 45,XY,der(13;14)(q10;q10) | seq(2) × 1[0.60],(6) × 1,(12) × 3,(14) × 3,(15) × 1 | arr(6) × 1,(12) × 3,(14) × 3,(15) × 1 |
| 4 | 46,XY,t(9;15)(q22;q22) | seq9q22.1q34.3(91710848_141017812) × 1,15q22.2q26.3(59561721_102100186) × 3 | arr9q22.1q34.3(91578102_141044489) × 1,15q22.2q26.3(59920197_102397836) × 3 |
| 5 | 46,XY,t(9;15)(q22;q22) | seq(15) × 1 | arr(15) × 1 |
| 6 | 46,XY,t(9;15)(q22;q22) | seq9p24.3q22.1(10291_91710847) × 3,15q11.1q22.2(20000001_59561720) × 1 | arr9p24.3q22.1(46587_91578102) × 3,15q11.1q22.2(22789021_59920197) × 1 |
| 7 | 46,XY,t(9;15)(q22;q22) | seq9q22.1q34.3(91710848_141017812) × 3,15q22.2q26.3(60317011_102100186) × 1 | arr9q22.1q34.3(91578102_141044489) × 3,15q22.2q26.3(59920197_102397836) × 1 |
| 8 | 46,XY,t(9;15)(q22;q22) | seq9q22.1q34.3(91710848_141017812) × 1,15q22.2q26.3(59561721_102100186) × 3 | arr9q22.1q34.3(91578102_141044489) × 1,15q22.2q26.3(59920197_102397836) × 3 |
| 9 | 46,XY,t(9;15)(q22;q22) | seq9q22.1q34.3(91710848_141017812) × 3,15q22.2q26.3(60317011_102100186) × 1 | arr9q22.1q34.3(91578102_141044489) × 3,15q22.2q26.3(59920197_102397836) × 1 |
| 10 | 45,XY,der(13;14)(q10;q10) | seq(14) × 3 | arr(14) × 3 |
| 11 | Advanced maternal age | seq(19) × 3 | arr(19) × 3 |
| 12 | Advanced maternal age | seq(16) × 3 | arr(16) × 3 |
| 13 | Advanced maternal age | seq(16) × 1 | arr(16) × 1 |
| 14 | Advanced maternal age | seq(X) × 1,(Y) × 2,(1-22) × 2 | arr(X) × 1,(Y) × 2,(1–22) × 2 |
| 15 | Advanced maternal age | seq(2) × 1 | arr(2) × 1 |
| 16 | Advanced maternal age | seq(1) × 1 | arr(1) × 1 |
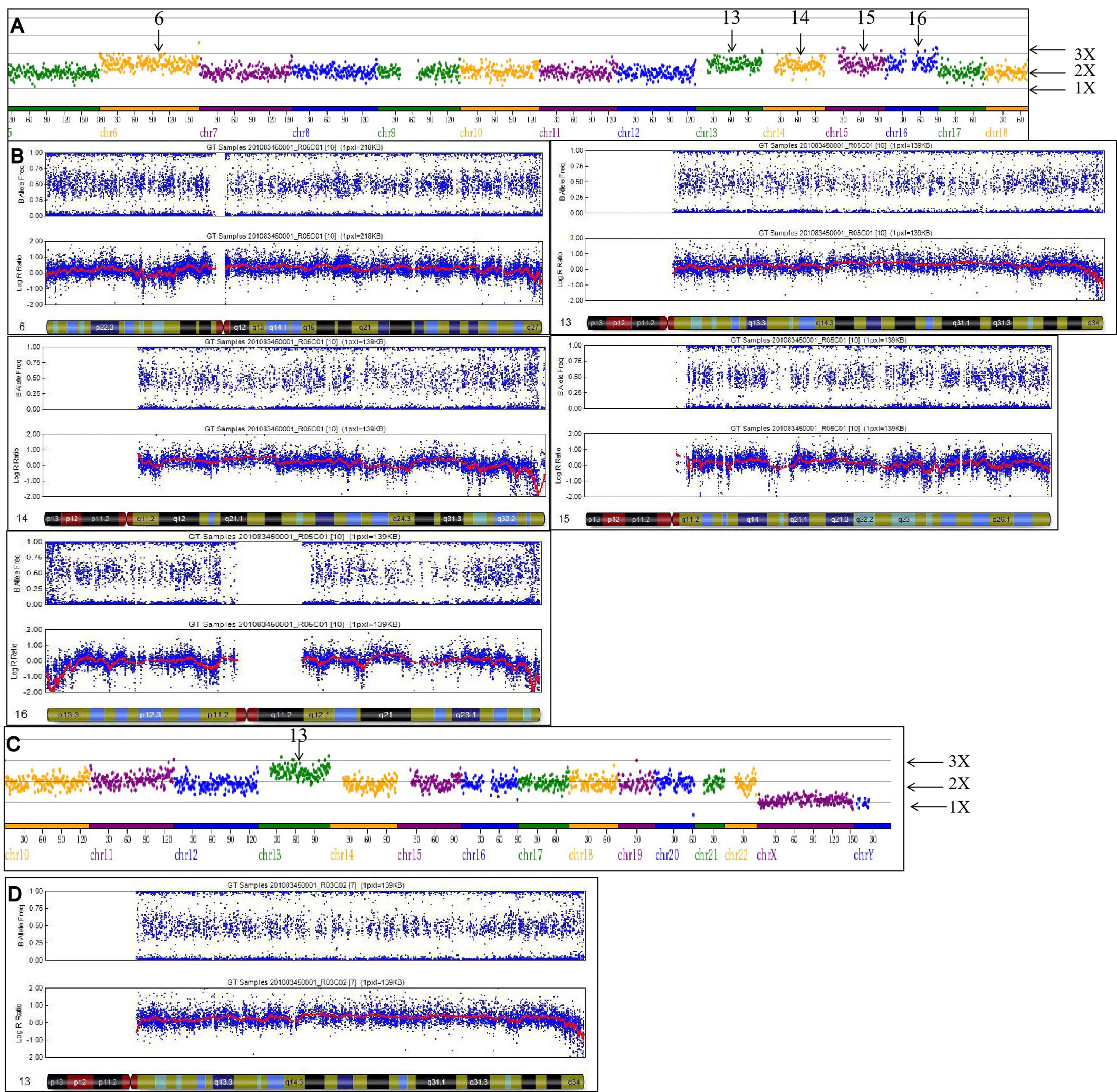 Fig. 2.
Fig. 2.The discordant results of two blastocysts detected by NGS and SNP. (A) The abnormality detected via NGS with low levels of mosaicism in chromosomes 6, 13, 14, 15, and 16. (B) The normal result detected via SNP array in chromosomes 6, 13, 14, 15, and 16. (C) The abnormality detected via NGS with a low level of mosaicism in chromosome 13. (D) The normal result detected via the SNP array in chromosome 13. 2X: chromosome with two copies (normal), 3X: three copies, 1X: one copy.
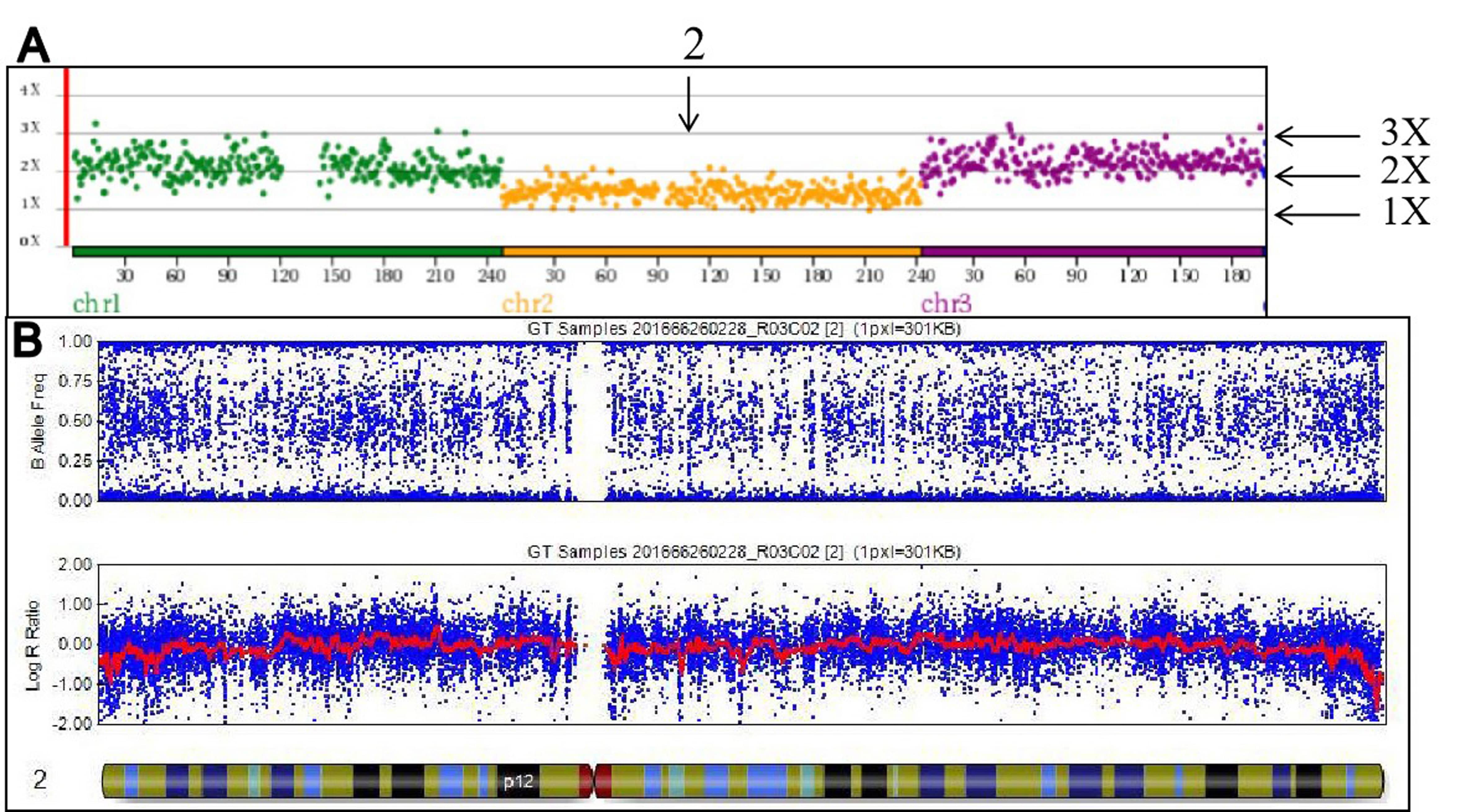 Fig. 3.
Fig. 3.The discordant results of a single blastocyst detected via NGS and SNP. (A) The abnormality detected by NGS with a low level of mosaicism in chromosome 2. (B) The normal result detected by SNP in chromosome 2. 2X: chromosome with two copies (normal), 3X: three copies, 1X: one copy.
In stage 2, 115 blastocysts from 18 couples were detected by NGS, followed by the SNP array. Thirty-nine blastocysts were considered normal from both techniques. Therefore, the concordance between NGS and SNP array was 100% (39/39).
Twenty-nine cycles (4 cycles in stage 1 and 25 cycles in stage 2) were transferred with a single blastocyst (4 blastocysts in stage 1 and 25 blastocysts in stage 2). The implantation rate was 82.8% (24/29), the clinical pregnancy rate was 75.9% (22/29), the ongoing pregnancy rate was 69.0% (20/29), and the live birth rate was 69.0% (20/29). Nineteen couples (20 babies) had healthy babies, and both their prenatal diagnosis results and karyotype analysis after delivery were concordant with the PGT results. Two cycles miscarried at 8 and 11 weeks of gestation, respectively, and the abortion villus showed the same karyotype as the PGT results.
TE biopsy combined with NGS for PGT-A/PGT-SR is now widely used in the clinical setting globally. It has been proven to increase the pregnancy rate of patients who are carriers of chromosome structure abnormalities, patients with advanced maternal age, and patients who have experienced recurrent miscarriages [20, 21]. The NGS technique has been shown to have more advantages over the SNP array, including lower cost, supporting its extensive use in selecting a normal embryo for implantation. NGS can also sensitively detect mosaicism [20]. Therefore, using NGS for PGT-A may increase the chances of having a healthy and live newborn child and lessen the stress of abortion [22, 23, 24]. In our study, we compared the use of NGS and SNP array in the same blastocyst from the clinic. We found high concordance between both technologies. When NGS revealed normal results, the SNP array confirmed the findings (the second stage of our study). Furthermore, in stage 1 of the study, there were two cases where NGS defined the blastocysts as abnormal, whereas the SNP array found them normal. Therefore, we conclude that NGS can reveal mosaicism when SNP analysis finds normal results.
As NGS is more sensitive for detecting mosaicism than the SNP array, we defined these two blastocysts as mosaic. We did not transfer these as the couples had four euploid blastocysts. A controversy regarding mosaicism exists as couples can still have healthy babies born after transferring mosaic embryos. Studies suggested that the level of mosaicism identified in the original TE biopsy is a predictor of miscarriage [25]. However, another study reported that mosaic embryos with low aneuploidy percentage have less chance to cause a miscarriage and can lead to a healthy birth [24]. Hence, transferring mosaic blastocysts is still controversial in clinical risk management and/or difficult counseling perspectives.
Both SNP arrays and NGS platforms have potential strengths and weaknesses. SNP array is a genotyping array that can detect uniparental disomic, identify the parental origin of chromosome abnormalities, and detect triploidy [14]. On the other hand, NGS can screen approximately 1 Mb of deletions or duplications in chromosomes and has a high sensitivity to detect low levels of mosaicism. Hence, NGS would detect more abnormalities than other methods and improve implantation. A study by Tan et al. [20] compared NGS to SNP array in PGT-A. Their results showed that NGS could detect some segmental imbalances that may be omitted by the SNP array. Therefore, the potential risks of false-negative results can be avoided by NGS. Furthermore, although the clinical outcome measures of the NGS cycles were the same as the SNP array, it appears that the implantation and clinical pregnancy rate in NGS were higher than that in the SNP array [14].
Here, according to the NGS and SNP array result, 23 blastocysts were transferred into the uterus with a single blastocyst, and 22 were implanted and developed. The karyotype was normal in the prenatal diagnosis or the blood from the newborns, indicating the accuracy of NGS and the SNP array in the PGT. Additionally, it illustrates that NGS is applicable for genetically high-risk populations, including carriers of Robertsonian and reciprocal translocations. However, three mosaic embryos could be detected by NGS but failed to be confirmed by SNP array. Therefore, with NGS, embryos with mosaic abnormalities can be selected and will not be used for transfer. This avoids abortion or newborns with prenatal abnormality caused by embryos with mosaic abnormalities, which could not be detected by SNP. Conversely, if false positives exist, it will lead to a waste of embryos. In our previous study, some embryos were classified as mosaic in the original analysis by NGS but were re-classified as chromosomally balanced after whole blastocyst sequencing. Hence, to maximize the number of embryos available for PGT-SR/A patients, we suggest that embryos with mosaic non-SR chromosomal rearrangement should be stored and considered for transfer after appropriate counseling [26]. Although PGT-A can detect most of the abnormal blastocysts, it suggests the need to confirm the PGT analysis with prenatal testing as it is a screening method, and results can never be used to definitively predict the chromosomal status of the embryo and fetus [27, 28].
There are some limitations to this study. Firstly, there were relatively few mosaic blastocysts in this research. Therefore, to better understand embryos with mosaic results, we need to acquire more samples. Secondly, all samples in the second stage of the study were normal. Ideally, we would like to compare these methods using both normal and abnormal embryos.
The results achieved in this study demonstrate the reliability of the NGS-based protocol for the detection of chromosome status in embryos.
ZHO, ZHC and LS—conception and design of the study. ZHO, YD and ZHC—data collection, statistical analysis, construction of figures and tables. ZHO, ZHC and YD—drafted the article. ZHO, YD, ZHC and LS—reviewed the manuscript and approved the version to be published.
All subjects provided informed consent for participating in the study. The study was conducted in accordance with the Declaration of Helsinki, and the study protocol was approved by the Ethics Committee of Guangzhou Women and Children’s Hospital (approval number: #2016-03).
Thanks to all the peer reviewers for their opinions and suggestions. We also thanks to the company of Jabrehoo from China for some technologic helps.
This study was supported by institutional funding of Health Science and technology project of Guangzhou (20201A011029 and 20211A011026), Guangzhou Women and Children’s Medical Center (1600057-04) and National Natural Science Foundation of China (81800110).
The authors declare no conflict of interest.
